Abstract
Prolamine and glutelin RNAs are localized to two subdomains of the cortical endoplasmic reticulum (ER), the protein body ER and the cisternal ER, in developing rice seeds. The addition of nearly full-length prolamine sequences at the 3′ untranslated region of a reporter RNA redirects its localization from the cisternal ER to the protein body ER. Deletion analysis of prolamine RNA sequences indicates the presence of two partially redundant cis elements required for protein body ER targeting. The addition of glutelin 3′ untranslated region to protein body ER cis sequences, however, redirects RNA localization to the cisternal ER. These results indicate that there are at least two regulated RNA transport pathways as well as a constitutive pathway to the cortical ER.
INTRODUCTION
The targeting of proteins to specific subcellular locations (e.g., vacuole, chloroplast, etc.) is directed by a variety of signal determinants. Many of these signals consist of amino acids arranged linearly, whereas others are arranged noncontiguously and constitute three-dimensional motifs. In addition to peptide-sorting signals, it is well established that the mRNA itself may have signals that direct it to discrete locations within the cell (Singer, 1993). Such RNA targeting provides a mechanism to synthesize proteins proximal to their normal intracellular locations. Because RNA localization also is evident to specific subdomains of the endoplasmic reticulum (ER), RNA sorting may provide a mechanism to target proteins to specific destinations within the endomembrane system (Pollock et al., 1990; Svoboda, 1991; Okita and Rogers, 1996; Deshler et al., 1997; Mowry and Cote, 1999).
RNA localization to the ER is a conspicuous feature in developing rice endosperm. This seed storage organ accumulates two major types of storage proteins, prolamines and glutelins, which are sequestered in separate endomembrane compartments. Prolamines accumulate and assemble to form a spherical intracisternal inclusion granule, 1 to 2 μm in diameter, within the ER lumen. Although glutelins are translocated initially to the ER lumen by a cotranslational import process, these proteins are transported to the Golgi and later to small storage vacuoles to form irregularly shaped, electron-dense protein bodies 2 to 3 μm in diameter. Li et al. (1993) obtained biochemical and in situ hybridization data demonstrating that the storage protein mRNAs of developing rice endosperm were not translated randomly on the rough ER but, instead, were enriched on morphologically distinct subdomains of this membrane complex. Prolamine mRNAs are localized to the prolamine protein bodies (PBs) and, specifically, to the ER membranes (PB-ER) that delimit the prolamine intracisternal inclusion granules, whereas glutelin RNAs predominate on the cisternal ER. Both membrane types constitute the cortical ER in developing rice endosperm (Muench et al., 2000). The segregation of these storage RNAs to distinct ER subdomains may facilitate the location of these storage proteins in separate endomembrane compartments. In particular, targeting of prolamine RNAs to the PB-ER concentrates newly synthesized prolamine polypeptides within this ER subdomain, thereby promoting their assembly into an intracisternal inclusion granule (Okita and Rogers, 1996).
The segregation of these storage protein RNAs to specific cortical ER subdomains was confirmed and extended by a recent study (Choi et al., 2000) using fluorescently labeled nucleotides and in situ RNA localization techniques. Localization of prolamine RNAs was shown not to be dependent on the synthesis of prolamine primary amino acid sequence, indicating that the RNA was targeted to the PB-ER by an RNA-based mechanism. The cis sequences required for prolamine RNA targeting to the PB-ER have yet to be identified, although at least part of the sorting determinant may reside in the 3′ untranslated region (UTR) as replacement of the prolamine 3′ UTR with corresponding sequences from glutelin RNA, resulting in displacement of the RNA to the cisternal ER (Choi et al., 2000).
Here, we defined the prolamine RNA sorting signals by studying the localization of RNAs coded by a series of green fluorescent protein (GFP)–prolamine cDNA genes. GFP RNAs, which normally are localized to the cisternal ER, are redirected to the PB-ER when prolamine RNA sequences are attached to the 3′ end. These results indicate the existence of a regulated prolamine RNA transport pathway to the PB-ER as well as a constitutive pathway to the cisternal ER. Deletion analysis showed that the regulated prolamine RNA transport pathway to the PB-ER requires two partially redundant cis elements, one located in the coding sequence and a second residing in the 3′ UTR. In addition, we provide evidence that glutelin RNAs are transported by a second regulated RNA transport pathway to the cisternal ER by a process that is dominant over the prolamine-regulated pathway.
RESULTS
Prolamine RNA Placed at the 3′ UTR Maintains Targeting Specificity
We demonstrated previously by in situ hybridization at the electron microscopy level (Li et al., 1993) that the rice storage protein mRNAs showed a polarized distribution pattern in developing endosperm tissue, with glutelin RNAs located predominantly on the cisternal ER and prolamine RNAs on the PB-ER. This initial conclusion was verified and extended at the light microscopy level, where the localization of prolamine RNAs to the PB-ER was found to occur by an RNA-based mechanism, as demonstrated by the targeting to the PB-ER of a β-glucuronidase RNA containing prolamine RNA sequences positioned at the 3′ UTR (Choi et al., 2000). These results prompted us to further define the cis elements, or “zip code” sequences (Singer, 1993), responsible for prolamine mRNA targeting to the PB-ER using a deletion approach.
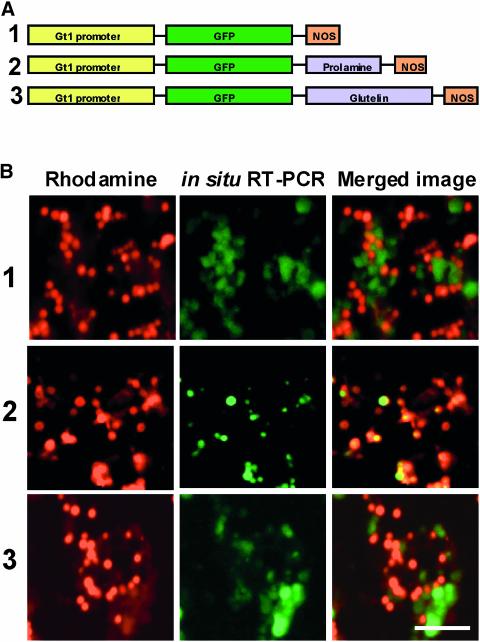
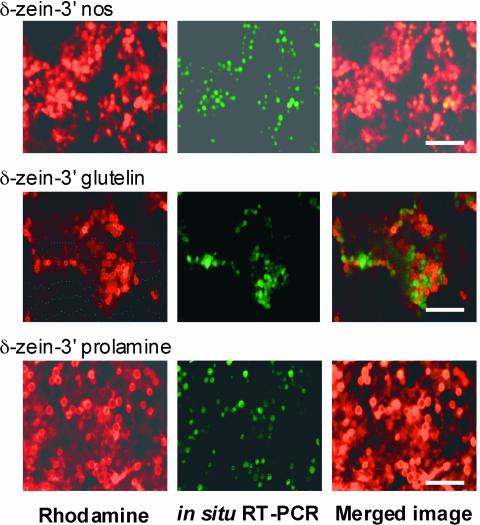
Before this study was initiated, we studied the expression and localization of a series of GFP storage protein RNAs (nearly full-length prolamine and glutelin RNA)–3′ nopaline synthase (nos) RNAs (Figure 1A) in developing rice endosperm to verify that the hybrid RNAs would be targeted to the same cortical ER subdomains as the native storage protein RNA species. RNAs containing prolamine and glutelin RNA sequences were ∼800 and 1800 nucleotides longer, respectively, than GFP–3′ nos RNAs (data not shown). Plants expressing detectable GFP levels, as determined by immunoblot analysis, were examined for the distribution of cognate RNAs by subjecting endosperm sections to in situ reverse transcriptase–mediated (RT) PCR in the presence of Oregon Green 488–dUTP. The endosperm sections then were poststained with rhodamine hexyl ester to label the prolamine PBs. Confocal microscopic analysis showed that GFP–3′ nos RNAs were localized to regions in close proximity to the prolamine PBs (Figure 1B, row 1). A similar distribution pattern was evident with GFP–glutelin–3′ nos RNAs (Figure 1B, row 2). By contrast, a prolamine containing hybrid RNA was localized to the prolamine PBs (Figure 1B, row 3) in a pattern similar, if not identical, to that of endogenous prolamine transcripts (Choi et al., 2000). These results indicate that sorting signals present in the prolamine RNA sequences positioned within the 3′ UTR of the hybrid RNA are able to direct RNA transport to the prolamine PBs. In the absence of any regulated transport signals, RNAs are transported to the cisternal ER located in the cortical region, presumably by a constitutive (default) pathway.
Figure 1.
Gene Expression and RNA Localization.
(A) Schemes of the GFP reporter genes expressed during rice endosperm development. The seed-specific glutelin Gt1 promoter was used to drive the expression of the GFP reporter. Polyadenylation and transcriptional termination signals were provided by the 3′ flanking region of the nos gene. Prolamine and glutelin cDNAs were inserted between the GFP reporter and the 3′ nos sequences.
(B) Localization of GFP RNAs in developing endosperm sections. Endosperm sections expressing the three genes depicted in (A) were subjected to in situ RT-PCR using GFP-specific primers to assess RNA localization (middle images). Prolamine PBs (1 to 2 μm in diameter) were visualized by staining the sections with rhodamine hexyl ester (left images). Note that GFP RNAs containing the prolamine sequences are localized to the PB-ER that bound the prolamine PBs, as indicated by the coincident labeling seen in the merged images (right), whereas the other two RNA species are localized to the cisternal ER. Bar = 10 μm.
Prolamine RNAs Contain Two cis Elements for PB-ER Targeting
To identify the cis-acting signal determinants of the prolamine mRNA, a series of 5′ and 3′ deletions of the prolamine transcribed gene sequences were constructed. Deletions were made by dividing the coding sequence, which spans nucleotides 205 to 689, into four segments; the 3′ UTR was divided into two segments for deletion (Figure 2). These various 5′ and 3′ deletions of the prolamine transcribed gene sequences then were cloned between the GFP reporter and 3′ nos UTR plus terminator sequences and expressed in developing endosperm in transgenic rice plants.
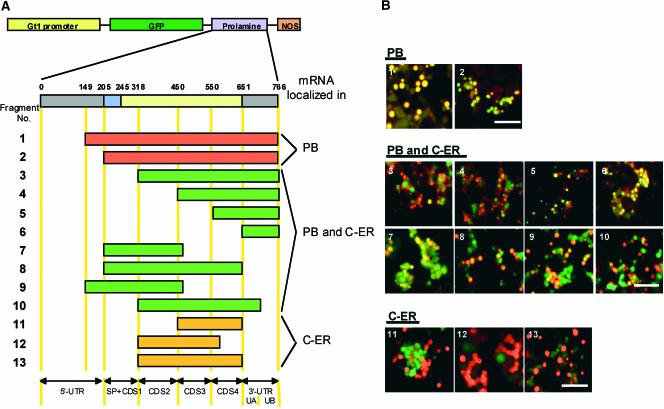
Figure 2.
Localization of GFP RNAs Containing Various Prolamine cDNA Segments.
(A) Schemes of GFP reporter genes (constructs 1 to 13) containing various segments of the prolamine cDNA inserted at the 3′ UTR of the hybrid GFP RNA. C-ER, cisternal ER.
(B) Visualization of GFP RNAs containing various prolamine cDNA segments (1 to 13), as detected by in situ RT-PCR, and their spatial relationships to prolamine PBs. Depicted are merged images of RNA localization (green channel) and prolamine PBs (red channel). Three RNA distribution patterns are evident: RNAs restricted to the PB-ER (constructs 1 and 2), RNAs localized to both the PB-ER and the cisternal ER (constructs 3 to 10), and RNAs localized only to the cisternal ER (constructs 11 to 13). Bar = 10 μm.
Confocal microscopic observation of endosperm sections expressing these gene constructs showed that hybrid RNAs lacking the prolamine 5′ UTR sequences remained restricted to the prolamine PBs (Figure 2, constructs 1 and 2) in a distribution pattern identical to that observed for wild-type prolamine mRNAs. Hence, cis-acting sorting determinants required for prolamine PB localization are located in the coding sequence and/or 3′ UTR. Removal of the signal peptide and coding sequence region 1 (CDS1; nucleotides 205 to 317) resulted in a change in RNA distribution pattern. Although the hybrid RNAs were observed on the prolamine PBs, they also were distributed on the cisternal ER as well. This distribution of RNAs to both the PB-ER and the cisternal ER also was observed for other prolamine 5′ deletions (constructs 4 to 6). The smallest 5′ deletion fragment containing only the 3′ UTR (construct 6; nucleotides 651 to 766) was sufficient for this dual RNA localization pattern. These observations suggest that the 3′ UTR contains one or more sorting signal determinants required for RNA localization to the PB-ER.
3′ deletion analysis confirmed the presence of a cis element in the prolamine 3′ UTR. Removal of the distal half of the 3′ UTR (construct 10) had no effect on the RNA distribution to both the PB-ER and the cisternal ER. By contrast, removal of the proximal 3′ UTR end (constructs 11 to 13) resulted in the total loss of RNA localization to the PB-ER. Hence, the proximal half of the prolamine 3′ UTR contains at least one cis element required for PB-ER localization.
As described above, 5′ deletion of the signal peptide and CDS1 (construct 3) resulted in only a partial localization of RNAs to the PB-ER, suggesting the presence of a cis element in this region. Because we had demonstrated previously that the removal of the signal peptide sequences had no effect on prolamine RNA targeting to the PB-ER (Choi et al., 2000), the cis element is likely located in CDS1 (nucleotides 245 to 317). This view was supported by the result that RNAs containing CDS1 but lacking the 3′ UTR (constructs 7 to 9) were distributed to both the PB-ER and the cisternal ER. By contrast, deletions of prolamine sequences containing CDS1 and the proximal 3′ UTR (constructs 11 to13) resulted in the localization of the RNA to the cisternal ER. Collectively, these results suggest that prolamine RNA contains at least two cis elements necessary for restricted localization to the PB-ER: one located at the 5′ end of the coding sequence (CDS1) and a second located in the proximal half (UA) of the 3′ UTR.
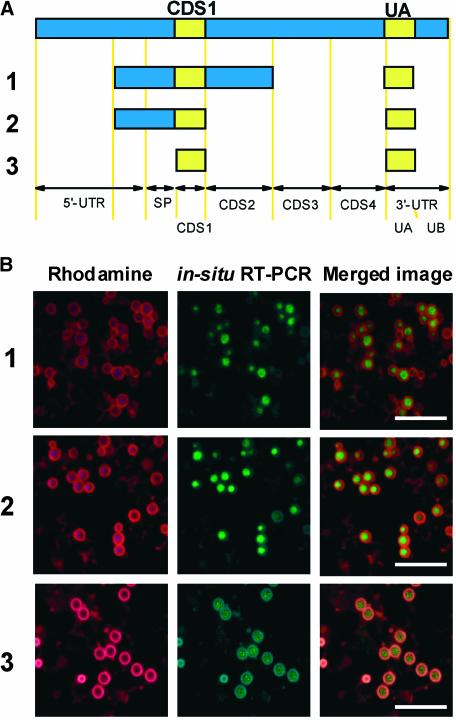
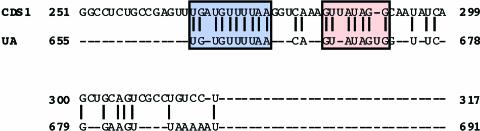
To confirm the presence of cis elements in CDS1 and UA, a series of internal deletions of the prolamine gene sequences were obtained (Figure 3A) and expressed as described above. All internal deleted mRNAs containing CDS1 and UA were targeted to the PB-ER (Figure 3B). These results demonstrate that CDS1 and UA are sufficient to direct RNA targeting to the prolamine protein body. A direct comparison of the RNA sequences within CDS1 and UA indicates considerable sequence similarity, especially when gaps are introduced to maximize alignment (Figure 4).
Figure 3.
Localization of GFP RNAs Containing Internal Deleted Prolamine Segments.
(A) Schemes of internally deleted prolamine fragments.
(B) Visualization of GFP RNAs containing various prolamine cDNA segments, as detected by in situ RT-PCR, and their spatial relationships to prolamine PBs. Bar = 10 μm.
Figure 4.
Putative cis Sorting Determinant Sequences Responsible for Prolamine RNA Localization.
CDS1 and UA sequences were aligned, and conserved regions are boxed.
Glutelin RNAs Are Transported to the Cisternal ER by a Regulated Pathway
The deletion studies demonstrate that PB-ER targeting of prolamine RNAs requires two partially redundant cis elements and that the removal of one of these elements (e.g., the 3′ UTR) results in a reduction in RNA localization to the PB-ER. However, results from a previous study (Choi et al., 2000) have shown that replacement of the prolamine 3′ UTR with the corresponding glutelin 3′ UTR led to the total displacement of the prolamine RNAs from the PB-ER to the cisternal ER. Because these prolamine RNAs contain at least one cis element that is able to confer partial localization to the PB-ER, these observations suggest that the glutelin 3′ UTR itself contains a signal determinant that targets RNAs to the cisternal ER.
To test for the presence of a glutelin signal determinant, the localization of a maize 10-kD zein RNA in developing rice endosperm was studied. A hybrid RNA containing the 10-kD zein coding sequence and the nos 3′ UTR was targeted to the PB-ER, indicating that the coding sequence of this maize storage protein RNA contains one or more cis elements functionally equivalent to those present in the prolamine RNA. Replacement of the nos 3′ UTR with the glutelin 3′ UTR, however, redirected the location of the RNAs mainly to the cisternal ER (Figure 5). Overall, the results described here (Figures 2 and 5) and those from a previous study (Choi et al., 2000) indicate that the glutelin 3′ UTR contains one or more cis elements that targets RNAs to the cisternal ER. Moreover, the glutelin signal determinant is dominant over the “prolamine-type” cis elements in its capacity to retarget the bulk of the rice prolamine and maize zein RNAs from the PB-ER to the cisternal ER.
Figure 5.
Localization of 10-kD Zein RNAs in Developing Rice Endosperm.
The 10-kD zein RNA is located on the PB-ER, indicating that it contains a cis sorting determinant in the coding sequence. The RNA is redirected to the cisternal ER when the nos 3′ UTR is replaced by the glutelin 3′ UTR. Bar = 10 μm.
DISCUSSION
We had demonstrated previously that prolamine RNAs were sorted to the PB-ER by an RNA-based mechanism (Choi et al., 2000). In this study, deletion analysis permitted us to localize two partially redundant cis elements involved in trafficking prolamine mRNAs to the cortical ER. One of the cis elements is located in the coding region of the prolamine RNA. Similarly, one or more cis elements are present in the 10-kD maize coding sequence as well. The location of cis elements in the coding region for the rice and maize RNAs places far more constraints for sequence specificity–based mechanisms than cis elements located in the 3′ UTR because of stringent codon usage, as demonstrated for the expression of the Bacillus thuringiensis toxin coding region in plants (Perlak et al., 1991). In view of this consideration, one would expect more complex interactions between these RNAs and different components of the RNA transport process and of the cortical subcellular architecture, the latter involving the protein translational machinery as well.
The results described here also show that there is a second regulated RNA transport pathway from the nucleus to the cortical region of the developing rice endosperm cell. The glutelin RNA transport pathway is evident by its apparent dominance over the prolamine pathway when the glutelin 3′ UTR is present (Choi et al., 2000) (Figure 5). Identification of the cis elements present in the glutelin 3′ UTR should reveal whether its dominant character is attributable to the existence of a hierarchy of signal determinants that direct RNAs to specific subdomains of the cortical ER and/or whether it is simply the result of the relative spatial location of these cis elements (i.e., position effects). Studies to discriminate between these two possibilities are under way.
The prolamine targeting domains, CDS1 and UA, share two small conserved sequences of ∼10 and 8 nucleotides (Figure 4), respectively, which may constitute at least part of the cis element responsible for RNA localization. This redundancy of signals is essential, because the cis elements present in either prolamine targeting domain are not sufficient by themselves to direct RNA localization exclusively to the PB-ER. Our deletion analysis does not exclude the possibility that other cis elements may be present, especially a signal determinant formed by two noncontiguous RNA sequences. If present, however, such signals are not essential for the restricted localization of RNAs to the PB-ER, as is evident for cis elements present in CDS1 and UA. The requirement for multiple redundant cis elements has been observed for the localization of Ash1 in budding yeast (Chartrand et al., 1999), Vg1 (Deshler et al., 1998; Mowry and Cote, 1999; Yaniv and Yisraeli, 2001) and Fatvg (Chan et al., 1999) RNAs in Xenopus oocytes, and Nanos RNA in Drosophila oocytes (Bergsten et al., 2001). The redundant cis elements may serve as enhancers to promote multiple protein–protein interactions between trans factors that interact with these cis elements and one or more other factors required for RNA transport and localization.
RNA localization signals can be both sequence and/or structural in nature (Serano and Cohen, 1995; Ferrandon et al., 1997; Chartrand et al., 1999; Jansen, 2001; Kloc et al., 2002). The MFOLD program for RNA secondary structure (Zucker, 1998) predicts that CDS1 and UA are capable of forming stem loops, although the latter structure would have only moderate stability at best (ΔG of −2.0 kcal for UA and −17.2 kcal for CDS1). This property, together with the fact that the conserved sequence motifs in CDS1 and UA form different parts of the two stem-loop structures (data no shown), suggest that the signal determinant is sequence dependent, although secondary and tertiary structural signals cannot be excluded completely. Interestingly, the 10-kD zein RNA, which also is targeted to the PB-ER, shares no obvious identity with CDS1 or UA. This lack of conservation between these RNAs suggests that the cis elements responsible for directing these RNAs to the PB-ER may be recognized by different RNA binding trans factors.
A question that arises from this study is why the storage protein RNAs are transported and localized to the cortical ER. In plant cells that have a large central vacuole, the transport, localization, and translation of RNAs to the cortical region would be expected, because the large vacuole would displace the cytoplasm to the peripheral regions of the cell. However, the young subaleurone cells of developing rice endosperm studied here lack large vacuoles. Instead, they have a dense cytoplasm containing small vacuoles (one type is the glutelin PBs) dispersed throughout the cell. Although the ER-associated nuclear envelope and the ER distributed throughout the cytoplasm are potential sites for the localization and translation of storage protein RNAs, they appear to be underused for protein synthesis, because our in situ RT-PCR analysis showed that these RNAs are restricted to the cortical ER. Hence, the transport and targeting of RNAs to the cortical region is not caused by a trivial condition but likely is a requisite step for efficient gene expression.
Closely associated with this ER domain is a prominent cytoskeleton network (Muench et al., 2000). The concentration of cytoskeleton and, in particular, actin filaments would serve two functions. First, it would serve as the anchoring site for RNAs transported from the nucleus (Singer, 1992; Bogucka-Glotzer and Ephrussi, 1996; Bloom and Beach, 1999; Muench et al., 2000). Indeed, evidence was obtained showing that prolamine RNAs are retained on prolamine PBs by detergent-resistant, salt-sensitive binding sites that likely are associated with the cytoskeleton (Muench et al., 1998). Several RNA binding activities have been identified in an enriched cytoskeleton PB fraction that may serve as this RNA-anchoring activity (Sami-Subbu et al., 2000, 2001). Second, actin filaments also have been suggested to serve as a scaffold for polysomes or as sites enriched for translational factors (Lenk et al., 1977; Hesketh, 1994; Abe et al., 2003). If the physical association of translational complexes with actin filaments is necessary for efficient protein synthesis, this requirement would be fulfilled by the cortical region, which contains the highest density of cytoskeletal elements in plant cells (Kost and Chua, 2002). Hence, nuclear RNA transport to the cortical region and its translation in this cellular region may be general features in plants.
RNAs that code for protein reporters such as GFP (this study) and β-glucuronidase (Choi et al., 2000) are localized to the cisternal ER located proximal to the prolamine PBs. This RNA distribution is observed irrespective of whether the RNA codes for a secretory protein containing an N-terminal signal peptide such as the chitinase signal peptide–GFP used here or for cytoplasm-localized proteins such as the β-glucuronidase reporter (Choi et al., 2000) or nucleus-localized MS2-GFP (Hamada et al., 2003). The localization of RNAs that code for nonsecretory proteins and nontranslatable RNAs to the cortical ER (Choi et al., 2000; Hamada et al., 2003) excludes the involvement in RNA targeting of the signal recognition particle (SRP), which mobilizes the translational complex to the ER. SRP may play a role in stabilizing the location of RNAs that code for secretory proteins (such as the chitinase signal peptide–GFP reporter used in this study) to the cortical ER but not in the initial transport and targeting of these RNAs to this cellular region.
In addition to these RNA transport pathways to the cortical region of the cell, others are likely to exist. Viral and plant RNAs are able to move intercellularly and even long distances between tissues. The entry point for this transport is the plasmodesmata, channels that connect the cytoplasm of neighboring cells (Lucas and Gilbertson, 1994; Lucas et al., 2001; Haywood et al., 2002; Okita and Choi, 2002). The inner space of the channels is occupied by the desmotubule, tightly furled membranes continuous with the cortical ER (Overall and Blackman, 1996). The selectivity of RNAs capable of intercellular and long-distance transport indicates the presence of cis elements responsible for specific sorting to this intercellular channel (Lucas et al., 2001; Haywood et al., 2002).
The segregation of prolamine and glutelin RNAs to the PB-ER and the cisternal ER, respectively, likely facilitates the targeting of the coded proteins to their respective intracellular protein bodies (Okita et al., 1994; Okita and Rogers, 1996; Okita and Choi, 2002). The identification of cis elements that direct regulated RNA transport to the PB-ER or cisternal ER now enables us to directly test this hypothesis relating RNA localization to protein localization within the plant endomembrane system. Results from ongoing studies show that the 10-kD zein can be localized to the storage vacuole when its RNA is targeted from the PB-ER to the cisternal ER (our unpublished data). Similar efforts are under way to redirect a normally cisternal ER-localized RNA that codes for a storage vacuole–localized protein to the PB-ER.
Our results support the existence of two RNA pathways to the cisternal ER: a regulated glutelin RNA transport pathway and a constitutive RNA pathway. The redundancy of multiple pathways to the cisternal ER raises the possibility that the regulated pathway targets RNAs to a destination site on the cortical ER that is not identical to those of RNAs engaged along the constitutive pathway. Efforts are under way to determine whether the cisternal ER is composed of subdomains that harbor unique RNAs.
METHODS
T-DNA Vector Construction and Rice Transformation
The T-DNA vector pCAMBIA 1301 was used for all expression studies in transgenic rice (Oryza sativa). The core vector (pYW502) contained a rice glutelin Gt1 2.5-kb promoter (Choi et al., 2000), which was cloned into the HindIII and BamHI sites, and a nopaline synthase (nos) 3′ transcriptional terminator sequence, which was inserted into the SacI and EcoRI sites of pCAMBIA 1301. pYW100R was obtained by placing the chitinase signal peptide from pBIN mgfp5ER (provided by J. Haseloff, MRC Laboratory of Molecular Biology, Cambridge, UK) into the BamHI and EcoRI sites of pET30c (Novagen, Madison, WI) followed by the insertion of GFP reporter sequences (Davis and Vierstra, 1998) in the EcoRI and SacI sites. Manipulated DNA fragments as well as those described below were verified by DNA sequencing.
Various prolamine cDNA sequences were obtained by PCR using the primers listed in Table 1. For plasmid constructs 1 to 13 (Figure 2), the sense primers contained a SacI site at the 5′ end and the downstream antisense primers contained BamHI-SacI sites at the 3′ end. Amplified prolamine cDNA sequences were purified, digested with SacI, and cloned 3′ to the chitinase signal peptide–GFP reporter sequences contained in pYW100R. The GFP-prolamine DNA sequences then were obtained by BamHI digestion and subsequently cloned into the corresponding site of pYW502. Plasmid DNAs containing correctly oriented chitinase signal peptide–GFP–prolamine cDNA sequences were identified by restriction enzyme mapping.
Table 1.
Primers Used for PCR Amplification of Prolamine cDNA Sequences and for RT-PCR of GFP Nucleotide Sequences
| Name | Nucleotides | Constructs | DNA Sequence |
|---|---|---|---|
| PF1 | 149 to 170 | 1 and 9 | 5′-AAGGGAGCTCGTTCACACAGTTCAAGCATTAT-3′ |
| PF3 | 205 to 227 | 2, 7, and 8 | 5′-AAGGGAGCTCATGAAGATCATTTTCGTCTTTGC-3′ |
| PF4-Bgl | 245 to 267 | 17 | 5′-CTAAGATCTCATGCAGGCCTCTGCCGAGTTTG-3′ |
| PF5 | 452 to 470 | 4 and 11 | 5′-AAGGGAGCTCCAACAATCTCGCTATCAGGA-3′ |
| PF7 | 550 to 572 | 5 | 5′-AAGGGAGCTCAAGCTCAAGCTCTATTGGCTTT-3′ |
| PF9 | 318 to 341 | 3, 12, and 13 | 5′-AAGGGAGCTCGCTACAGCAACAGGTGCTTAGCCC-3′ |
| PF11 | 651 to 674 | 6 | 5′-AAGGGAGCTCTAATGTGTTTTAACAGTATAGTGG-3′ |
| PF11-Xba | 651 to 674 | 14 and 15 | 5′-AGGTCTAGATAATGTGTTTTAACAGTATAGTGGT-3′ |
| PF12 | 245 to 267 | 17 | 5′-CTAAGATCTCATGCAGGCCTCTGCCGAGTTTG-3′ |
| PFM | 655 to 676 | 16 and 17 | 5′-CGGGTCGACTGTGTTTTAACAGTATAGTGGT-3′ |
| PR12-Xba | 317 to 293 | 15 | 5′-AGGTCTAGAGGACAGGCGACTGCAGCTGATATT-3′ |
| PR11-Xba | 449 to 429 | 14 | 5′-AGGTCTAGACCACCAGCCTGATGTTGCCAG-3′ |
| PR10 | 572 to 550 | 12 | 5′-AAGGGAGCTCGGATCCAAAGCCAATAGAGCTTGAGCTTG-3′ |
| PR8 | 467 to 450 | 7 and 9 | 5′-AAGGGAGCTCGGATCCTGATAGCGAGATTGTTGG-3′ |
| PR6 | 650 to 633 | 8, 11, and 13 | 5′-AAGGGAGCTCGGATCCAAGACACCGCCAAGGGTG-3′ |
| PR4 | 690 to 667 | 10 | 5′-AAGGGAGCTCGGATCCTTTTTAACTTCCGAACCACTATAC-3′ |
| PR4-Bam | 691 to 665 | 17 | 5′-CGGGGATCCATTTTTAACTTCCGAACCACTATACTG-3′ |
| PR2 | 766 to 739 | 1 to 6, 14, and 15 | 5′-AAGGGAGCTCGGATCCAAATATGAAAGGCAACTTTATTTCTATT-3′ |
| PR1 | 691 to 665 | 16 and 17 | 5′-CGGGGATCCATTTTTAACTTCCGAACCACTATACTG-3′ |
| PRM | 312 to 297 | 16 and 17 | 5′-AAGGTCGACAGGACAGGCGACTGCAGCTGA-3′ |
| PF-GFP | 84 to 99 | 5′-TTTCACTGGAGTTGTC-3′ | |
| PR-GFP | 779 to 763 | 5′-TCTTTGTATAGTTCATC-3′ |
Sense (PF) and antisense (PR) primers were used to amplify specific prolamine cDNA regions, which were cloned subsequently downstream of a chitinase signal peptide–GFP coding sequence, and the resulting DNA was inserted between the rice Gt1 promoter and the 3′ nos transcriptional termination sequences in pYW502 to form almost all of the constructs listed in Figures 2 and 3. PF-GFP and PR-GFP are the sense and antisense primers used in RT-PCR to visualize the location of RNAs in endosperm sections.
Plasmid constructs 14 and 15 containing internal deletions of the prolamine cDNA sequences were obtained by PCR amplification of individual DNA fragments that flanked the 5′ and 3′ ends of the deleted DNA segment and subsequent ligation. Both plasmid constructs 14 and 15 contained the proximal half (nucleotides 651 to 691) of the 3′ untranslated region (UTR), which was amplified using PF11-Xba and PR4-Bam primers for PCR amplification. The amplified 3′ UTR fragment was digested with XbaI and BamHI and then cloned into the complementary sites of pBluescript II KS− (Stratagene) to give pSB33. The upstream regions were amplified with PF1 in combination with PR12-Xba or PF11-Xba. The resulting DNA fragments then were digested with SacI and XbaI and cloned into the corresponding sites of pSB33 to give pSB36 and pSB37, respectively. These prolamine DNA sequences were digested with SacI and HindIII and inserted into the corresponding sites of pYW100R, which places the prolamine sequences 3′ to the chitinase signal peptide–GFP reporter gene to give pSB36-1 (construct 15) and SB37-1 (construct 14). These DNAs then were digested with BamHI, and the chitinase signal peptide–GFP–prolamine sequences were cloned into pYW502.
Plasmid construct 16 was prepared as follows. pYW103, which contains the chitinase signal peptide–GFP–prolamine sequences (construct 1) cloned in pBluescript II KS− was digested with SalI, filled in with Klenow, and ligated to remove the SalI site in the polylinker region. The resulting plasmid was used in PCR to remove prolamine sequence 313 to 654 using primers PFM and PRM. The resulting DNA then was digested with SalI and ligated to give pCW78. This plasmid was used as a template for PCR amplification using PF3-Bgl and PR4-Bam or PF4-Bgl and PR4-Bam. These amplified DNA fragments were cloned into the filled-in SacI site of pYW500R to form pCW79. pYW500R contains the chitinase signal peptide–GFP sequences cloned in the BamHI and SacI sites of pYW502.
T-DNA vectors containing the 10-kD δ-zein were constructed as follows. The zein genomic clone pZ10-H3 (Kirihara et al., 1988) was digested with XbaI and then blunt-ended by fill-in reaction with Klenow. The reaction was incubated with NcoI, which releases the 10-kD zein coding sequence. The 453-nucleotide DNA fragment was cloned into the NcoI and BamHI (filled-in) sites of pTO158, which positions the 10-kD zein coding sequence between the 2.3-kb Gt1 promoter and the 3′ nos transcriptional termination sequences. Two other plasmids were constructed containing the 3′ end of a glutelin Gt2 (Okita et al., 1989) or prolamine Prol4a (Kim and Okita, 1988) gene in lieu of the nos transcriptional terminator sequences. The glutelin Gt2 sequences, which were obtained by PCR amplification, contained 310 nucleotides of the coding sequence and 208 nucleotides of the 3′ UTR and transcriptional termination sequences. Prolamine contained 356 nucleotides of the coding sequence, 107 nucleotides of the 3′ UTR, and 153 nucleotides of the 3′ flanking region. The resulting plasmids then were inserted into the HindIII site of pCAMBIA1301.
The various T-DNA vectors were transferred to Agrobacterium tumefaciens strain AgL1, which was used to transform rice as described previously (Choi et al., 2000). Regenerated transgenic rice plants were grown in a controlled-environment growth chamber under an 11-h, 26°C day/13-h, 22°C night growing regime.
RNA and Protein Gel Blot Analysis
RNA gel blot analysis was performed using 32P-labeled GFP DNA as a probe prepared by random-hexamer priming. Immunoblot analysis was performed using affinity-purified anti-GFP antibody generated in the laboratory.
In Situ Reverse Transcriptase–Mediated PCR and Microscopy
Cryosectioning, fixation, and washing of 10- to 14-day-old developing rice seed sections that were prepared and subjected to reverse transcriptase–mediated (RT) PCR were performed as described previously. RT-PCR was performed in the presence of 10 μM Oregon Green 488–dUTP and GFP primers (Table 1). After washing, the sections were incubated with rhodamine B hexyl ester (final concentration of 0.1 μM) to label the prolamine protein bodies.
Confocal microscopy was performed on a Zeiss 410 series laser scanning confocal microscope (Jena, Germany) and a Bio-Rad View Scan DVC-250 laser scanning confocal microscope using the fluorescein and rhodamine filter sets. Image processing was performed using Adobe Photoshop (Mountain View, CA), NIH Image, or Microsoft Powerpoint (Redmond, WA).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact T.W. Okita, okita@mail.wsu.edu.
Acknowledgments
This work was supported by grants from the National Science Foundation awarded to T.W.O. and by Department of Energy Grant DOE DE-FG05-95ER20194 to J.M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013821.
References
- Abe, S., Azama, K., Sugimoto, H., and Davies, E. (2003). Protein accumulation in the maize endosperm: Role of polyribosomes and the cytoskeleton. Plant Physiol. Biochem. 41, 125–131. [Google Scholar]
- Bergsten, S.E., Huang, T., Chatterjee, S., and Gavis, E.R. (2001). Recognition and long-range interactions of a minimal nanos RNA localization signal element. Development 128, 427–435. [DOI] [PubMed] [Google Scholar]
- Bloom, K., and Beach, D.L. (1999). mRNA localization: Motile RNA, asymmetric anchors. Curr. Opin. Microbiol. 2, 604–609. [DOI] [PubMed] [Google Scholar]
- Bogucka-Glotzer, J., and Ephrussi, A. (1996). mRNA localization and the cytoskeleton. Semin. Cell Dev. Biol. 7, 357–365. [Google Scholar]
- Chan, A.P., Kloc, M., and Etkin, L.D. (1999). fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development 126, 4943–4953. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.H., Singer, R.H., and Long, R.M. (1999). Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333–336. [DOI] [PubMed] [Google Scholar]
- Choi, S.-B., Wang, C., Muench, D.G., Ozawa, K., Franceschi, V.R., Wu, Y., and Okita, T. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407, 765–767. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Abramson, T., and Schnapp, B.J. (1998). A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8, 489–496. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., and Schnapp, B.J. (1997). Localization of Xenopus Vg1 mRNA by vera protein and the endoplasmic reticulum. Science 276, 1128–1131. [DOI] [PubMed] [Google Scholar]
- Ferrandon, D., Koch, I., Westhof, E., and Nusslein-Volhard, C. (1997). RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, S., Ishiyama, K., Choi, S.-B., Wang, C., Singh, S., Kawai, N., and Okita, T.W. (2003). The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15, 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.), S303.–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh, J. (1994). Translation and the cytoskeleton: A mechanism for targeted protein synthesis. Mol. Biol. Rep. 19, 233–243. [DOI] [PubMed] [Google Scholar]
- Jansen, R.P. (2001). mRNA localization: Message on the move. Nat. Rev. Mol. Cell Biol. 2, 247–256. [DOI] [PubMed] [Google Scholar]
- Kim, W.T., and Okita, T.W. (1988). Structure, expression, and heterogeneity of the rice seed prolamines. Plant Physiol. 88, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirihara, J.A., Petri, J.B., and Messing, J. (1988). Isolation and sequence of a gene encoding a methionine-rich 10-kDa zein protein from maize. Gene 71, 359–370. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Zearfoss, N.R., and Etkin, L.D. (2002). Mechanisms of subcellular mRNA localization. Cell 108, 533–544. [DOI] [PubMed] [Google Scholar]
- Kost, B., and Chua, N.H. (2002). The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 108, 9–12. [DOI] [PubMed] [Google Scholar]
- Lenk, R., Ransom, L., Kaufmann, Y., and Penman, S. (1977). A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell 10, 67–78. [DOI] [PubMed] [Google Scholar]
- Li, X., Franceschi, V.R., and Okita, T.W. (1993). Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72, 869–879. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., and Gilbertson, R.L. (1994). Plasmodesmata in relation to viral movement within leaf tissues. Annu. Rev. Phytopathol. 32, 387–411. [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2, 849–857. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L., and Cote, C.A. (1999). RNA sorting in Xenopus oocytes and embryos. FASEB J. 13, 435–445. [DOI] [PubMed] [Google Scholar]
- Muench, D.G., Chuong, S.D.X., Franceschi, V.R., and Okita, T.W. (2000). Developing prolamine protein bodies are associated with the cortical cytoskeleton in rice endosperm cells. Planta 211, 227–238. [DOI] [PubMed] [Google Scholar]
- Muench, D.G., Wu, Y., Coughlan, S.J., and Okita, T.W. (1998). Evidence for a cytoskeleton-associated binding site in prolamine mRNA localization to the protein bodies in rice endosperm. Plant Physiol. 116, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita, T.W., and Choi, S.-B. (2002). mRNA localization in plants: Targeting to the cell's cortical region and beyond. Curr. Opin. Plant Biol. 5, 553–559. [DOI] [PubMed] [Google Scholar]
- Okita, T.W., Hwang, Y.S., Hnilo, J., Kim, W.T., Aryan, A.P., Larsen, R., and Krishnan, H.B. (1989). Structure and expression of the rice glutelin multigene family. J. Biol. Chem. 264, 12573–12581. [PubMed] [Google Scholar]
- Okita, T.W., Li, X., and Roberts, M.W. (1994). Targeting of mRNAs to domains of the endoplasmic reticulum. Trends Cell Biol. 4, 91–96. [DOI] [PubMed] [Google Scholar]
- Okita, T.W., and Rogers, J.C. (1996). Compartmentation of proteins in the endomembrane system of plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 327–350. [DOI] [PubMed] [Google Scholar]
- Overall, R.L., and Blackman, L.M. (1996). A model of the macromolecular structure of plasmodesmata. Trends Plant Sci. 1, 307–311. [Google Scholar]
- Perlak, F.J., Fuchs, R.L., Dean, D.A., McPherson, S.L., and Fischoff, D.A. (1991). Modification of the coding sequence enhances plant expression of insect control protein genes. Proc. Natl. Acad. Sci. USA 88, 3324–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, J.A., Ellisman, M.H., and Benzer, S. (1990). Subcellular localization of transcripts in Drosophila photoreceptor neurons: Chaoptic mutants have an aberrant distribution. Genes Dev. 4, 806–821. [DOI] [PubMed] [Google Scholar]
- Sami-Subbu, R., Choi, S.-B., Wu, Y., Wang, C., and Okita, T.W. (2001). Identification of a cytoskeleton-associated 120 kDa RNA binding protein in developing seeds. Plant Mol. Biol. 46, 79–88. [DOI] [PubMed] [Google Scholar]
- Sami-Subbu, R., Muench, D.G., and Okita, T.W. (2000). A cytoskeleton-associated RNA-binding protein binds to the untranslated regions of prolamine mRNA and to poly(A). Plant Sci. 152, 115–122. [Google Scholar]
- Serano, T.L., and Cohen, R.S. (1995). A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development 121, 3809–3818. [DOI] [PubMed] [Google Scholar]
- Singer, R.H. (1992). The cytoskeleton and mRNA localization. Curr. Opin. Cell Biol. 4, 15–19. [DOI] [PubMed] [Google Scholar]
- Singer, R.H. (1993). RNA zipcodes for cytoplasmic addresses. Curr. Biol. 3, 719–721. [DOI] [PubMed] [Google Scholar]
- Svoboda, K.K.H. (1991). Intracellular localization of types I and II collagen mRNA and endoplasmic reticulum in embryonic corneal epithelia. J. Cell Sci. 100, 23–33. [DOI] [PubMed] [Google Scholar]
- Yaniv, K., and Yisraeli, J.K. (2001). Defining cis-acting elements and trans-acting factors in RNA localization. Int. Rev. Cytol. 203, 521–539. [DOI] [PubMed] [Google Scholar]
- Zucker, M. (1998). On finding all suboptimal foldings of an RNA molecule. Science 244, 48–52. [DOI] [PubMed] [Google Scholar]