Abstract
Phytochromes regulate various light responses through their interactions with different signaling proteins, such as phytochrome interacting factor 3 (PIF3). However, the physiological functions of PIF3 in light signaling are not yet fully understood. To increase our understanding of these roles, we characterized a T-DNA insertional pif3 mutant and transgenic plants overexpressing the full-length PIF3. Transgenic overexpressing lines displayed longer hypocotyls and smaller cotyledons under red light and reduced cotyledon opening under both red and far-red light, whereas the pif3 mutant showed the opposite phenotypes. The accumulation of anthocyanin and chlorophyll further indicated complicated features of PIF3 function. The accumulation of anthocyanin was increased and the content of chlorophyll was decreased in the overexpression lines. Our data indicate that PIF3 plays complex roles depending on the type of light response and the light conditions.
INTRODUCTION
Phytochromes, which are the red and far-red light receptors of plants, regulate various light responses, including seedling photomorphogenesis, shade avoidance, and flowering (Quail et al., 1995; Neff et al., 2000; Quail, 2002). In Arabidopsis, five phytochromes have been identified (PHYA to PHYE) and categorized as photolabile (PHYA) or photostable (PHYB to PHYE) (Sharrock and Quail, 1989; Clack et al., 1994). Regardless of their photostability, all phytochromes undergo the same red and far-red light–dependent photoisomerization between the Pr and Pfr forms. During photomorphogenesis, the photolabile PHYA acts as a photoreceptor to mediate the very-low-fluence response and the far-red high-irradiance response; the photostable PHYB acts as a typical red light receptor to mediate the low-fluence response and the red light high-irradiance response (Somers et al., 1991; Nagatani et al., 1993; Parks and Quail, 1993; Reed et al., 1993; Shinomura et al., 1996; Yanovsky et al., 1997). The molecular mechanism for the action of PHYA as a far-red light receptor may hinge on its ability to translocate into the nucleus even under far-red light, whereas PHYB translocates into the nucleus only under red light (Kircher et al., 1999; Yamaguchi et al., 1999).
Molecular and genetic approaches have identified many phytochrome signaling components, including PHYA- or PHYB-specific signaling components, signaling components that are common to both, and components that interact directly with the phytochromes (Hudson, 2000; Quail, 2000). Among the phytochrome-interacting proteins, the basic helix-loop-helix (bHLH) protein phytochrome interacting factor 3 (PIF3) is the most extensively characterized to date. PIF3 was identified originally as a phytochrome-interacting protein using yeast two-hybrid screening (Ni et al., 1998). Characterization of transgenic plants expressing an N-terminal truncated PIF3 gene and antisense PIF3 has shown that PIF3 is a positive regulator of PHYB signal transduction. In a related study, poc1 mutant plants were found to have decreased PIF3 expression when grown in the dark and increased expression when grown under red light. These plants had shorter hypocotyls, which was interpreted as being associated with PIF3 overexpression (Halliday et al., 1999). Biochemical analysis indicated that the Pfr form of phytochrome interacts reversibly with PIF3 bound to the G-box element of various promoters (Ni et al., 1999; Martinez-Garcia et al., 2000), such as those of CCA1 and LHY. The decreased light inducibility of these two genes in PIF3 antisense transgenic plants corroborated the notion that phytochrome signals are targeted directly to the promoters of light-inducible genes through PIF3.
Currently, PIF3 is thought to be a positive regulator of PHYB-mediated light signal transduction. However, this hypothesis is based largely on the phenotypes associated with PIF3 antisense transgenic plants and those with altered PIF3 expression. This fact raises three cautions. First, the expression of antisense versions of genes that belong to multigene family members (such as PIF3) could perturb the expression of homologous family members. The phenotypes shown in such antisense transgenic plants could be attributable to the downregulation of some or all of the homologous genes. Second, the poc1 mutant, which has low PIF3 expression in the dark and higher expression in red light, may not simply be a PIF3-overexpressing mutant, as was claimed originally (Halliday et al., 1999). The phenotype could be the result of altered expression patterns rather than of the higher expression level under red light. Third, it has not been proven that an N-terminal truncated PIF3 is functionally equivalent to the full-length PIF3. The lack of a clearly visible phenotype in PIF3 sense transgenic plants reported in the literature (Ni et al., 1998) could be attributable to the overexpression of nonfunctional PIF3 caused by the N-terminal truncation. Overall, given the central importance of PIF3 in phytochrome signal transduction, it is necessary to firmly establish the physiological role of PIF3 using a conventional loss-of-function mutant and transgenic plants expressing the full-length PIF3.
Accordingly, we sought to determine the physiological role of PIF3 by characterizing a T-DNA insertional pif3-deficient mutant and transgenic plants expressing the full-length PIF3. Contrary to the current view, we found that PIF3 plays complex roles depending on the type of light response and the light conditions.
RESULTS
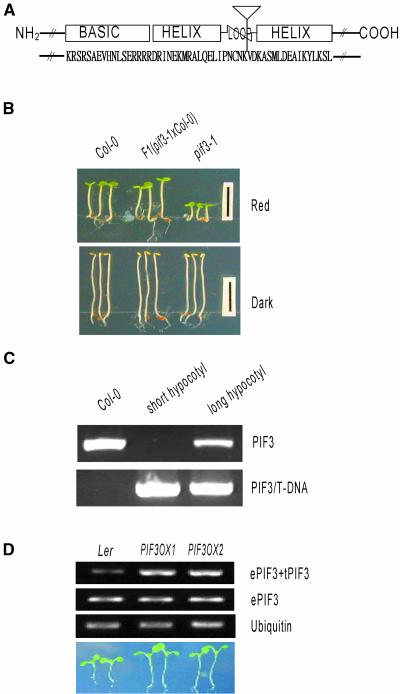
To investigate the physiological role of PIF3, we used a T-DNA insertional pif3 knockout line, pif3-1 (Salk_030753; Arabidopsis Stock Center) that was originally generated and characterized by J. Ecker's group at the Salk Institute (signal.salk.edu). The T-DNA was inserted into the fifth intron of the PIF3 gene (Figure 1A), leading to the deletion of the C-terminal 149 amino acids, including the second helix motif in the bHLH domain. Because both helices are required for DNA binding and dimerization (Ferre-D'Amare et al., 1993; Ellenberger et al., 1994; Brownlie et al., 1997; Shimizu et al., 1997), the insertion of T-DNA into the loop region is likely to disrupt the function of PIF3. Furthermore, the mutant phenotype (short hypocotyl length under red light) was recessive and cosegregated with the T-DNA, suggesting that the mutation is a loss-of-function mutation caused by a T-DNA insertion (Figures 1B and 1C). To further investigate the physiological role of PIF3, we generated transgenic plants expressing full-length PIF3 under the control of the 35S promoter of Cauliflower mosaic virus. After the creation of the transgenic line (PIF3OX), we confirmed the expression of both endogenous and transgenic PIF3 by reverse transcriptase–mediated PCR using primers that were capable of distinguishing between the two alleles (Figure 1D).
Figure 1.
Phenotypes of the pif3-1 Mutant and PIF3OX.
(A) Diagram of the PIF3 protein and the T-DNA insertion site. The inverted triangle indicates the T-DNA insertion site, and uppercase letters indicate the amino acid sequence corresponding to the bHLH region in PIF3.
(B) Hypocotyl phenotypes of wild type Columbia (Col-0), pif3-1, and heterozygous PIF3 [F1(pif3-1×Col-0] plants.
(C) Cosegregation analysis. Col-0 indicates genomic DNA of the wild-type plant, short hypocotyl indicates a genomic pool of 40 seedlings that had short hypocotyls and larger cotyledons, and long hypocotyl indicates a genomic pool of 126 seedlings that had long hypocotyls and smaller cotyledons. PIF3 indicates the PCR product of full-length PIF3, and PIF3/T-DNA indicates the PCR product of the PIF3–T-DNA hybrid fragment.
(D) Overexpression of PIF3 in transgenic plants. ePIF3 indicates endogenous PIF3, and tPIF3 indicates transgenic PIF3 in the PIF3-overexpressing lines (PIF3OX1 and PIF3OX2). The photograph at bottom shows the hypocotyl lengths of wild-type Landsberg erecta (Ler) and PIF3OX plants grown under white light.
PIF3 Negatively Regulates PHYB- but Not PHYA-Mediated Inhibition of Hypocotyl Elongation
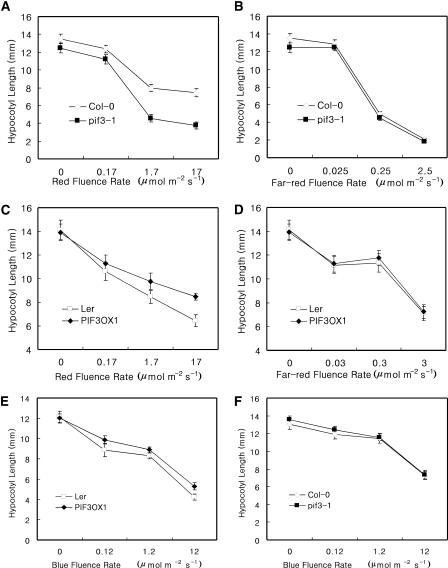
Hypocotyl elongation is regulated by both PHYA and PHYB. To determine whether PIF3 mediates the phytochrome-dependent inhibition of hypocotyl elongation, we measured the hypocotyl lengths of both mutant and transgenic plants grown under various fluence rates of either red or far-red light. As shown in Figures 2A and 2B, pif3-1 responded hypersensitively to red light but normally to far-red light for the inhibition of hypocotyl elongation. Consistent with the mutant phenotype, the overexpression of full-length PIF3 caused a hyposensitive response to red light and a normal response to far-red light (Figures 2C and 2D). Together, these data suggest that PIF3 negatively regulates PHYB- but not PHYA-mediated inhibition of hypocotyl elongation.
Figure 2.
Red and Far-Red Fluence Rate Response Curves for Hypocotyl Lengths.
(A) Red light fluence rate response curve of pif3-1 plants.
(B) Far-red light fluence rate response curve of pif3-1 plants.
(C) Red light fluence rate response curve of PIF3OX plants.
(D) Far-red light fluence rate response curve of PIF3OX plants.
(E) Blue light fluence rate response curve of PIF3OX plants.
(F) Blue light fluence rate response curve of pif3-1 plants.
The difference in hypocotyl lengths between the wild type (Landsberg erecta) and PIF3OX was more pronounced under white light than under red light. To determine whether this was the result of a blue light effect, we measured hypocotyl lengths in plants of both lines grown in blue light. As shown in Figure 2E, PIF3OX showed slightly longer hypocotyls than did the wild type in blue light. This result suggests that PIF3 also may act as a negative regulator for the blue light–mediated inhibition of hypocotyl elongation. An opposing effect, however, was not observed in pif3-1 plants grown under blue light, suggesting the presence of compensating components in blue light (Figure 2F).
PIF3 Negatively Regulates Both PHYA- and PHYB-Mediated Cotyledon Expansion
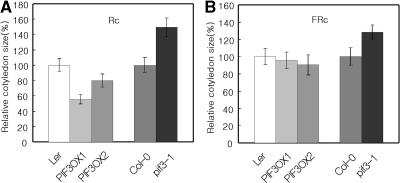
During our experiments, we noted that cotyledon sizes differed between the experimental lines and the wild type. To determine whether PIF3 regulates PHY-mediated cotyledon expansion, we measured areas of cotyledons from plants grown in either red or far-red light. As shown in Figure 3, PIF3OX plants grown in red light had smaller cotyledons than did wild-type plants, whereas pif3-1 plants had larger than normal cotyledons under the same conditions. In far-red light, PIF3OX plants had nearly wild-type-sized cotyledons, whereas pif3-1 plants had larger cotyledons. Because the insertion mutation causes increased cotyledon size in far-red light, the lack of a cotyledon size phenotype in PIF3OX plants may imply that PIF3 is not a rate-limiting factor for cotyledon expansion under far-red light. Together, these results suggest that PIF3 negatively regulates both PHYA- and PHYB-mediated cotyledon expansion.
Figure 3.
PHY-Induced Cotyledon Expansion in Both pif3-1 and PIF3OX Plants.
(A) Red light–induced cotyledon expansion.
(B) Far-red light–induced cotyledon expansion.
FRc, samples grown in far-red light; Rc, samples grown in red light.
PIF3 Negatively Regulates Both PHYA- and PHYB-Mediated Cotyledon Opening
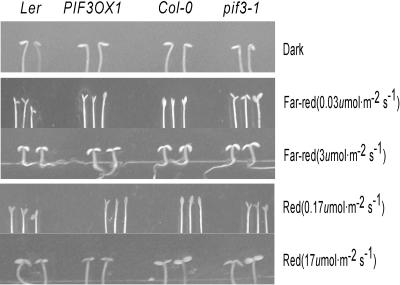
Cotyledon opening also is mediated by phytochrome signaling. To determine whether PIF3 plays a role in PHYA- or PHYB-mediated cotyledon opening, we observed cotyledon opening of pif3-1 and PIF3OX plants grown under low fluence rates of either red (0.17 μmol·m−2·s−1) or far-red (0.03 μmol·m−2·s−1) light. As shown in Figure 4, cotyledons of the pif3-1 mutant plants opened slightly more than did control (wild-type) cotyledons under both red and far-red light, whereas cotyledons of the PIF3OX plants opened slightly less than did control cotyledons in both conditions. The altered cotyledon opening was not caused by an abnormality in the cotyledons, as indicated by the normal closed state of the cotyledons in the dark and the appropriate opening response at higher fluence rates of both red and far-red light. These results suggest that PIF3 negatively regulates both PHYA- and PHYB-mediated cotyledon opening.
Figure 4.
PHY-Induced Cotyledon Opening in pif3-1 and PIF3OX Plants.
Phenotypic differences can be seen at the lower fluence rates of red and far-red light.
PIF3 Positively Regulates Both PHYA- and PHYB-Mediated CHS Induction
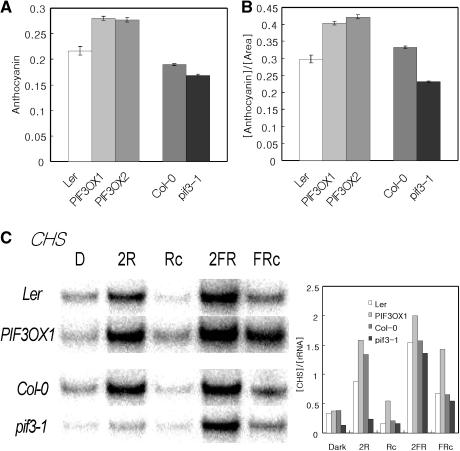
Phytochrome signaling modulates various physiological processes by regulating sets of light-response genes (Ma et al., 2001; Tepperman et al., 2001; Wang et al., 2002). The production of anthocyanin is one such response. To determine whether PIF3 has any role in anthocyanin biosynthesis, we quantified the content of anthocyanin in mutant, transgenic, and wild-type seedlings grown under continuous far-red light. As shown in Figure 5A, the anthocyanin content was slightly higher in the overexpression lines and marginally lower in the mutant plants compared with the wild-type control. Because anthocyanin was accumulated mainly in the upper part of hypocotyls and cotyledons and PIF3 had little effect on hypocotyl length in far-red light, we normalized the anthocyanin content by cotyledon size to control for the various cotyledon phenotypes. As shown in Figure 5B, after normalization, the anthocyanin content per area remained higher in the PIF3OX plants and lower in the mutant plants.
Figure 5.
Accumulation of Anthocyanin and Expression of CHS in pif3-1 and PIF3OX Plants.
(A) Accumulation of anthocyanin in pif3-1 and PIF3OX plants.
(B) Anthocyanin content normalized by cotyledon size.
(C) RNA gel blot analysis of CHS. Samples were grown for 4 days. The graph indicates the levels of CHS normalized to that of 18s rRNA. D, samples grown in the dark; 2R, samples transferred from dark to red light for 2 h; Rc, samples grown in red light; 2FR, samples transferred from dark to far-red light for 2 h; FRc, samples grown in far-red light.
The expression of the chalcone synthase gene (CHS), which encodes the first enzyme in the anthocyanin biosynthesis pathway, is modulated by light. To determine whether PIF3 regulates the expression of CHS, we used RNA gel blot analysis to determine the expression of CHS. As shown in Figure 5C, the expression level of CHS was lower in pif3-1 plants and higher in PIF3OX plants under both red and far-red light conditions. This expression pattern of CHS in pif3-1 and PIF3OX plants is consistent with the anthocyanin content. Together, these data suggest that PIF3 is a positive component for both PHYA- and PHYB-mediated induction of CHS.
Regulation of Chlorophyll Accumulation by PIF3
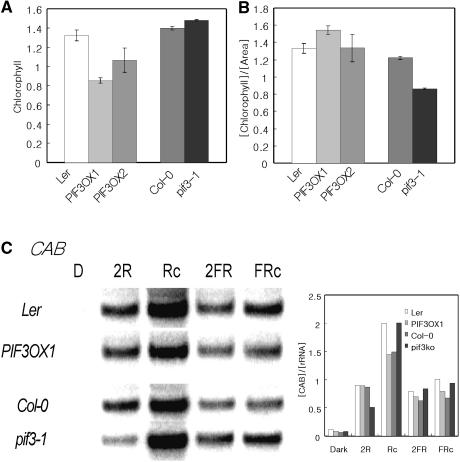
One of the main physiological functions regulated by light is photosynthesis. To determine if PIF3 plays any role in chlorophyll accumulation, we first determined the content of chlorophyll in seedlings grown under continuous red light. As shown in Figure 6A, PIF3-overexpressing plants contained a lower amount of chlorophyll, whereas the mutant plants had levels similar to those of wild-type controls. Because the majority of chlorophyll is found in cotyledons, however, it was possible that the apparent difference in chlorophyll content was the result of the different sizes of the respective cotyledons. When chlorophyll levels were normalized by cotyledon size, PIF3OX and pif3-1 plants contained chlorophyll amounts comparable to those of the wild-type controls (Figure 6B). The expression of CAB also was not much different among plants. These results imply that although PIF3 negatively regulates PHYB-mediated chlorophyll accumulation at the seedling level, this negative action is largely the result of the difference in cotyledon size.
Figure 6.
Accumulation of Chlorophyll and the Expression Pattern of CAB in pif3-1 and PIF3OX Plants.
(A) Accumulation of chlorophyll in pif3-1 and PIF3OX plants.
(B) Chlorophyll content normalized by cotyledon size.
(C) RNA gel blot analysis of CAB. Samples were grown for 4 days. The graph indicates levels of CAB normalized to that of 18s rRNA. D, samples grown in the dark; 2R, samples transferred from dark to red light for 2 h; Rc, samples grown in red light; 2FR, samples transferred from dark to far-red light for 2 h; FRc, samples grown in far-red light.
DISCUSSION
Reevaluating the Function of PIF3 in Phytochrome Signaling
We characterized the physiological function of PIF3 using a T-DNA insertional pif3-deficient mutant and transgenic plants overexpressing the full-length PIF3. Contrary to previous reports, our data showed that PIF3 acts as either a positive or a negative component for phytochrome signaling, depending on the nature of the applied light. We showed that PIF3 is a negative regulator of PHYB- but not PHYA-mediated inhibition of hypocotyl elongation and a negative regulator of both PHYA- and PHYB-induced cotyledon expansion and opening. For the induction of CHS, PIF3 acted as a positive component for both PHYA and PHYB signal transduction. Together, our data suggest that PIF3 modulates phytochrome-mediated light responses both positively and negatively depending on the light condition and the type of phytochrome.
The contradictory data on the physiological function of PIF3 might be attributable to the plants used in each of the studies. Here, we used transgenic plants expressing full-length PIF3 and a T-DNA insertional pif3-deficient mutant, whereas the previous studies used transgenic plants expressing an antisense PIF3, an N-terminal truncated PIF3, or the mutant poc1 (Ni et al., 1998; Halliday et al., 1999). However, these previous reports have three major limitations that suggest that they be interpreted with caution. First, the expression of antisense PIF3 could cause the suppression of homologous genes in addition to PIF3. A Basic Local Alignment Search Tool (BLAST) search with PIF3 showed that a short sequence encoding the helix motif was a good match with fragments of at least two other bHLH genes (At2g20180 and At4g36930). Because a short stretch of shared nucleotide sequence (∼23 bp) is capable of suppressing homologous genes in transgenic plants (Hamilton and Baulcombe, 1999; Thomas et al., 2001), the phenotypes of the antisense transgenic lines could be the result of the suppression of not only PIF3 but also other homologous genes.
Second, the functional equivalence between full-length PIF3 and the N-terminal (63 amino acids) deleted PIF3 used in the previous study (Ni et al., 1998) has not been proven. Thus, the transgenic phenotypes shown in the previous study do not necessarily reflect the native function of full-length PIF3. Third, the poc1 mutant, which was interpreted as a PIF3 overexpressor mutant (Halliday et al., 1999), showed altered expression of PIF3, not simple overexpression. The poc1 mutant showed markedly decreased PIF3 expression in the dark and increased expression in red light, as determined by semiquantitative reverse transcriptase–mediated PCR (Halliday et al., 1999). Although the shorter hypocotyl length of the poc1 mutant may be caused by the increased expression of PIF3 under red light, the exact nature of the poc1 mutant remains unclear. We believe that the characterization of transgenic lines expressing the full-length PIF3 and the conventional loss-of-function mutant, both discussed here, are more robust methods for establishing the function of PIF3.
Positive and Negative Control of Light Responses by PIF3
Our results showed that PIF3 acts as either a positive or a negative regulator of light responses, although the exact mechanism responsible for this is not clear. Because PIF3 is a transcription factor, a simple way to explain the differential regulation is to assume that in the presence of the appropriate light, PIF3 activates the transcription of both positive and negative factors involved in light responses. In this explanation, PIF3 activates transcription in both cases, and the differential regulation is attributable to the nature of the transcribed positive or negative factors. Alternatively, PIF3 may function as either a transcriptional activator or a repressor, depending on the specific promoters of the different target genes.
Overall, the fact that responses are regulated either positively or negatively by a single phytochrome-interacting protein (PIF3) suggests that members of the phytochrome signaling pathway cannot be classified simply as either positive or negative signaling components. We showed that PIF3 is a positive component for CHS induction, whereas it is a negative component for the inhibition of hypocotyl elongation, cotyledon opening, and cotyledon expansion. The apparent positive and negative regulation of light responses by a single factor is not unprecedented. Among the previously characterized signaling components, ELF3, CCA1, LHY, and HFR1 showed both positive and negative effects on light responses.
ELF3 is a PHYB-interacting protein that is involved in circadian clock function. The mutation in ELF3 caused longer hypocotyls and enhanced acute induction of CAB by light, whereas ELF3 overexpression caused shorter hypocotyls (Zagotta et al., 1996; Liu et al., 2001). CCA1 and LHY, both of which are MYB-like transcription factors, also are involved in circadian clock function. The overexpression of either CCA1 or LHY caused longer hypocotyls and induction of CAB (Wang et al., 1997; Schaffer et al., 1998; Mizoguchi et al., 2002). Functional characterization of an hfr1-deficient mutant showed that HFR1 is necessary for PHYA- but not PHYB-induced inhibition of hypocotyl elongation and CAB expression (Fairchild et al., 2000; Soh et al., 2000). Microarray analysis, however, showed that HFR1 regulates the expression of the PHYA-inducible genes both positively and negatively (Wang et al., 2002). Because phyA and phyB mutants show longer hypocotyls and decreased expression of CAB and CHS in red and far-red light, respectively, the phenotypes caused by those genes clearly indicate that a single signaling component can act either positively or negatively depending on light responses. These data suggest that the apparent light responses are the result of actions exerted by various signaling components, each of which can act either positively or negatively depending on the light condition.
However, it is unclear why a single signaling component has opposite effects on different light responses. One possibility is that such a signaling component is not used solely in light signaling. Hypocotyl length and leaf expansion are regulated not only by light but also by various other signals, including plant hormones (Lincoln et al., 1990; Jensen et al., 1998; Friedrichsen et al., 2000; Choe et al., 2001; Tseng et al., 2001; Borevitz et al., 2002). Similarly, anthocyanin production and chloroplast function also are regulated by various signals (Ivanov et al., 1995; Weatherwax et al., 1996; Laby et al., 2000; Winkel-Shirley, 2001). Although light signals lead to shortened hypocotyl length and increased anthocyanin production, it is feasible that other signals may regulate those processes differentially. To integrate these various signals, plants might have evolved the ability to use the sum of signals from various components rather than from one or two master signaling components.
Negative Regulation of PHYB- but Not PHYA-Induced Inhibition of Hypocotyl Elongation by PIF3
The regulation of PHYB- but not PHYA-induced hypocotyl elongation by PIF3 is intriguing. We have shown that both PIF3 overexpression and deficiency affect PHYB- but not PHYA-induced inhibition of hypocotyl elongation, whereas they affect both PHYA- and PHYB-induced cotyledon expansion, cotyledon opening, and CHS expression. This is puzzling, considering that PIF3 is able to bind the G-box and interact with both PHYA and PHYB (Martinez-Garcia et al., 2000). One possible explanation is that the inhibition of hypocotyl elongation by phytochromes requires a stronger interaction than do the other light responses. It has been shown that PIF3 interacts more strongly with PHYB than with PHYA in vitro (Zhu et al., 2000). If the inhibition of hypocotyl elongation requires a stronger interaction than other light responses, this may explain why PIF3 does not have any effect on PHYA-induced hypocotyl elongation even though it affects other PHYA-induced light responses.
Another possibility is the presence of excessive amounts of other components only under far-red light, leading to suppression of the PHYA-induced inhibition of hypocotyl elongation. Some phytochrome signaling components involved in hypocotyl elongation are expressed differentially under red and far-red light. HY5, which regulates both PHYA- and PHYB-induced inhibition of hypocotyl elongation, is induced preferentially by far-red light (Wang et al., 2002). Similarly, HFR1, which regulates only PHYA-induced inhibition of hypocotyl elongation, is induced preferentially by far-red light (Fairchild et al., 2000; Soh et al., 2000). Furthermore, it was shown that the hypocotyls of hy5 and hfr1 double knockout mutants grown in far-red light were only marginally shorter than the hypocotyls of phyA mutant plants grown under the same light conditions (Kim et al., 2002). These results suggest that HY5 and HFR1 are two dominant components that regulate the PHYA-induced inhibition of hypocotyl elongation in far-red light. If the apparent absence of PIF3 function represents the presence of excessive amounts of other far-red light–specific components, it will be informative to determine the role of PIF3 in the PHYA-induced inhibition of hypocotyl elongation in the hy5 or the hfr1 mutant background.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a growth room with a 16-h-light/8-h-dark cycle at 22 to 24°C for general growth and seed harvesting. The T-DNA insertional mutant, phytochrome interacting factor 3-1 (pif3-1), was obtained from the Arabidopsis Stock Center (Salk_030753). The mutant line was backcrossed to the wild type (Columbia), and five homozygous lines were established from the progeny. Because plants from all lines showed the same hypocotyl phenotype, members from a single line were used for all other analyses. To test the recessive nature of pif3-1 segregation, >20 mutant carpels were pollinated with wild-type (Columbia) pollen, and hypocotyl lengths of the F1 seeds were compared with those of both the wild type and pif3-1 homozygotes. For cosegregation analysis, 800 F2 seeds were plated on Murashige and Skoog (1962) (MS) agar plates and grown for 4 days under red light.
Among seedlings, 40 seedlings with clearly short hypocotyls and larger cotyledons and 126 seedlings with clearly long hypocotyls and smaller cotyledons were selected. Genomic DNA was extracted from the two seedling pools, and the full-length PIF3 (5′-GAGGGATCCAAAATGCCTCTGTTTGAGCTTTTC-3′ and 5′-GAGGGATCCTCACGACGATCCACAAAACTG-3′) and the PIF3–T-DNA hybrid (5′-GAGGGATCCAAAATGCCTCTGTTTGAGCTTTTC-3′ and 5′-GCGTGGACCGCTTGCTGCAACT-3′) were amplified by PCR. To generate transgenic plants expressing the full-length PIF3, full-length PIF3 was amplified using specific primers (5′-GAGGGATCCAAAATGCCTCTGTTTGAGCTTTTC-3′ and 5′-GAGGGATCCTCACGACGATCCACAAAACTG-3′), cloned into the BamHI site of a pBI121 vector from which the β-glucuronidase gene had been deleted, and transformed into Arabidopsis ecotype Landsberg erecta. Five independent homozygous lines were established, two of which were used for further analysis.
The expression of endogenous PIF3 was determined by amplifying the gene with endogenous gene-specific primers (5′-GGTTACTATCTGCCACCGGCG-3′ and 5′-TGCTAACAAATAAACAATACATC-3′), whereas the expression of total PIF3 (transgene plus endogenous PIF3) was determined by amplification with a PIF3-specific primer set (5′-GAGGAATTCATGCCTCTGTTTGAGCTTTTC-3′ and 5′-TCACTCGAGCGACGATCCACAAAACTGATC-3′).
Phenotype Analysis
To measure the hypocotyl length, seeds were plated on MS agar (half-strength MS, 0.7% phytoagar, and 0.05% Mes, pH 5.7), cold-treated for 3 days at 4°C in the dark, induced to germinate by illumination with white light (17 μmol·m−2·s−1) for 4 h, and grown under various fluence rates of either red or far-red light for 4 days. Growth chambers having red diodes (660 nm), far-red diodes (730 nm), and blue diodes (450 nm) were used (VS-9108M-LED; Vision, Seoul, Korea) to induce the different light conditions. For measurements, hypocotyl lengths of the 50 longest of 100 seedlings were measured for each sample. Similarly, cotyledon sizes were determined by measuring the areas of 40 cotyledons after digitizing the image.
For RNA gel blot analysis, seedlings were grown on MS agar plates (full-strength MS, 0.7% phytoagar, 1% sucrose, and 0.05% Mes, pH 5.7) under various light conditions and total RNA was purified with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's guidelines. PCR amplifications were performed for CAB3 (5′-ATGAGGAAGACTGTTGCCAAG-3′ and 5′-TCACTTTCCGGGAACAAAGTTG-3′), CHS (5′-GTCGTCTTCTGCACTACCTC-3′ and 5′-CACCATCCTTAGCTGACTTC-3′), and 18S rRNA (5′-CCTGCGGCTTAATTTGACTC-3′ and 5′-ACCGGATCATTCAATCGGTA-3′), and the resulting fragments were cloned into pTOPOII (Invitrogen, Carlsbad, CA). For the probe, the appropriate DNA fragments were excised from the vector and labeled using α-32P-dCTP and the Rediprime II Random Prime Labeling System (Amersham, Buckinghamshire, UK) as directed by the manufacturer's guidelines. Ten micrograms of total RNA was separated on a formaldehyde agarose gel and transferred to a Hybond N+ membrane (Amersham). Blots were probed overnight at 65°C in QuikHyb buffer (Stratagene, La Jolla, CA) and then washed at 65°C with 0.5× SSC containing 1% SDS. The intensity of the signal was quantified with a PhosphorImager (Storm 860; Molecular Dynamics, Sunnyvale, CA) and normalized against 18S rRNA expression.
For chlorophyll and anthocyanin measurements, seedlings were grown on MS agar plates for 4 days under either red light (17 μmol·m−2·s−1) or far-red light (3 μmol·m−2·s−1). Chlorophyll was extracted by incubating 30 seedlings in 500 μL of 95% ethanol overnight at 4°C in the dark. The contents of chlorophyll a and b were calculated as A664.2 + A648.6. The anthocyanin was extracted by incubating 50 seedlings in 300 μL of extraction solution (methanol plus 1% HCl) overnight at 4°C. After the extraction, 200 μL of water and 200 μL of chloroform were added, and the mixture was centrifuged to remove the seedlings. The amount of anthocyanin was calculated as A530 − 0.33A657. The contents of chlorophyll and anthocyanin for each line were measured in four independent samples.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Giltsu Choi, gchoi@kaist.ac.kr.
Accession Numbers
The GenBank accession numbers for the genes mentioned in this article are as follows: CAB3, X03908; CHS, BT000596; and 18S rRNA, X16077.
Acknowledgments
Insertion mutant information was obtained from the SIGnAL website at http://signal.salk.edu. We thank Korea Advanced Institute of Science and Technology Plant Developmental Biology laboratory members for their helpful discussion. This work was supported in part by grants from the Kumho Petrochemical Co., the Plant Metabolism Research Center funded by the Korea Science and Engineering Foundation (to Giltsu Choi), the National Research Laboratory by Korea Institute of Science and Technology Evaluation and Planning (to P.S.-S.), and Biogreen21 (to P.S.-S.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014498.
References
- Borevitz, J.O., Maloof, J.N., Lutes, J., Dabi, T., Redfern, J.L., Trainer, G.T., Werner, J.D., Asami, T., Berry, C.C., Weigel, D., and Chory, J. (2002). Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie, P., Ceska, T., Lamers, M., Romier, C., Stier, G., Teo, H., and Suck, D. (1997). The crystal structure of an intact human Max-DNA complex: New insights into mechanisms of transcriptional control. Structure 5, 509–520. [DOI] [PubMed] [Google Scholar]
- Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Ellenberger, T., Fass, D., Arnaud, M., and Harrison, S.C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980. [DOI] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare, A.R., Prendergast, G.C., Ziff, E.B., and Burley, S.K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Hudson, M., Ni, M., Qin, M., and Quail, P.H. (1999). poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 96, 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hudson, M.E. (2000). The genetics of phytochrome signalling in Arabidopsis. Semin. Cell Dev. Biol. 11, 475–483. [DOI] [PubMed] [Google Scholar]
- Ivanov, A.G., Krol, M., Maxwell, D., and Huner, N.P. (1995). Abscisic acid induced protection against photoinhibition of PSII correlates with enhanced activity of the xanthophyll cycle. FEBS Lett. 371, 61–64. [DOI] [PubMed] [Google Scholar]
- Jensen, P.J., Hangarter, R.P., and Estelle, M. (1998). Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.M., Woo, J.C., Song, P.S., and Soh, M.S. (2002). HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 30, 711–719. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2000). Phytochrome-interacting factors. Semin. Cell Dev. Biol. 11, 457–466. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shimizu, T., Toumoto, A., Ihara, K., Shimizu, M., Kyogoku, Y., Ogawa, N., Oshima, Y., and Hakoshima, T. (1997). Crystal structure of PHO4 bHLH domain-DNA complex: Flanking base recognition. EMBO J. 16, 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh, M.S., Kim, Y.M., Han, S.J., and Song, P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Sharrock, R.A., Tepperman, J.M., and Quail, P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3, 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.L., Jones, L., Baulcombe, D.C., and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Tseng, T.S., Swain, S.M., and Olszewski, N.E. (2001). Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 126, 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H., and Deng, X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32, 723–733. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2001). Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Luppi, J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 12, 659–667. [DOI] [PubMed] [Google Scholar]
- Zagotta, M.T., Hicks, K.A., Jacobs, C.I., Young, J.C., Hangarter, R.P., and Meeks-Wagner, D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]