Abstract
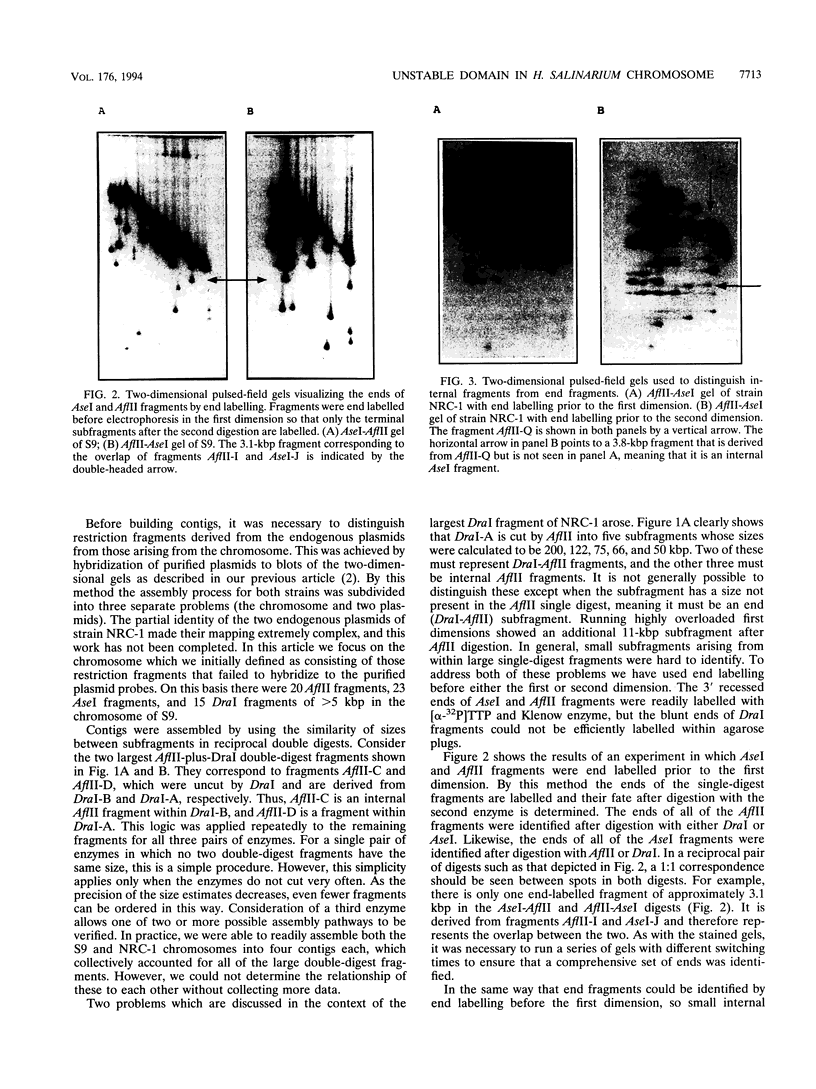
Phenotypic variants of Halobacterium salinarium NRC-1 arise at a frequency of 10(-2). These result from transpositions of halobacterial insertion sequences and rearrangements mediated by halobacterial insertion sequences. We have tested the hypothesis that such mutations are confined to only a portion of the genome by comparing the chromosomal restriction map of H. salinarium NRC-1 and that of the derivative S9, which was made in 1969. The two chromosomes were mapped by using two-dimensional pulsed-field gel electrophoresis and the restriction enzymes AflII, AseI, and DraI. A comparison of the two deduced maps showed a domain of about 210 kbp to be subject to many rearrangements, including an inversion in S9 relative to NRC-1. However, the rest of the chromosome was conserved among NRC-1, S9, and an independent Halobacterium isolate, GRB, previously mapped by St. Jean et al. (A. St. Jean, B. A. Trieselmann, and R. L. Charlebois, Nucleic Acids Res. 22:1476-1483, 1994). This concurs with data from eubacteria suggesting strong selective forces maintaining gene order even in the face of rearrangement events occurring at a high frequency.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Betlach M., Friedman J., Boyer H. W., Pfeifer F. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 1984 Oct 25;12(20):7949–7959. doi: 10.1093/nar/12.20.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassarma S., Halladay J. T., Jones J. G., Donovan J. W., Giannasca P. J., de Marsac N. T. High-frequency mutations in a plasmid-encoded gas vesicle gene in Halobacterium halobium. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6861–6865. doi: 10.1073/pnas.85.18.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Betlach M. Genome organization in Halobacterium halobium: a 70 kb island of more (AT) rich DNA in the chromosome. Mol Gen Genet. 1985;198(3):449–455. doi: 10.1007/BF00332938. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Blaseio U., Horne M. Genome structure of Halobacterium halobium: plasmid dynamics in gas vacuole deficient mutants. Can J Microbiol. 1989 Jan;35(1):96–100. doi: 10.1139/m89-015. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Blaseio U. Insertion elements and deletion formation in a halophilic archaebacterium. J Bacteriol. 1989 Sep;171(9):5135–5140. doi: 10.1128/jb.171.9.5135-5140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Tümmler B. The impact of two-dimensional pulsed-field gel electrophoresis techniques for the consistent and complete mapping of bacterial genomes: refined physical map of Pseudomonas aeruginosa PAO. Nucleic Acids Res. 1991 Jun 25;19(12):3199–3206. doi: 10.1093/nar/19.12.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Rose M. R., Doolittle W. F. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature. 1982 Sep 9;299(5879):182–185. doi: 10.1038/299182a0. [DOI] [PubMed] [Google Scholar]
- Simsek M., DasSarma S., RajBhandary U. L., Khorana H. G. A transposable element from Halobacterium halobium which inactivates the bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7268–7272. doi: 10.1073/pnas.79.23.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- St Jean A., Trieselmann B. A., Charlebois R. L. Physical map and set of overlapping cosmid clones representing the genome of the archaeon Halobacterium sp. GRB. Nucleic Acids Res. 1994 Apr 25;22(8):1476–1483. doi: 10.1093/nar/22.8.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckenius W., Lozier R. H., Bogomolni R. A. Bacteriorhodopsin and the purple membrane of halobacteria. Biochim Biophys Acta. 1979 Mar 14;505(3-4):215–278. doi: 10.1016/0304-4173(79)90006-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., Wertman K. F. Host strains that alleviate underrepresentation of specific sequences: overview. Methods Enzymol. 1987;152:173–180. doi: 10.1016/0076-6879(87)52017-1. [DOI] [PubMed] [Google Scholar]







