Abstract
Arabidopsis stt3a-1 and stt3a-2 mutations cause NaCl/osmotic sensitivity that is characterized by reduced cell division in the root meristem. Sequence comparison of the STT3a gene identified a yeast ortholog, STT3, which encodes an essential subunit of the oligosaccharyltransferase complex that is involved in protein N-glycosylation. NaCl induces the unfolded protein response in the endoplasmic reticulum (ER) and cell cycle arrest in root tip cells of stt3a seedlings, as determined by expression profiling of ER stress–responsive chaperone (BiP-GUS) and cell division (CycB1;1-GUS) genes, respectively. Together, these results indicate that plant salt stress adaptation involves ER stress signal regulation of cell cycle progression. Interestingly, a mutation (stt3b-1) in another Arabidopsis STT3 isogene (STT3b) does not cause NaCl sensitivity. However, the stt3a-1 stt3b-1 double mutation is gametophytic lethal. Apparently, STT3a and STT3b have overlapping and essential functions in plant growth and developmental processes, but the pivotal and specific protein glycosylation that is a necessary for recovery from the unfolded protein response and for cell cycle progression during salt/osmotic stress recovery is associated uniquely with the function of the STT3a isoform.
INTRODUCTION
Sensory and signal transduction mechanisms that perceive ionic and osmotic stress and control cellular homeostasis are the focus of much current research activity (Hasegawa et al., 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2001; Zhu, 2002). Furthermore, many of the transport systems and the biochemical determinants responsible for ion homeostasis and compatible solute accumulation have been identified, and dissection of their functions in stress adaptation is ongoing (Hasegawa et al., 2000; Zhu, 2002). Despite these substantial gains, our understanding of the adaptive processes that are required for the homeostasis of basic cellular and metabolic processes in stress environments remains rudimentary. For example, secretory and cell division processes must readjust in response to stress imposition to establish cellular homeostasis. The secretory system is implicated in osmotic stress adaptation because genes that encode secretory and vacuolar proteins are induced by hypersaline stress (Gong et al., 2001). Alleles that render components of the secretory machinery dysfunctional also cause osmotic sensitivity (Zhu et al., 2002). Transport proteins that are targeted through the secretory system are essential for ion homeostasis (Shi et al., 2003). Although osmotic stress is implicated in the cell cycle arrest of plants (Burssens et al., 2000) as it is in yeast (Alexander et al., 2001), the mechanism(s) that coordinates osmosensing and plant cell cycle regulation has not been identified; consequently, its role in plant adaptation is not understood.
Protein N-glycosylation occurs in the lumen of the endoplasmic reticulum (ER) and in the secretory system. Protein glycosylation steps that occur predominantly in the ER can be separated into distinct processes (Silberstein and Gilmore, 1996; Helenius and Aebi, 2001). First, the dolicol-linked core oligosaccharide (Glc3Man9GlcNac2) is assembled on the cytosolic side of the ER membrane and then translocated to the ER lumenal surface by a flippase (Helenius et al., 2002). This assembly process is the target of the antibiotic tunicamycin, which inhibits UDP-GlcNac:dolichol phosphate GlcNac-P transferase activity. Subsequently, the oligosaccharyltransferase (OST) transfers the core oligosaccharide via N-linkage to an Asn residue of a nascent peptide. The third stage, glycan trimming, facilitates proper protein folding. Removal of glucose residues, by α-glucosidase I and II, from the N-linked core oligosaccharide produces the monoglucosylated form (GlcMan9GlcNac2) of the glycoprotein that can bind to the ER-localized lectins calnexin and calreticulin, which are associated, in animal cells, with the cochaperone ERp57 (Oliver et al., 1997). Interference with any of these processes results in the accumulation of misfolded proteins in the ER lumen and triggers an unfolded protein response (UPR).
UPR induces the expression of genes whose products, such as calnexin, calreticulin, binding protein (BiP), and peptide disulfide isomerase, facilitate proper protein folding in the ER (Travers et al., 2000; Martinez and Chrispeels, 2003). In mammals, UPR inhibits protein synthesis (Harding et al., 1999) and leads to cell cycle arrest at the G1-phase (Brewer et al., 1999). These coordinated responses are likely stress-induced checkpoints that trigger cells to reestablish homeostasis (Kaufman, 1999). Glycan modifications of the N-linked oligosaccharide that occur in the Golgi apparatus usually are crucial for cellular function in mammals. However, this processing may not be essential in plants. Inactivation of N-acetyl glucosaminyl transferase I (i.e., the first enzyme in protein glycan modification) by the cgl1 mutation prevents glycan modification of proteins but has no apparent effect on Arabidopsis growth (von Schaewen et al., 1993).
The heteromeric OST complex (Silberstein and Gilmore, 1996; Helenius and Aebi, 2001) catalyzes en bloc transfer of the core oligosaccharide from a membrane-bound dolichylpyrophosphate precursor to the N group of the Asn-X-Ser/Thr motif of a newly synthesized peptide (Yan and Lennarz, 1999). To date, nine subunit genes of the OST complex (OST1 to OST6, SWP1, WBP1, and STT3) have been identified in Saccharomyces cerevisiae (Knauer and Lehle, 1999). Temperature-sensitive mutations in many OST subunit genes decrease protein glycosylation. In many instances, these mutations are lethal at restrictive temperatures but also cause the UPR even at permissive temperatures (Knauer and Lehle, 1999). Arabidopsis DAD1 (Defender Against Death) is an ortholog of yeast OST2 and mammalian DAD1 and is the only characterized OST subunit of plant origin (Gallois et al., 1997). AtDAD1 functions as a genetic suppressor of the dad1 mutation in Chinese hamster cells. However, the in planta function of AtDAD1 or of any other OST subunit is not known.
T-DNA insertion mutant alleles of two members of the Arabidopsis STT3 family, encoding STT3a and STT3b OST subunits, were identified by a forward genetic screen for NaCl-sensitive mutants and by a computerized search. T-DNA insertions in STT3a and STT3b resulted in nonsense mutations with expression of truncated transcripts that encode proteins without the essential C-terminal lumenal domain in STT3 proteins. stt3a, but not stt3b, plants were sensitive to salt (NaCl and KCl) and to nonionic hyperosmotic stress. Decreased protein glycosylation, BiP hyperexpression, and reduced CycB1;1 expression in stt3a seedlings indicate that osmotic stress disturbs the cell cycle progression of stt3a root meristematic cells that is linked to improper glycosylation and protein folding. No obvious phenotype was detected for stt3b plants. However, the stt3a stt3b double mutation was gametophytic lethal, indicating that these OST isoforms have overlapping and essential functions. stt3a-1 and stt3a-2 now are established to cause decreased protein glycosylation. These results indicate that the STT3a subunit isoform mediates specific protein glycosylation steps that are necessary for cell cycle progression during osmotic stress adaptation.
RESULTS
Isolation of the Salt/Osmotic Stress–Sensitive stt3a Mutants
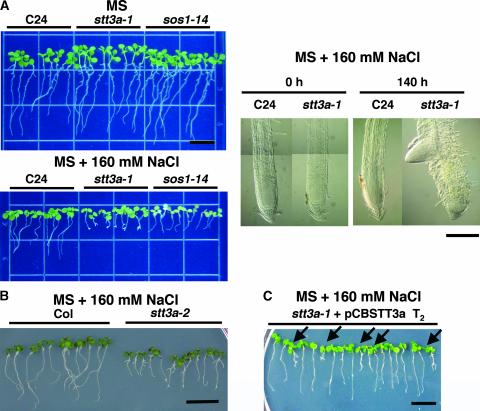
A salt/osmotic stress–sensitive mutant (stt3a-1; see below for explanation) was identified in a screen of an Arabidopsis ecotype C24 T-DNA insertion mutant population on medium supplemented with 160 mM NaCl (Zhu et al., 2002) (Figure 1A). The salt sensitivity of stt3a-1 was associated with root tip growth arrest and swelling and the induction of lateral roots (Figure 1). Thermal asymmetric interlaced PCR analysis identified a T-DNA insertion in the 18th exon of At5g19690 (at position 1986), which is annotated to encode an INTEGRAL MEMBRANE PROTEIN1/OST STT3 (Figure 2). Analysis of F2 progeny from a cross between stt3a-1 and C24 indicated that the mutation is recessive: 68 of 284 F2 plants tested exhibited NaCl sensitivity (χ2 = 0.12, P = 0.729 for a 3:1 segregation ratio). The T-DNA insertion is linked tightly to the mutation responsible for the salt-sensitive phenotype within 1 centimorgan, because all 48 randomly chosen NaCl-sensitive F2 plants were homozygous for the stt3a-1 allele based on PCR analysis (data not shown). Another salt-sensitive line (Figure 1B) that contains a T-DNA insertion in the STT3a locus (stt3a-2) was identified (SALK_058814) by analysis of the SIGnAL database (Columbia [Col] ecotype) (Figure 2). Transformation of stt3a-1 plants with a genomic fragment from BAC T29J13 (positions 39716 to 49044) that contains only STT3a as a full open reading frame resulted in a wild-type phenotype (Figure 1C). These results demonstrate that STT3a is a hyperosmotic/saline adaptation determinant of Arabidopsis.
Figure 1.
Salt-Hypersensitive stt3a Mutants Identified by Forward Genetic and Computerized Screening.
(A) At left, photographs of seedlings that were grown on MS agar medium for 1 week and then transferred to MS agar medium without (top) or with (bottom) 160 mM NaCl and incubated for 8 additional days: wild type (C24), stt3a-1, or C24 sos1 allele (sos1-14) seedlings. At right, root tip morphology of stt3a-1 before (0 h) and after (140 h) seedlings were exposed to NaCl. Seedlings were treated with NaCl, cleared with chloral hydrate, and observed with a microscope equipped with Nomarski differential interference contrast optics. Bars = 0.2 mm.
(B) Seedlings of the stt3a-2 mutant, identified by computerized screening, also are NaCl hypersensitive. Bar = 10 mm.
(C) An STT3a genomic fragment complements the salt-sensitive phenotype of stt3a-1 seedlings. T2 seedling progeny of stt3a-1 plants were transformed with pCBSTT3a, which contains a 9.3-kb genomic fragment containing the STT3a locus. Seedlings were transferred to medium containing 160 mM NaCl. Arrows indicate seedlings with a mutant phenotype (azygotes). Bar = 10 mm.
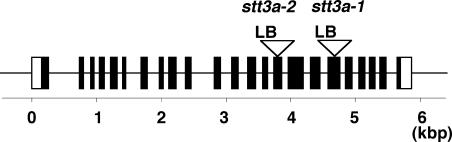
Figure 2.
stt3a-1 and stt3a-2 Alleles Contain T-DNA Insertions in Different STT3a Exons.
Scheme illustrates the location of the T-DNA insertions in STT3a (At5g19690) of the independent alleles. Exons (closed boxes) were deduced from the cDNA sequence, and open boxes indicate the 5′ and 3′ untranslated regions. Open triangles indicate T-DNA insertion sites, and the orientation of the T-DNA left border (LB) is indicated. stt3a-1 and stt3a-2 alleles contain T-DNA insertions in the 18th and 15th exons, respectively.
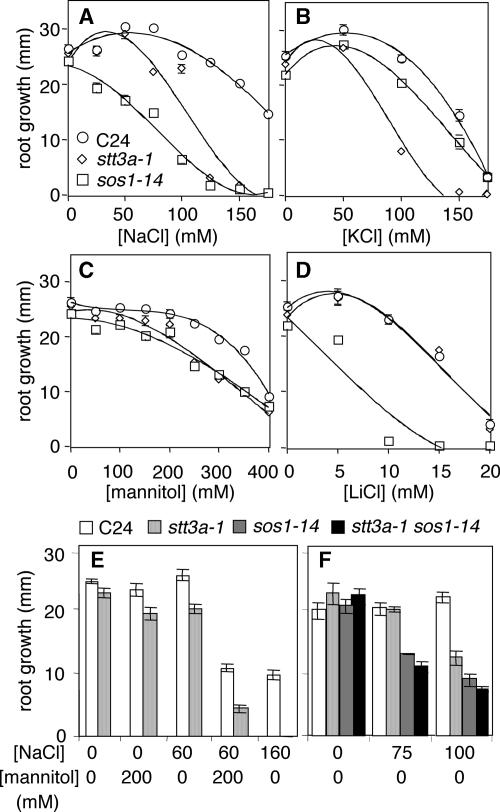
Root growth of stt3a-1 seedlings was hypersensitive to NaCl, KCl, and mannitol but not to LiCl (Figure 3) and K2SO4 (data not shown). Median inhibitory concentrations for wild-type and stt3a-1 seedlings were 175 and 110 mM NaCl, 150 and 90 mM KCl, 380 and 300 mM mannitol, 15 and 15 mM LiCl, and 62 and 62 mM K2SO4, respectively. These results indicate that stt3a-1 seedlings are sensitive to hyperosmotic stress and perhaps Cl− but are not sensitive to specific cations. Hyperosmotic sensitivity of stt3a-1 seedlings was confirmed, because root growth inhibition was increased substantially on medium supplemented with a combination of mannitol and NaCl at concentrations that individually cause minimal inhibition (Figure 3E). In addition, sos1-14 stt3a-1 seedlings were not more NaCl sensitive than sos1-14 seedlings, indicating that there is no additive effect on salt sensitivity (Figure 3F).
Figure 3.
Root Growth of stt3a-1 Seedlings after Salt and/or Osmotic Stress Treatment.
stt3a seedling root growth compared with that of the wild type (C24), sos1-14, or sos1-14 stt3a-1 according to the procedure described for Figure 1. One-week-old seedlings were transferred to medium with or without various amounts of NaCl (A), KCl (B), mannitol (C), LiCl (D), or NaCl and/or mannitol ([E] and [F]). Root growth (i.e., increase in length after transfer) was determined after 8 days. Error bars indicate standard errors.
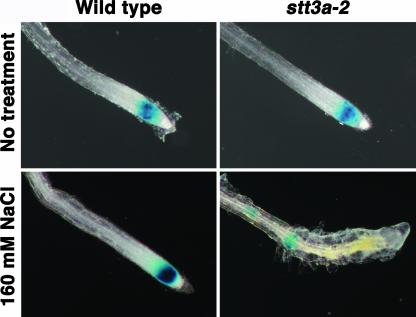
The salt/osmotic stress sensitivity of stt3a seedlings was associated with root tip swelling and enhanced lateral root development (Figure 1). This phenotype is unlike that of sos1 seedlings, for which salt/osmotic stress sensitivity is associated with the death of root meristematic cells (Huh et al., 2002). Instead, the salt/osmotic stress–responsive phenotype of stt3a seedlings resembles the phenotype caused by the mutation to the RSW3 locus (rsw3-1) that encodes α-glucosidase II (Burn et al., 2002). At restrictive temperatures, rsw3-1 causes the inhibition of cell division (Burn et al., 2002). To determine whether salt/osmotic stress inhibits root cell mitotic activity in stt3a seedlings, the expression of CycB1;1 was monitored (Figure 4). CycB1;1 is expressed during the G2/M-phase transition and is a marker for cell division activity (Shaul et al., 1996; Burssens et al., 2000). Col stt3a-2 plants were crossed to a ColCycB1;1-GUS (Colón-Carmóna et al., 1999) reporter line, and homozygous stt3a-2 F3 plants that exhibited salt/osmotic stress sensitivity were analyzed for β-glucuronidase (GUS) expression. CycB1;1 expression in the root tips of stt3a-2 seedlings was substantially lower after NaCl stress treatment than in the root tips of wild-type seedlings, indicating that mitotic activity was reduced substantially (Figure 4). Furthermore, CycB1;1 expression detected in stt3a-2 seedlings occurred in cells above rather than in the apex. The terminal meristem appeared disorganized and dysfunctional, which may contribute to lateral root initiation as a result of the lack of apical dominance (Figure 4). The NaCl-induced swelling of stt3a seedling root tips (Figures 1 and 4) was attributable primarily to radial cell expansion and not division. These results indicate that STT3 functions in salt stress adaptation to maintain the mitotic activity of root cells.
Figure 4.
CycB1;1 Expression in stt3a-2 Seedling Roots Is Disturbed by NaCl Treatment.
NaCl sensitivity of F3 seedlings from a FA4 (CycB1;1-GUS) × stt3a-2 cross was tested as described for Figure 1 (i.e., stt3a-2 homozygotes are salt sensitive). GUS activity in the root tip was detected 4 days after transfer to medium with or without 160 mM NaCl.
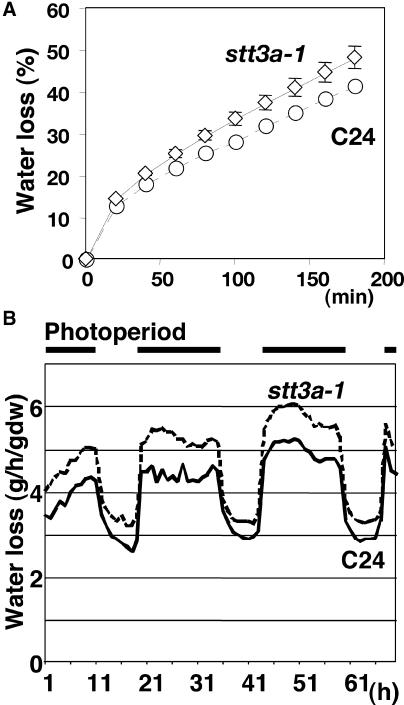
stt3a-1 plants exhibited greater water loss than wild-type plants (Figure 5), indicating that the stomatal response of stt3a-1 plants to lower water potential is impaired. Water loss of stt3a-1 plants also was substantially greater than that of wild-type plants during a light/dark cycle (Figure 5B). The altered stomatal regulation of stt3a-1 plants suggests that one possible mechanism for osmotic stress sensitivity is that the stt3a-1 mutation affects abscisic acid signaling, which controls both root and shoot responses to stress (Zhu, 2002). However, expression of RD29a, which is cold, abscisic acid, and NaCl controlled through ABRE (abscisic acid–responsive element) and DRE (desiccation-responsive element) (Yamaguchi-Shinozaki and Shinozaki, 1994; Ishitani et al., 1997), was not affected by the stt3a-1 mutation (data not shown). These results indicate that the function of STT3a in salt/osmotic adaptation does not involve an ABRE- or DRE-mediated signal pathway.
Figure 5.
stt3a-1 Plants Have Greater Transpirational Water Loss Than Wild-Type Plants (C24).
(A) Shoots were detached from greenhouse-grown 1-month-old Arabidopsis plants and dehydrated on filter paper at 25°C and ∼50% humidity. The fresh weight of each shoot was measured at the time indicated (after detachment). Data shown are averages from 10 shoots of C24 wild-type plants or 11 shoots of stt3a-1 plants. Error bars indicate standard errors of the mean; note that the small error bars for C24 are invisible on the graph.
(B) Transpirational water loss during a diurnal photoperiod (intensity of 140 μmol·m−2·s−1) at 25°C. Bars at top indicate the light period. The soil surface of pots in which plants were growing was sealed with plastic film to reduce evaporation. Plant water loss was measured by the gravimetric method using continuous monitoring of plant weight over a 70-h period. Each data point represents average results from nine plants. dw, dry weight.
STT3a and STT3b Encode Proteins Homologous with the Endomembrane-Localized Yeast OST Subunit STT3
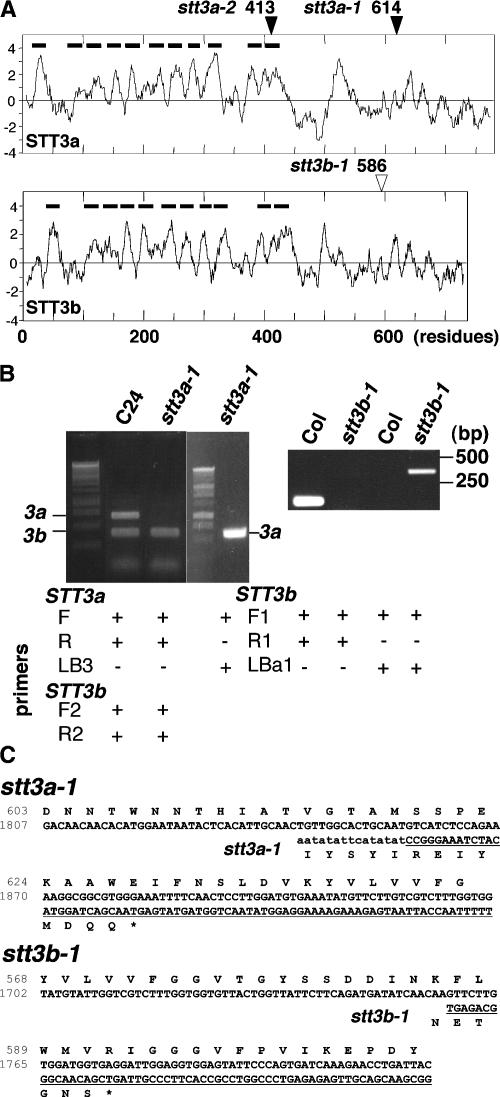
The At5g19690 product is similar in sequence to the S. cerevisiae protein STT3 (staurosporin and temperature sensitive) (Figure 6). Another STT3-like gene, At1g34130, exists in the Arabidopsis genome and is now referred to as STT3b. Arabidopsis STT3a (AtSTT3a) and AtSTT3b are 52% identical to each other and 43 and 44% identical, respectively, to S. cerevisiae STT3 (ScSTT3). A mutant with a T-DNA insertion in STT3b (stt3b-1) was identified in the SIGnAL database (SALK_033391). Comparison of hydrophobicity profiles indicates that AtSTT3a and AtSTT3b have the same topology as ScSTT3, in which the N and C termini are located in the ER lumenal space (Figure 7A). STT3a and STT3b contain a N-terminal signal peptide and 10 putative transmembrane regions. Reverse transcriptase–mediated (RT) PCR analyses indicated that both stt3a-1 and stt3b-1 plants produce chimeric transcripts consisting of ∼2 kb of STT3a or STT3b and a T-DNA border sequence (Figures 7B and 7C). Thus, stt3a-1 and stt3b-1 could encode peptides that have C-terminal truncations after the transmembrane region (Figure 7C). Several yeast stt3 alleles have been mapped to the C-terminal domain that would not be present in the truncated proteins (Spirig et al., 1997), and based on the yeast paradigm, Arabidopsis stt3a-1 and stt3b-1 are predicted to result in dysfunctional proteins. RT-PCR analysis indicated that STT3a expression occurs in the root and the shoot, whereas STT3b transcript is more abundant in the root than in the shoot (Figure 8). NaCl treatment did not induce STT3a or STT3b expression in wild-type plants. STT3b expression was not altered in stt3a-1 plants by NaCl treatment, indicating that there is no transcriptional compensation because of the dysfunctional allele.
Figure 6.
Arabidopsis STT3a and STT3b Are Homologous with S. cerevisiae STT3.
The predicted peptides of STT3a (At5g19690), STT3b (At1g34130), and STT3 from S. cerevisiae (NP_011493) were aligned using the CLUSTAL V analysis method. Identical residues in STT3 peptides are shaded in black, and residues with amino acid similarity are shaded in light gray. Closed and open arrowheads indicate the positions for truncation caused by the stt3a and stt3b mutations, respectively.
Figure 7.
stt3a-1 and stt3b-1 Produce Chimeric Transcripts.
(A) Kyte-Doolittle hydropathy profiling of STT3a (top) and STT3b (bottom). Bars indicate putative signal peptides and transmembrane regions, and arrowheads indicate the positions of mutations caused by T-DNA insertion. Note that the hydrophobic peaks at ∼500 are not conserved in yeast STT3.
(B) RT-PCR analysis of STT3a and STT3b transcripts in stt3a-1 (left gel) and stt3b-1 (right gel) seedlings. Five micrograms of total RNA extracted from C24, stt3a-1, Col, or stt3b-1 seedlings were used for reverse transcription. One-twentieth of the first-strand cDNA was used as a template to perform PCR. Primers STT3aF1 (F), STT3aR1 (R), and LB3 were used to analyze stt3a-1 seedling RNA, and STT3bF1 (F1), STT3bR1 (R1), and LBa1 were used to analyze stt3b-1 seedling RNA. STT3b primers STT3bF2 (F2) and STT3bR (R2) were used as positive controls for the analysis of stt3a-1 seedlings. Both stt3a-1 and stt3b-1 seedlings produced chimeric transcripts that were amplified with forward primer and T-DNA left border primer but not with primers for wild-type transcripts.
(C) Chimeric stt3a-1 (top) and stt3b-1 (bottom) RT-PCR product and predicted amino acid sequences compared with wild-type sequences. The underlined sequence indicates the transcript region that originates from the inserted T-DNA.
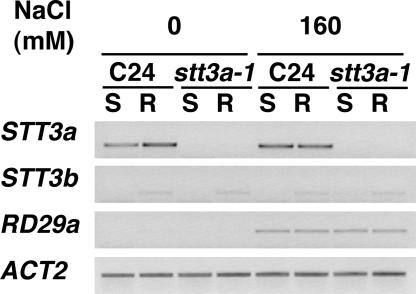
Figure 8.
Expression of STT3 Genes Is Not Regulated by Salt Stress.
C24 and stt3a-1 plants inoculated onto medium with or without 160 mM NaCl for 4 days. Transcript levels for STT3a, STT3b, RD29a (a salt-responsive control gene), and ACT2 (a loading control) were analyzed by RT-PCR using 10 μg of total RNA extracted from shoots (S) or roots (R).
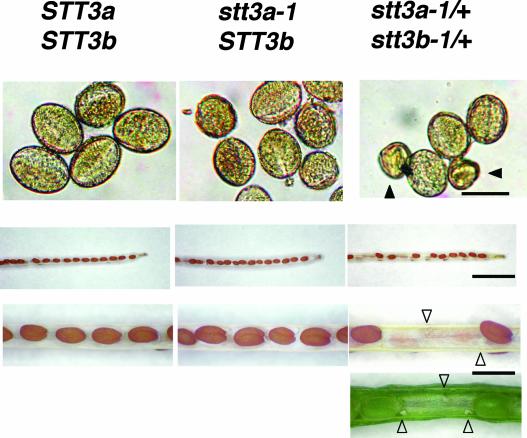
Unlike stt3a-1 or stt3a-2, the stt3b-1 mutation does not cause salt sensitivity (data not shown) or any other obvious specific phenotypic abnormality. Because neither STT3a nor STT3b is essential, an evaluation of their functional redundancy was conducted, and F2 segregants from a genetic cross (stt3a-1 × stt3b-1) were analyzed. Of 184 F2 plants, only 14 NaCl-sensitive plants were identified, and these were homozygous for the stt3a-1 T-DNA insertion. Surprisingly, all 14 of these plants were homozygous for the wild-type allele STT3b. F2 seedlings that are heterozygous at both loci exist within the same population, which is to be expected if crossing is successful and because STT3a and STT3b are on different chromosomes. The inability to recover homozygous stt3a-1 plants with a stt3b-1 allele indicates the lethality of stt3a-1 stt3b-1 gametes. Consistent with this hypothesis, stt3a-1/+ stt3b-1/+ plants produced flowers with normal and abnormal (inviable) pollen (Figure 9) at a ratio of 216:37 (normal:abnormal). A relatively lower frequency of abnormal pollen than expected from a 3:1 segregation (χ2 = 13.97, P = 0.00018 for a 3:1 segregation ratio) may be caused by an inability to recover all of the abnormal pollen during harvesting.
Figure 9.
The stt3a-1 stt3b-1 Double Mutant Is Lethal at the Gamete Stage.
The pollen and siliques produced from self-pollination of C24 wild-type (STT3a STT3b), stt3a-1 (stt3a-1 STT3b), and stt3a-1/+ stt3b-1/+ heter- ozygous plants were analyzed by light microscopy. Abnormal pollen found in stt3a-1/+ stt3b-1/+ pollen are indicated by closed arrowheads. Aborted ovules that failed to develop normally are labeled with open arrowheads. Bars = 0.025, 3, and 1 mm from top to bottom.
Examination of siliques in which there was incomplete seed development (Figure 9) revealed that 41 of 179 ovules (22.9%) did not produce mature seeds (χ2 = 0.32, P = 0.572 for a 3:1 segregation ratio), whereas all of the ovules produced mature seeds in siliques of wild-type (n = 154 ovules) and stt3a-1 (n = 158 ovules) plants. Together, these results indicate that both male and female stt3a-1 stt3b-1 gametes are not viable (i.e., only STT3a STT3b, stt3a-1 STT3B, or STT3a stt3b-1 gametes can survive). Because the stt3a-1 stt3b-1 gametes are not viable, a ratio of salt sensitive (stt3a-1):salt tolerant (STT3a) of 1:8 is predicted, and no plants should contain stt3a-1 or stt3b-1 alleles when the reciprocal mutation is homozygous. Because at least one functional wild-type STT3a or STT3b allele is required for gamete survival of progeny from the cross stt3a-1 × stt3b-1, it follows that the C-terminal lumenal domain in STT3 isoforms may be essential.
STT3a Functions in Protein N-Glycosylation
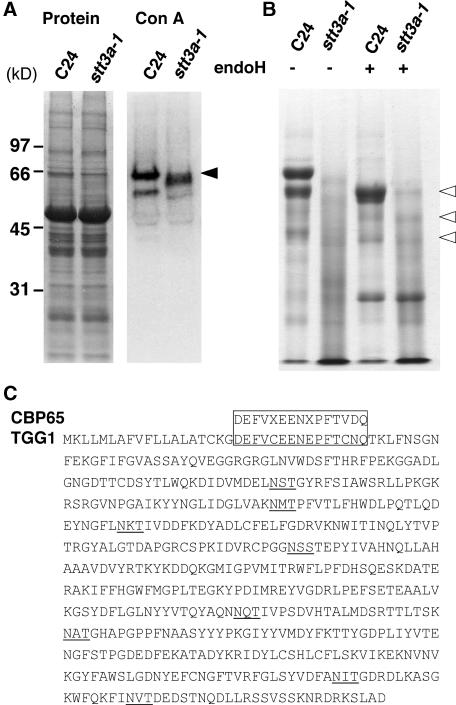
Yeast STT3 and orthologous proteins in other organisms function as an OST subunit (Karaoglu et al., 1997; Spirig et al., 1997). Neither AtSTT3a nor AtSTT3b cDNA suppressed stt3 mutations in S. cerevisiae (data not shown). When protein extracts from wild-type and stt3a-1 plants were separated by SDS-PAGE and detected with concanavalin A (Con A) lectin (which binds to the terminal mannose residue of N-linked oligosaccharides), a major 65-kD peptide (CBP65; Con A binding protein) was identified in the extract from wild-type but not from stt3a-1 plants (Figure 10A). A less distinct band of ∼60 kD was detected from stt3a-1 plants. A similar Con A binding peptide profile was observed for stt3a-2 but not for stt3b-1 or their parental Col plants (data not shown), indicating that this specific glycosylation defect is associated with stt3a but not with stt3b mutations. For further characterization of CBP65, extracts of both wild-type and stt3a-1 plants were fractionated using Con A–Sepharose binding affinity purification, and the proteins were separated by SDS-PAGE. The peptide profile of the Con A–purified fraction from stt3a-1 plants was smeared (Figure 10B), which is characteristic of proteins that are abnormally glycosylated. When the Con A binding fractions from extracts of both wild-type and stt3a-1 plants were treated with endoglycosidase H, to cleave N-linked glycans, and resolved by SDS-PAGE, a similar peptide profile (e.g., a 60-kD band) was detected (Figure 10B), indicating that the difference observed in the glycoprotein profiles (Figures 10A and 10B) between the genotypes is attributable to N-linked glycosylation differences.
Figure 10.
stt3a-1 Plants Are Defective in Protein Glycosylation.
(A) Glycosylated proteins in crude extracts of wild-type (C24) and stt3a-1 plants were resolved by SDS-PAGE and detected by Con A lectin blot analysis in total proteins (left gel) and Con A binding proteins (CBPs; right gel). A major 65-kD protein (CBP65) in wild-type seedlings is identified by the arrowhead. The Con A blot was exposed longer to visualize a faint 60-kD band detected in the stt3a-1 plant extract.
(B) Endoglycosidase H treatment resolves differences in protein glycosylation in wild-type (C24) and stt3a-1 plants. Proteins from crude extracts were absorbed onto Con A–Sepharose (Pharmacia Amersham), and the bound protein was recovered by elution with α-methylmannopyranoside. Fifteen micrograms of CBPs with or without deglycosylation treatment by endoglycosidase H (endoH) was analyzed by SDS-PAGE. Open arrowheads identify bands that are common in wild-type and stt3a-1 CBP fractions after endoglycosidase H treatment.
(C) Alignment of the CBP65 N-terminal sequence with the Arabidopsis TGG1 (At5g26000) peptide sequence. Potential N-glycosylation motifs are underlined.
The N-terminal sequence of the CBP65 protein that was obtained by Con A affinity purification was determined by Edman degradation analysis. The sequence of the first 15 amino acid residues of the peptide is nearly identical to the sequence of a myrosinase encoded by TGG1 in Arabidopsis (Figure 10C). TGG1 is a vacuolar protein (Andreasson et al., 2001) that contains eight potential N-glycosylation sites (Figure 10C). These results also indicate that STT3a functions in specific protein N-glycosylation.
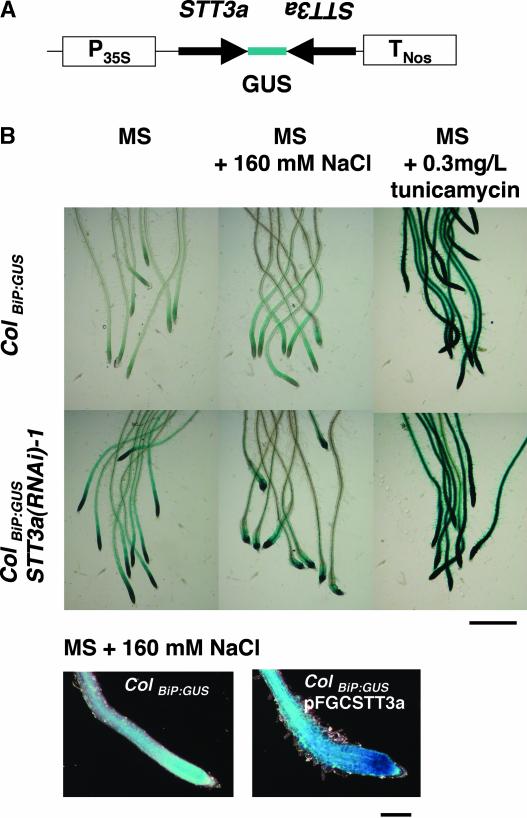
We evaluated stt3a plants for N-glycosylation defects in planta by monitoring the expression of the gene that encodes the ER isoform of the HSP70 chaperone, BiP, which is induced by incomplete protein folding caused by N-glycosylation defects in the ER (Oh et al., 2003). ColBiP-GUS plants (Oh et al., 2003) were transformed with a cassette expressing a hairpin double-stranded RNA using a 0.5-kb fragment of STT3a cDNA (pFGCSTT3a) (Figure 11). Introduction of pFGCSTT3a to wild-type C24 and Col plants caused a salt-sensitive phenotype indistinguishable from that of stt3a-1 or stt3a-2 (data not shown). Seven independent T1 plants transformed with pFGCSTT3a were confirmed for salt sensitivity, and all three ColBiP-GUS RNA interference (RNAi) lines that were tested showed reduced protein glycosylation and constitutively increased overall BiP-GUS expression, particularly in the root tip elongation zone, even in the absence of salt/hyperosmotic stress. This result confirms that inactivation of STT3a is sufficient to trigger the UPR pathway. Furthermore, BiP-GUS expression was even stronger in the root tip region of these STT3a-suppressed plants after NaCl treatment. Thus, cell cycle arrest, UPR, and abnormal enlargement of root tip cells occur in seedlings with a STT3a after NaCl/osmotic stress treatment (Figures 1, 4, and 11). No difference in BiP-GUS expression was detected in roots of wild-type or transgenic plants transformed with a RNAi cassette for STT3b (data not shown). As expected, the glycosylation inhibitor tunicamycin strongly induced BiP-GUS expression in root tips of both wild-type and pFGCSTT3a seedlings. These results demonstrate that salt/osmotic stress activates the UPR in cells of the root tip and link the STT3a isoform to the UPR, presumably through its function in protein glycosylation.
Figure 11.
RNAi of STT3a Activates the UPR Based on Activation of the BiP Promoter.
(A) Scheme of the STT3a gene-silencing cassette in pSTT3aR that was transformed into the ColBIP-GUS line.
(B) GUS activity is illustrated in the root tip of wild-type (ColBIP-GUS) and pSTT3aR transgenic [ColBiP-GUS STT3a(RNAi)-1] plants. T2 transformants were subjected to 160 mM NaCl stress treatment as described for Figure 1 and then stained for GUS activity after 48 h. Tunicamycin treatment was administered by inoculating seedlings onto MS agar medium plus 0.3 mg/L tunicamycin 24 h before the GUS reaction was initiated. Bars = 1 mm (top) and 0.2 mm (bottom).
DISCUSSION
Here, we provide evidence that the plant OST complex functions in salt/osmotic stress adaptation and that the expression of a functional OST subunit is essential for gametophytic development. Previously, OST subunit function in plants has been implicated only through heterologous complementation of the Chinese hamster cell dad1 mutant by AtDAD1 (Gallois et al., 1997). We show that mutations in a gene encoding an ortholog of STT3 cause salt/osmotic stress hypersensitivity in Arabidopsis. Unlike the single-copy and essential yeast STT3 gene (Zuffery et al., 1995), the Arabidopsis STT3 subunit is encoded by two genes, STT3a and STT3b. None of the three stt3 mutations (stt3a-1, stt3a-2, or stt3b-1) individually causes lethality. However, the stt3a-1 stt3b-1 double mutant is lethal at the gametophyte stage. Therefore, the STT3 subunit is essential for plant development and survival, and STT3a and STT3b have overlapping functions. Null mutations to genes that encode α-glucosidase I (gcs1) or II (rsw3-Ds), which are responsible for protein glycan trimming of N-linked glycosides, do not affect gamete viability but cause embryonic lethality (Boisson et al., 2001; Burn et al., 2002). Salt/osmotic stress–induced root swelling and enhanced lateral root initiation of stt3a seedlings strongly resemble the phenotypes associated with the temperature-sensitive mutant rsw3-1 at restrictive temperatures (Burn et al., 2002). However, the phenotypes caused by stt3a are a specific response to salt/osmotic stress.
STT3 Genes Have Isoform-Specific Functions
Notwithstanding STT3 redundancy in plant development, only mutations in the STT3a gene cause salt/osmotic stress sensitivity. This finding implies that the glycosylation of specific proteins is mediated by the OST enzyme complex that contains STT3a and that these proteins are required for salt adaptation, because STT3 is a likely catalytic subunit of the OST complex that recognizes the glycosylation motif of nascent peptides (Yan and Lennarz, 2002; Nilsson et al., 2003). Indeed, in vitro, the ScSTT3 subunit confers broad oligosaccharide substrate specificity to the OST complex (Zuffery et al., 1995). An OST complex with a functional ScSTT3 subunit can transfer even partially assembled core oligosaccharide to proteins, whereas upon depletion of functional STT3, OST activity decreases significantly and only fully assembled core oligosaccharide is a functional OST substrate, albeit with very low efficiency (Zuffery et al., 1995). Several proteins require N-glycosylation to express their normal function, including meprin A metalloproteinase (Kadowaki et al., 2000), GD3 synthase (CMP-NeuAc:GM3 α-2,8-sialyltransferase) (Martina et al., 1998), GM2 synthase (UDP-GalNAc:GM3 β-1,4-N-acetylgalactosaminyltransferase) (Haraguchi et al., 1995), and high-affinity immunoglobulin E receptor (Letourneur et al., 1995).
Lack of N-glycosylation negatively affects enzymatic activity, protein–protein interaction, and stability as well as folding. In plants, the glycosylated form of the ER-localized 3-hydroxy-3-methylglutaryl-CoA reductases appears to produce phytoalexin, whereas the unglycosylated form produces sterols (Denbow et al., 1996). In view of these important functions of glycosylation in phenotype determination, it is plausible that only subtle changes in the glycosylation of specific proteins resulting from stt3a mutations lead to the disturbance of specific salt adaptation responses. Another possible explanation for the unique salt/osmotic stress–adaptive function for STT3a is its differential expression in cells of the root tip. Although the expression of both STT3a and STT3b is detected constitutively in roots and shoots (Figure 8), it remains possible that a more detailed expression analysis will reveal that STT3a is expressed preferentially in cells of the root that are critical to salt adaptation.
Alternatively, STT3a may represent the prevalent enzymatic activity of the two isoforms. Indeed, the transcript abundance detected by RT-PCR analysis implies that STT3a may be the major isoform. Constitutive reduction of protein glycosylation and increased BiP-GUS expression in stt3a cells but not in stt3b cells indicate that STT3a is the major isoform of the two and suggest that stress-induced de novo synthesis of ER-targeted proteins may overload the already suboptimal protein glycosylation capacity of stt3a mutant cells. BiP-GUS expression and morphological abnormality were most profound in the root tip region, where cell division and elongation require active vesicular trafficking and deposition of cell wall constituents, membrane materials, and proteins that are important for adaptation to high salinity (Zhu et al., 2002; Shi et al., 2003). In the stt3a mutant background, the function of the minor isoform STT3b may not be sufficient to meet the requirements imposed by the fast-growing root tip.
The UPR May Regulate Cell Cycle Progression during Osmotic Stress Adaptation
Several lines of evidence indicate that there is integration of salt/osmotic stress, ER stress (i.e., the UPR), and cell cycle signaling networks. In yeast, protein N-glycosylation is required for cell cycle progression through the G1-phase (Arnold and Tanner, 1982), and the UPR causes G1 phase arrest (Carlberg and Larsson, 1993). Cell cycle arrest and growth inhibition caused by the UPR can be rescued by the overexpression of yeast G1 cyclins (Benton et al., 1996). Similarly, overexpression of cyclin D can rescue mammalian cells from cell cycle arrest when the UPR is activated by tunicamycin (Brewer et al., 1999). In both instances, the overexpression of cyclins permitted cell cycle progression under activated UPR, although protein glycosylation defects were not corrected (Benton et al., 1996; Brewer et al., 1999). In Arabidopsis, CycB1;1-GUS is expressed at G2-phase and is degraded at M-phase (Colón-Carmóna et al., 1999). The cessation of CycB1;1:GUS activity in stt3a root tip cells of salinized plants (Figure 4) indicates that cell cycle arrest occurs during G1- to S-phase. In stt3a but not in wild-type plants, salinity-induced cessation of CycB1;1-GUS was concomitant with enhanced BiP-GUS expression. Together, these results are in agreement with the occurrence of UPR-induced cell cycle arrest and the involvement of STT3a in cell cycle progression during salt adaptation. Even in wild-type plants, severe osmotic stress can induce BiP expression (Cascardo et al., 2000) and temporal decline in cyclin expression (Burssens et al., 2000). Therefore, recovery from the UPR and the resumption of cell cycle progression appears to be an integral part of plant salt/osmotic stress adaptation, and the stt3a mutation disturbs these essential adaptation mechanisms.
In yeast, the survival of ER and osmotic/salt stresses involves Ca2+ and calcineurin as common signaling components. Misfolded protein accumulation in both ER and osmotic stress trigger Ca2+ influx through the plasma membrane channel Cch1-Mid1, and this Ca2+ influx is critical for both cell survival under ER stress (Bonilla et al., 2002) and salt adaptation in yeast cells (Matsumoto et al., 2002). However, TCN1, the transcription factor that transduces calcineurin signaling into the activation of the ENA1 Na+-ATPase and salt tolerance, is dispensable for ER stress survival, even though calcineurin itself is required. Therefore, only upstream components of the calcineurin pathway are shared by both ER stress and salt stress signaling. Similarly, protein glycosylation defects activate FUS1 transcription through Sho1, Ste20/Ste50, Ste11, Ste7, Kss1, and Ste12, which overlap with signaling intermediaries in the HOG (mitogen-activated protein kinase) pathway, a principal osmotic stress–signaling pathway in yeast (Cullen et al., 2000). A functional HOG pathway also is essential for the survival of glycosylation-defective yeast cells, indicating a fundamental overlap between ER stress and salt/osmotic stress adaptation mechanisms.
A similar linkage may exist in plants. In Arabidopsis, osmotic stress induces a Ca2+ transient (Knight et al., 1998) that is presumed to activate the Ca2+-dependent salt overly sensitive (SOS) pathway (Zhu, 2002). Ca2+ activates the myristoylated calcium binding protein (SOS3), which in turn activates and recruits the protein kinase SOS2 to the plasma membrane, where the kinase activates the Na+/H+ antiporter SOS1 to enhance Na+ extrusion from cells (Quintero et al., 2002). Increased activation of the UPR pathway by salt is a result of the stt3a mutation. It remains to be determined if regulating the consequences of UPR signaling can modulate adaptation. However, water stress tolerance of BiP-overexpressing tobacco plants suggests that this is plausible (Alvim et al., 2001). Dissection of the networking UPR (Koizumi et al., 2001), SOS, and mitogen-activated protein kinase pathways (Mizoguchi et al., 2000) under salt/osmotic stress will provide further insights regarding integrated plant mechanisms that can lead to tolerance.
METHODS
Plant and DNA Materials
Arabidopsis thaliana ecotype C24RD29a-LUC (Ishitani et al., 1997) and ecotype ColumbiaCycB1;1-GUS line FA4 (Colón-Carmóna et al., 1999) were provided by J.-K. Zhu (University of Arizona, Tucson) and P. Doerner (University of Edinburgh, UK), respectively. The ColumbiaBiP-GUS line was prepared using a 1.5-kb Arabidopsis genomic promoter fragment of AtBiP2 (Oh et al., 2003). Columbia T-DNA insertion mutants were identified using the SIGnAL World Wide Web site at http://signal.salk.edu and were obtained from the ABRC (Columbus, OH). sos1-14 stt3a-1, stt3a-1 stt3b-1, and ColumbiaCycB1;1-GUS stt3a-2 genotypes were obtained by genetic crossing. BAC clone T29J13 was obtained from the ABRC.
Isolation of Salt-Hypersensitive Mutants and Growth Measurements
Preparation of the T-DNA–tagged Arabidopsis ecotype C24RD29a-LUC population and root-bending assay identification of salt-sensitive mutants were described previously (Koiwa et al., 2002; Zhu et al., 2002). Briefly, Arabidopsis seeds were sown onto MS agar medium (Murashige and Skoog, 1962) (1× MS salts, 30 g/L sucrose, and 16 g/L agar, pH 5.7), stratified for 4 days, and then incubated at 25°C for 1 week. Seedlings were transferred to basal medium supplemented with 160 mM NaCl or various concentrations of other compounds, and root growth was scored 8 days later. RD29a-LUC expression was monitored with a charge-coupled device imaging system (Roper Scientific, Trenton, NJ) after seedlings were subjected to cold (0°C, 2 days), abscisic acid (100 μM, 4 h), or NaCl (300 mM, 4 h) treatment (Ishitani et al., 1997; Koiwa et al., 2002).
Measurement of Transpirational Water Loss
Four-week-old C24 and stt3a-1 plants were grown in 3-inch pots in a greenhouse. The soil surface of each pot was covered with plastic wrap to determine transpirational water loss by the gravimetric method. The pots with plants were placed under light intensity of 140 μmol·m−2·s−1 at 25°C, and the weight was recorded every hour. Weight loss values are expressed as a function of shoot dry weight as determined after the completion of the experiment. Dehydration of detached shoots was measured as described previously (Zhu et al., 2002).
Molecular Genetic Analysis of stt3 T-DNA Insertion Alleles
Genomic sequence flanking the T-DNA insertion was determined with the thermal asymmetric interlaced PCR procedure of Liu et al. (1995) using primers corresponding to nested regions internal to the T-DNA left border and degenerate primers (Koiwa et al., 2002). Genetic cosegregation analysis was performed using an F2 population after backcrossing stt3a-1 to wild-type C24. F2 seedlings exhibiting salt/osmotic stress sensitivity (such as stt3a-1) were genotyped by PCR analysis. Similarly, salt/osmotic stress–sensitive seedlings from the SALK_058814 pool were genotyped. PCR-based genotypic analysis was performed as described (Koiwa et al., 2002) using primers STT3aF1 (5′-GGACAACACTTCCACGAACA-3′), STT3aR1 (5′-TCCAATACGAACCATCCACA-3′), and LB3 (Koiwa et al., 2002) for stt3a-1 and primers STT3aF2 (5′-ATTGCAAGTGTCAGTGAACATCAAC-3′), STT3aR2 (5′-CCTTGTCAGTCTTACCAGCAGAA-3′), and LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) for SALK_058814 (stt3a-2). stt3b-1 T-DNA alleles were identified by PCR screening of several plants obtained from the SALK_033391 seed stock using PCR with primers STT3bF1 (5′-ACTTGATGTGAACTATGTATTGGTCG-3′), STT3bR (5′-GGTTCTTTGATCACTGGGAATACTC-3′), and LBa1. Reverse transcriptase–mediated PCR analysis was performed as described (Koiwa et al., 2002) using primer pairs STT3aF1/STT3aR1 and STT3aF1/LB3 for STT3a transcript analysis and STT3bF1/STT3bR, STT3bF1/LBa1, and STT3bF2 (5′-TGTCTTAGCTGCAAGAGGAGC-3′)/STT3bR for STT3b transcript analysis. Two sets of primer pairs were designed to distinguish intact STT3a and STT3b transcripts and truncated transcripts caused by T-DNA insertions. A third set of STT3b primers was used with the STT3a primer set STT3aF1 and STT3aR1 for simultaneous detection of STT3a and STT3b.
Genetic Complementation and RNA Interference Gene Silencing
A genomic fragment containing the STT3a locus in BAC T29J13 was subcloned into pCB301 (Xiang et al., 1999). First, an 8-kb ClaI fragment was purified after digesting T29J13 and ligated into ClaI-digested pCB301 to obtain pBSTT3a-1. Then, a 1.4-kb AatII-BglII fragment from T29J13 was ligated into the AatII-BamHI site of pBSTT3a-1 to obtain pBSTT3a-2. Finally, the hygromycin resistance gene cassette from pPCVICEn4HPT (Hayashi et al., 1992) was inserted by blunt-end ligation into the KpnI site of pBSTT3a-2 to obtain pBSTT3a.
For construction of the STT3a RNA interference plasmid, a STT3a coding sequence was amplified using primers RiSTT3aF (5′-GGACTAGTGGCGCGCCGCTTCATTGTCATAGCTACCAGC-3′) and RiSTT3aR (5′-CGGGATCCATGGCGGCTCTAGAAAGCCCAC-3′) and integrated into the plasmid pFGC1008 (http://www.chromdb.org/strategy.html) to obtain pFGCSTT3a.
The binary plasmids were introduced into Agrobacterium tumefaciens GV3101. Arabidopsis plants were transformed using a floral spray procedure (Chung et al., 2000). T1 transformants were selected on one-quarter-strength MS medium containing 30 mg/L hygromycin, 100 mg/L carbenicillin, and 8 g/L agar.
Histochemical Analysis
BiP-GUS T2 plants transformed with pFGCSTT3a were treated with 160 mM NaCl as described above. Plants were harvested after 48 h of NaCl treatment, and GUS activity was detected in situ as described (Jefferson, 1987). Care was taken to ensure that wild-type and transgenic plants were salt treated, harvested, and stained for GUS activity simultaneously. Similarly, F3 plants from an FA4 × stt3a-2 cross were treated with 160 mM NaCl, harvested after 140 h, and analyzed for GUS activity.
Protein Analysis
For concanavalin A (Con A) staining, 15 μg of total soluble protein from Arabidopsis leaves was resolved on a 12% SDS-PAGE gel and transferred onto a nitrocellulose filter. The filter was incubated in blocking solution (1% BSA in PBS buffer) for 30 min and then in blocking solution containing 1 mg/L Con A–alkaline phosphatase conjugate (Roche, Mannheim, Germany) and 0.05% Tween 20 for 1 h. Filters then were washed three times with PBS containing 0.05% Tween 20 for 5 min each, rinsed once with alkaline phosphatase buffer (100 mM Tris, pH 9.3, and 100 mM NaCl), and then incubated with 25 μM CDP-star (Roche Applied Science, Indianapolis, IN) in alkaline phosphatase buffer. Con A binding glycoproteins were detected by chemiluminescence using a charge-coupled device camera imaging system (Princeton Instruments, Trenton, NJ).
Con A binding proteins (CBPs) were isolated from protein extracts of 1-month-old wild-type or stt3a-1 plants using Con A–Sepharose (Amersham Bioscience). Plant material (∼30 g fresh weight) was homogenized in liquid nitrogen and extracted in extraction buffer (20 mM Tris-HCl, pH 7.0, and 20 mM β-mercaptoethanol). Homogenate was filtered through Miracloth and centrifuged at 10,000g for 10 min. Proteins in cleared extracts were precipitated by 80% saturation of ammonium sulfate and centrifugation at 10,000g for 10 min at 4°C. Protein precipitates were resuspended in column buffer (20 mM Tris-HCl, pH 7.0, and 500 mM NaCl), centrifuged, and loaded onto Con A–Sepharose (1-mL bead volume) equilibrated with column buffer.
After washing with column buffer, bound proteins (CBPs) were eluted with column buffer containing 200 mM α-methylmannopyranoside. Eluates containing proteins were concentrated by trichloroacetic acid precipitation and resuspended in 1% SDS. For N-terminal sequencing by Edman degradation, a 30-μg CBP fraction was resolved by 12% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane, and the appropriate protein band was excised. For endoglycosidase H analysis, 10-μg CBP fractions were diluted in endoH buffer (50 mM Na citrate, pH 5.5, 0.5 mM phenylmethylsulfonyl fluoride, 0.1% Triton X-100, and 100 mM β-mercaptoethanol), incubated with 250 milliunits of endoglycosidase H (Roche) at 37°C for 7 h, and concentrated by trichloroacetic acid precipitation. Concentrated, dried proteins were resolved by 12% SDS-PAGE and detected using a Coomassie Brilliant Blue R 250 staining protocol.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Hisashi Koiwa, koiwa@neo.tamu.edu.
Accession Numbers
The GenBank accession number for STT3a (At5g19690) is AY056191. The accession number for Arabidopsis TGG1 (At5g26000) is P37702.
Acknowledgments
We thank J.-K. Zhu for providing the C24RD29a-LUC line and for helpful discussions, K. Okada and M. Ueda for their resourceful discussions and advice, P. Doerner for providing the CycB1;1-GUS line, and R. Jorgensen for pFGC1008 plasmid. We also thank for ABRC and the Salk Institute for T-DNA–tagged lines and a BAC clone used in this research. This work was supported by National Science Foundation Plant Genome Award DBI-9813360, by the College of Agriculture and Life Sciences at Texas A&M University and by Grant B1O2000-0938 from the Spanish Ministry of Science and Technology (to J.M.P). Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013862.
References
- Alexander, M.R., Tyers, M., Perret, M., Craig, M., Fang, K.S., and Gustin, M. (2001). Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell 12, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim, F.C., Carolino, S.M., Cascardo, J.C., Nunes, C.C., Martinez, C.A., Otoni, W.C., and Fontes, E.P. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, E., Bolt Jorgensen, L., Hoglund, A.S., Rask, L., and Meijer, J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127, 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, E., and Tanner, W. (1982). An obligatory role of protein glycosylation in the life cycle of yeast cells. FEBS Lett. 148, 49–53. [DOI] [PubMed] [Google Scholar]
- Benton, B.K., Plump, S.D., Roos, J., Lennarz, W.J., and Cross, F.R. (1996). Over-expression of S. cerevisiae G1 cyclins restores the viability of alg1 N-glycosylation mutants. Curr. Genet. 29, 106–113. [PubMed] [Google Scholar]
- Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20, 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla, M., Nastase, K.K., and Cunningham, K.W. (2002). Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21, 2343–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, J.W., Hendershot, L.M., Sherr, C.J., and Diehl, J.A. (1999). Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA 96, 8505–8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, J.E., Hurley, U.A., Birch, R.J., Arioli, T., Cork, A., and Williamson, R.E. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32, 949–960. [DOI] [PubMed] [Google Scholar]
- Burssens, S., Himanen, K., van de Cotte, B., Beeckman, T., Van Montagu, M., Inze, D., and Verbruggen, N. (2000). Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211, 632–640. [DOI] [PubMed] [Google Scholar]
- Carlberg, M., and Larsson, O. (1993). Role of N-linked glycosylation in cell-cycle progression and initiation of DNA synthesis in tumor-transformed human fibroblasts. Anticancer Res. 13, 167–171. [PubMed] [Google Scholar]
- Cascardo, J.C.M., Almeida, R.S., Buzeli, R.A.A., Carolino, S.M.B., Otoni, W.C., and Fontes, E.P.B. (2000). The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J. Biol. Chem. 275, 14494–14500. [DOI] [PubMed] [Google Scholar]
- Chung, M.H., Chen, M.K., and Pan, S.M. (2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9, 471–476. [DOI] [PubMed] [Google Scholar]
- Colón-Carmóna, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Cullen, P.J., Schultz, J., Horecka, J., Stevenson, B.J., Jigami, Y., and Sprague, G.F., Jr. (2000). Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denbow, C.J., Lang, S., and Cramer, C.L. (1996). The N-terminal domain of tomato 3-hydroxy-3-methylglutaryl-CoA reductases: Sequence, microsomal targeting, and glycosylation. J. Biol. Chem. 271, 9710–9715. [DOI] [PubMed] [Google Scholar]
- Gallois, P., Makishima, T., Hecht, V., Despres, B., Laudie, M., Nishimoto, T., and Cooke, R. (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J. 11, 1325–1331. [DOI] [PubMed] [Google Scholar]
- Gong, Z., Koiwa, H., Cushman, M.A., Ray, A., Bufford, D., Kore-eda, S., Matsumoto, T.K., Zhu, J., Cushman, J.C., Bressan, R.A., and Hasegawa, P.M. (2001). Genes that are uniquely stress-regulated in salt overly sensitive (sos) mutants. Plant Physiol. 126, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, M., Yamashiro, S., Furukawa, K., Takamiya, K., and Shiku, H. (1995). The effects of the site-directed removal of N-glycosylation sites from beta-1,4-N-acetylgalactosaminyltransferase on its function. Biochem. J. 312, 273.–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., Zhu, J.-K., and Bohnert, H.J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. [DOI] [PubMed] [Google Scholar]
- Hayashi, H., Czaja, I., Lubenow, H., Schell, J., and Walden, R. (1992). Activation of a plant gene by T-DNA tagging: Auxin-independent growth in vitro. Science 258, 1350–1353. [DOI] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2001). Intracellular functions of N-linked glycans. Science 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- Helenius, J., Ng, D.T., Marolda, C.L., Walter, P., Valvano, M.A., and Aebi, M. (2002). Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450. [DOI] [PubMed] [Google Scholar]
- Huh, G.-H., Damsz, B., Matsumoto, T.K., Reddy, P.M., Rus, A., Ibeas, J.I., Narasimhan, M.L., Bressan, R.A., and Hasegawa, P.M. (2002). Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J. 29, 649–659. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.-K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid–dependent and abscisic acid–independent pathways. Plant Cell 9, 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kadowaki, T., Tsukuba, T., Bertenshaw, G.P., and Bond, J.S. (2000). N-linked oligosaccharides on the meprin A metalloprotease are important for secretion and enzymatic activity, but not for apical targeting. J. Biol. Chem. 275, 25577–25584. [DOI] [PubMed] [Google Scholar]
- Karaoglu, D., Kelleher, D.J., and Gilmore, R. (1997). The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J. Biol. Chem. 272, 32513–32520. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- Knauer, R., and Lehle, L. (1999). The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta 1426, 259–273. [DOI] [PubMed] [Google Scholar]
- Knight, H., Brandt, S., and Knight, M.R. (1998). A history of stress alters drought calcium signalling pathways in Arabidopsis. Plant J. 16, 681–687. [DOI] [PubMed] [Google Scholar]
- Koiwa, H., et al. (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 99, 10893–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Martinez, I.M., Kimata, Y., Kohno, K., Sano, H., and Chrispeels, M.J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127, 949–962. [PMC free article] [PubMed] [Google Scholar]
- Letourneur, O., Sechi, S., Willette-Brown, J., Robertson, M.W., and Kinet, J.P. (1995). Glycosylation of human truncated Fc epsilon RI alpha chain is necessary for efficient folding in the endoplasmic reticulum. J. Biol. Chem. 270, 8249–8256. [DOI] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Oosumi, T., and Whittier, R. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Martina, J.A., Daniotti, J.L., and Maccioni, H.J. (1998). Influence of N-glycosylation and N-glycan trimming on the activity and intracellular traffic of GD3 synthase. J. Biol. Chem. 273, 3725–3731. [DOI] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, T.K., Ellsmore, A.J., Cessna, S.G., Low, P.S., Pardo, J.M., Bressan, R.A., and Hasegawa, P.M. (2002). An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 277, 33075–33080. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Ichimura, K., Yoshida, R., and Shinozaki, K. (2000). MAP kinase cascades in Arabidopsis: Their roles in stress and hormone responses. In Results and Problems in Cell Differentiation, H. Hirt, ed (Heidelberg, Germany: Springer-Verlag), pp. 29–38. [DOI] [PubMed]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nilsson, I., Kelleher, D.J., Miao, Y., Shao, Y., Kreibich, G., Gilmore, R., von Heijne, G., and Johnson, A.E. (2003). Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 161, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, D.-H., Kwon, C.-S., Sano, H., Chung, W.-I., and Koizumi, N. (2003). Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301, 225–230. [DOI] [PubMed] [Google Scholar]
- Oliver, J.D., van der Wal, F.J., Bulleid, N.J., and High, S. (1997). Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science 275, 86–88. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J., Ohta, M., Shi, H., Zhu, J.K., and Pardo, J.M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 99, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul, O., Mironov, V., Burssens, S., Van Montagu, M., and Inze, D. (1996). Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 93, 4868–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Silberstein, S., and Gilmore, R. (1996). Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 10, 849–858. [PubMed] [Google Scholar]
- Spirig, U., Glavas, M., Bodmer, D., Reiss, G., Burda, P., Lippuner, V., te Heesen, S., and Aebi, M. (1997). The STT3 protein is a component of the yeast oligosaccharyltransferase complex. Mol. Gen. Genet. 256, 628–637. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258. [DOI] [PubMed] [Google Scholar]
- von Schaewen, A., Sturm, A., O'Neill, J., and Chrispeels, M. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.K. (2001). Abiotic stress signal transduction in plants: Molecular and genetic perspectives. Physiol. Plant. 112, 152–166. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q., and Lennarz, W.J. (1999). Oligosaccharyltransferase: A complex multisubunit enzyme of the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 266, 684–689. [DOI] [PubMed] [Google Scholar]
- Yan, Q., and Lennarz, W.J. (2002). Studies on the function of oligosaccharyl transferase subunits: Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692–47700. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Gong, Z., Zhang, C., Song, C.-P., Damsz, B., Inan, G., Koiwa, H., Zhu, J.-K., Hasegawa, P.M., and Bressan, R.A. (2002). OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid–mediated and non-abscisic acid–mediated responses to abiotic stress. Plant Cell 14, 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuffery, R., Knauer, R., Burda, P., Stagljar, I., te Heesen, S., Lehle, L., and Aebi, M. (1995). STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14, 4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]