Abstract
The maize MuDR/Mu transposable elements are highly aggressive, and their activities are held in check by host developmental and epigenetic mechanisms. The Mutator regulatory element, MuDR, produces both sense and antisense transcripts. We have investigated the impact of the presence of antisense transcripts on the abundance of the corresponding sense messages and on the regulation of Mutator activities. We report that internal deletions in MuDR arise frequently in somatic tissues; preferential loss of the 3′ untranslated region of mudrA and/or mudrB containing the intergenic region is correlated with chimeric sense mudrA/antisense mudrB and sense mudrB/antisense mudrA transcripts. Heritable internal deletions are extremely frequent (>10−2 per element), and the resulting defective MuDR elements also encode antisense transcripts. Expression of endogenous or additional transgene-encoded antisense transcripts neither decreases sense transcript levels nor inhibits Mutator excision activity over the three generations examined. We propose that antisense transcripts produced by MuDR deletions are not dominant-negative regulators of Mutator activities.
INTRODUCTION
MuDR/Mu transposable elements of maize are extremely active in transposition, increasing mutation frequency by 50- to 100-fold above the spontaneous level (Walbot, 1992). MuDR elements in active Mutator plants encode functional MURA transposase (Eisen et al., 1994) and a MURB helper protein (Lisch et al., 1999) that excise diverse, multicopy MuDR/Mu elements and insert them into new loci throughout the maize genome (reviewed by Walbot and Rudenko, 2002).
One host control of Mutator activities is that MuDR/Mu transposition is restricted to cells undergoing the terminal cell divisions of tissue development, minimizing sector size and the number of gametes with each new mutation (Levy and Walbot, 1990; Raizada and Walbot, 2000; Raizada et al., 2001c). A counter strategy is that MuDR/Mu switches from “cut-and-paste” excision and insertion in the soma to a “replicative” outcome in the germinal cells and gametophytes. Therefore, Mu elements increase in copy number (Walbot and Rudenko, 2002). A second host control is that epigenetic loss of Mutator activity is very common (Walbot, 1986; Martienssen and Baron, 1994). The initiation of silencing coincides with the nuclear retention of nonpolyadenylated RNA derived from MuDR and MuDR homologs (hMuDR elements) (Rudenko et al., 2003). Epigenetic silencing is correlated with the methylation of the terminal inverted repeats (TIRs) (Chandler and Walbot, 1986) and the absence of MuDR transcripts (Hershberger et al., 1991).
Multiple levels of regulation have been proposed to explain the complex developmental and epigenetic regulation of Mutator activities. Both MuDR genes, mudrA and mudrB, encode alternatively spliced transcripts. Hence, it is possible that different MURA and MURB proteins program specific components of Mutator activities (Hershberger et al., 1995). Other mechanisms proposed to downregulate Mutator activities or maintain the silenced status include post-translational modifications of MURA or MURB (Walbot and Rudenko, 2002), poison proteins encoded by the homologs of MuDR (Rudenko and Walbot, 2001), host proteins that interfere with MURA binding to the TIRs until late in development (Benito and Walbot, 1997; Raizada et al., 2001b), developmentally progressive retention of MuDR/hMuDR RNA inside of the nucleus (Walbot and Rudenko, 2002), expression of the Mop1 gene (Lisch et al., 2002), introduction of a dominant silencing factor, MuKiller (Lisch, 2002), and the balance of sense and antisense MuDR transcripts (Hershberger et al., 1995; Lisch et al., 1999).
Antisense transgenes are used routinely to eliminate or reduce endogenous gene expression in plants (Bourque, 1995). Antisense RNAs are hypothesized to exert negative regulation by annealing to their complementary sense transcripts. The resulting double-stranded RNA structure may directly affect mRNA maturation, transport to the cytoplasm, RNA stability, or translation (Terryn and Rouze, 2000). Alternatively, double-stranded RNA can trigger small RNA–mediated post-transcriptional gene silencing (reviewed by Waterhouse et al., 2001). In some cases, antisense RNAs encode small proteins (Knee et al., 1997), and some of these serve as signal molecules (Bisseling, 1999).
There are several cases in which antisense RNAs are negative regulators of transposons. For example, Escherichia coli Tn10 transposon is inhibited by element-encoded antisense RNA, which interferes with translation by pairing with the 5′ end of the transposase mRNA (Simons and Kleckner, 1988). The micropia retrotransposon in Drosophila hydei encodes a testis-specific antisense RNA complementary to the reverse transcriptase and RNase H coding regions. This antisense transcript is hypothesized to control the germ line expression of the corresponding sense transcripts (Lankenau et al., 1994).
mudrA and mudrB are transcribed convergently from promoters in the TIRs, and their polyadenylation sites are separated by only a 225-bp intergenic region composed of numerous short repetitive elements (Figure 1A) (Hershberger et al., 1995). Recently characterized hMuDR elements are transcriptionally active but produce messages with numerous nucleotide polymorphisms, and these elements cannot program Mutator activities (Rudenko and Walbot, 2001). In active Mutator plants, RNase protection assays identified sense transcripts as well as antisense transcripts that are collinear with the MuDR genes (Hershberger et al., 1995). As determined by in situ hybridization, antisense mudrA transcripts colocalized with sense mudrA and mudrB transcripts, whereas antisense mudrB transcripts were difficult to detect in most tissues (Joanin et al., 1997). Interestingly, the detection of all MuDR transcripts by in situ hybridization increased significantly when RNA was denatured before hybridization. This finding suggests that the sense and antisense transcripts may have been paired in vivo.
Figure 1.
RT-PCR Amplification and Characterization of Somatic Antisense MuDR Transcripts from a Standard Mutator Plant Carrying Multicopy MuDR Elements.
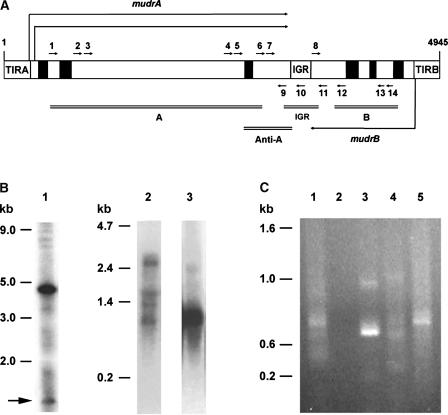
(A) Scheme of the 4.9-kb MuDR element, which encodes two genes (mudrA and mudrB) that are transcribed convergently from the TIRA and TIRB promoters. An AT- and repeat-rich intergenic region (IGR) separates these genes (Hershberger et al., 1995). Closed boxes represent introns, and open boxes indicate exons. The positions and directions (5′ to 3′) of the RT-PCR primers used in this study are shown by short arrows. Each primer has a number corresponding to the 5′ base according to Hershberger et al. (1991), and A, B, or IGR at the beginning of each primer name indicates that the primer resides within mudrA, mudrB, or the intergenic region, respectively: 1, A493; 2, A1621; 3, A1813; 4, A2298; 5, A2713; 6, A2907; 7, A2952; 8, IGR3501; 9, A3276; 10, IGR3469; 11, B3843; 12, B3950; 13, B4334; and 14, B4473. MuDR regions recognized by hybridization probes are indicated by double lines and labeled as follows: A for a probe detecting mudrA; B for a probe detecting mudrB; Anti-A for a region of mudrA used for the generation of antisense mudrA transgenic plants; and IGR for the intergenic region.
(B) Heritable MuDR derivatives in the standard Mutator plant SK26-1 and characterization of MuDR transcripts. DNA gel blots prepared with SstI-digested DNA were probed with a DNA fragment that detects both mudrA and mudrB (A and B) as shown in (A). The band of 4.7 kb represents an intact, unmethylated MuDR element. MuDR fragments >4.7 kb are methylated elements and/or hMuDR elements that lack the SstI site in the TIRs (Rudenko and Walbot, 2001). Bands <4.7 kb indicate deleted MuDR derivatives and are marked with an arrow (lane 1). In lanes 2 and 3, total RNA was extracted from the same SK26-1 plant, and RNA gel blots were hybridized with double-stranded mudrA (A) or mudrB (B) probes to detect mudrA and/or antisense mudrA (lane 2) or mudrB and/or antisense mudrB (lane 3).
(C) RT-PCR amplification of antisense MuDR transcripts from RNA fractions (see Methods) extracted from plant SK26-1. Different sizes of chimeric mudrA/antisense mudrB transcripts were amplified using an antisense mudrB-specific primer (primer 11 in [A]) for RT, followed by an amplification of the cDNA with primer 2 (lanes 1, 2, 4, and 5). In lane 2, RNA was pretreated with RNase A before reverse transcription. In lane 3, primer 2 was used as the RT primer and primer 11 was used for cDNA amplification. Total (lanes 1, 2, and 3), poly(A+) (lane 4), or poly(A−) (lane 5) preparation was used for the RT-PCR analysis. Forty cycles of PCR amplification were used. PCR products were fractionated on a 1% agarose gel and stained with ethidium bromide.
Diverse antisense MuDR transcript types have been characterized in this study. We propose that somatic deletions within MuDR elements generate derivatives that encode substantial and diverse chimeric transcripts. These chimeric constructs consist of sense mudrA and antisense mudrB or sense mudrB and antisense mudrA and presumably resulted from the failure of transcription termination within the intergenic region. Molecular and genetic analysis of heritable MuDR deletions producing antisense transcripts and of transgenic maize expressing specific forms of antisense RNA demonstrated that neither endogenous nor transgene-encoded antisense transcripts affect the levels of MuDR sense transcripts or Mu excision activity over three generations. Although these results do not prove that Mutator is inherently insensitive to antisense RNA, they do indicate that Mutator activities are tolerant of the types and levels of antisense transcripts produced by somatic and germinal deletion derivatives, both of which arise at a high frequency in active Mutator lines. Therefore, the possibility that these antisense MuDR transcripts are immediate and effective negative regulators of Mutator activities is excluded by this study.
RESULTS
Cloning and Characterization of Somatic Antisense MuDR Transcripts
As determined by RNA gel blot analysis using single-stranded RNA or double-stranded DNA probes to detect MuDR transcripts in a survey of 65 plants, we found that most active Mutator plants produce transcripts with a mixture of discrete sizes plus a background smear; a subset of transcripts is antisense (data not shown). One plant was examined in detail to identify the origin of antisense transcripts. As shown in lane 1 of Figure 1B, the active Mutator plant SK26-1 contains multiple, intact 4.9-kb MuDR elements as well as a heritable, unmethylated 1.2-kb MuDR deletion derivative (ΔMuDR). RNA gel blot hybridizations demonstrate that this Mutator plant expressed not only the expected 2.8-kb mudrA (lane 2) and 1.0-kb mudrB (lane 3) transcripts encoded by intact MuDR elements but also three smaller transcripts detected by a double-stranded mudrA probe (lane 2). These short transcripts could be either sense or antisense transcripts. We did not detect a full-length “read-through” transcript of 4.7 kb that would represent full-length sense mudrA/antisense mudrB or sense mudrB/antisense mudrA transcripts.
Reverse transcription (RT) PCR was tested as an alternative to recover individual chimeric sense/antisense transcripts using total RNA isolated from a seedling leaf of SK26-1 (Figure 1C). Chimeric transcripts (sense mudrB/antisense mudrA) of varying lengths were amplified using primer 11 as the RT primer and primer 2 as the PCR amplification primer (Figure 1C, lane 1, Table 1). The amplification products were not derived from DNA templates, because no products were amplified when the RNA preparation was pretreated with RNase (Figure 1C, lane 2). Also, different product sizes were obtained when primer 2 was used as the RT primer (Figure 1C, lane 3) to amplify sense mudrA/antisense mudrB transcripts. We found both polyadenylated (Figure 1C, lane 4) and nonpolyadenylated (Figure 1C, lane 5) antisense mudrB/sense mudrA transcripts. Supporting evidence for the existence of poly(A)-tailed antisense mudrB transcripts was obtained by DNA sequence analysis of RT-PCR fragments amplified using a poly(dT)-anchored adaptor primer and an antisense mudrB-specific primer. Interestingly, one type of polyadenylated antisense mudrB cDNA had several single–base pair mismatches and an insertion of a nonhomologous DNA fragment, implying that this transcript originated from a hMuDR element (Figure 2).
Table 1.
Sequence Analysis of Antisense MuDR Transcripts Recovered from a Leaf Patch of SK26-1
| Primer Sets a | Transcript Type b | Case Number c | Deletion (bp) [Position] d | Flanking Sequence at the Deletion End Points e | Insertion (bp) [Position] f |
|---|---|---|---|---|---|
| 7/10 | B and αA | 1 | None | None | |
| 2 | 98 [3344 to 3441] | TTATTTCagttttatt—gcaattgtCGCCCAG | None | ||
| 7/14 | B and αA | 3A | 963 [3189 to 4151] | GTATTTGttgtaaga—agtttagaTGCCTCC | None |
| 3B | 72 [4226 to 4297] | AAGATCCtgtgcaa—atccatacCACATTC | None | ||
| 11/1 | A and αB | 4A | 7 [612 to 618] | TTAGttgtaagACTG | None |
| 4B | 2909 [743 to 3651] | ATTATAGtatcagat—aacaaacACTACGG | 2 [743] | ||
| 5A | 86 [595 to 680] | AGTTGGAagactgct—acattgtcTGTAATA | None | ||
| 5B | 2303 [1252 to 3554] | TTGGGTCgcattcca—acactgagCCATTAG | 289 [1252] | ||
| 11/2 | A and αB | 6 | 1562 [1928 to 3489] | GTATTTGaacaccat—gaaatagaGCGCAGA | None |
| 7 | 1771 [1964 to 3734] | TTCACAAgattgctg—caaaatgaGACACCA | 17 [1964] | ||
| 8 | 1193 [2608 to 3800] | TCTCGAGggtgggag—ggtcgtttATCTCTT | 30 [2608] | ||
| 9 | 1370 [1912 to 3281] | GGCCTATaggagaga—cagtgtatTTGTTGT | None | ||
| 11/6 | A and αB | 10 | 441 [3090 to 3530] | CTAGTGAagaaaatc—gataacatATAACAC | 17 [3090] |
| 11 | 554 [3171 to 3724] | TGTGCTCagaacaac—atcccagaCAGACCA | None | ||
| 14/6 | A and αB | 12 | 1180 [3155 to 4334] | GCTAAGAacaaccac—ccttctccACGGCAA | None |
| 13 | 805 [3033 to 3837] | GCCGATGaaccagcc—gatagtgcTCTTCGA | 27 [3033] |
RT primer/PCR amplification primer. See Figure 1A for the positions and directions of primers.
Chimeric transcript of mudrB (B) and antisense mudrA (αA) or mudrA (A) and antisense mudrB (αB).
Individual RT-PCR products were amplified using the primer set listed in the first column. Three RT-PCR products (cases 3, 4, and 5) have two deletions in each cDNA, and these deletions are noted as items A and B.
Length of the missing MuDR sequence for each RT-PCR product. The 5′ to 3′ positions indicated in brackets are based on MuDR numbering according to Hershberger et al. (1991).
Sequences of flanking deletion end points, expressed as 5′ to 3′. The bases extending to each side of the deletions are shown in uppercase letters. Missing MuDR sequences are shown in either lowercase letters or dashes when deleted nucleotides are not shown. A direct repeat of 3 bp near the junction is underlined.
Length of filler DNAs in base pairs and their positions relative to MuDR.
Figure 2.
Nucleotide Alignment of Polyadenylated Antisense mudrB Transcripts Along with the Corresponding MuDR Sequence.
A two-nucleotide insertion and three single nucleotide mismatches in cDNA#3 are shown in boldface and italic type, respectively. The stars indicate polyadenylation starts, and the numbers represent their positions according to MuDR numbering for each antisense cDNA (Hershberger et al., 1991). All cDNA fragments were retrieved multiple times.
Using several primer sets complementary to different regions of mudrA or mudrB, 13 chimeric sense/antisense transcripts of diverse lengths were retrieved and sequenced (Table 1). Most transcripts were unique; 12 transcripts were found only once, and 1 transcript (case 5) was retrieved multiple times. One sequence (case 1) was completely collinear with MuDR throughout its length of 517 bp. By contrast, 12 of 13 of the antisense transcripts were collinear with only portions of MuDR; each transcript was missing one or two different segments of MuDR (7 bp in case 4A up to 2909 bp in case 4B). Three transcripts (cases 3 through 5) were missing two segments of MuDR, one within the intergenic region and one within a gene (Table 1). These missing segments could result from RNA splicing or from the transcription of deleted templates. Sequence inspection indicated that the borders of the missing segments did not match even relaxed consensus splice sites (Brendel et al., 1998). In a previous study, four mudrA transcripts lacking internal segments were inferred to arise from ΔMuDR rather than from RNA splicing (Hershberger et al., 1995). Although the intergenic region is only 225 bp (positions 3301 to 3525) of the 4.9-kb MuDR element, 10 antisense transcript types were missing all (cases 3, 4, 5, 7, 8, 11, and 12) or a portion (cases 2, 6, and 10) of the intergenic region. As determined by gel blot analysis, the source plant SK26-1 had only one detectable truncated MuDR element and only three detectable short mudrA transcripts (Figure 1B). Therefore, we infer that most chimeric antisense transcripts are encoded by diverse somatic (substoichiometric) ΔMuDR elements.
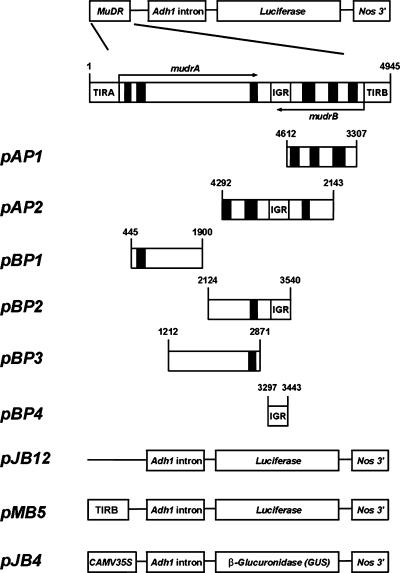
Using transient expression experiments, fragments of MuDR were examined for promoter activity that could result in an antisense transcript of one gene with or without chimeric continuation to the sense transcript of the other gene. Schemes of the antisense promoter constructs are shown in Figure 3, and transient analysis data are presented in Table 2. Weak promoter activities that could result in sense mudrA/antisense mudrB transcripts were detected in constructs harboring the distal half of mudrA and the intergenic region (pBP2; ∼10.1% of the control TIRB promoter activity derived from pMB5) and the intergenic region itself (pBP4; ∼15.1% of the TIRB activity). No significant promoter activity was detected in other coding regions in either reading frame. These results, considered together with the structures of the antisense transcripts, suggest that it is termination failure rather than internal antisense promoters that result in chimeric transcripts. ΔMuDR elements lacking the intergenic region appear to permit read-through transcription of mudrA or mudrB messages initiated from the TIR promoters.
Figure 3.
Schemes of the Constructs Used in Experiments Testing Antisense Promoter Activities.
Numbers on each scheme indicate the positions in MuDR. Drawings are not to scale. CaMV, Cauliflower mosaic virus; Nos, nopaline synthase.
Table 2.
Transient Analysis for Antisense Promoter Activity of Different MuDR Regions in Maize Black Mexican Sweet Protoplasts
| Promoter Constructs a | Luciferase Expression b |
|---|---|
| Antisense mudrA activity | |
| pAP1 [4612 to 3307] | 88 ± 18 |
| pAP2 [4292 to 2143] | 75 ± 13 |
| Antisense mudrB activity | |
| pBP1 [445 to 1900] | 58 ± 16 |
| pBP2 [2124 to 3540] | 185 ± 21 |
| pBP3 [1212 to 2871] | 62 ± 15 |
| pBP4 [3297 to 3443] | 275 ± 18 |
| Expression control | |
| pJB12 | 32 ± 8.5 |
| pMB5 | 1823 ± 160 |
Schemes of the constructs used in this experiment are shown in Figure 3. The Cauliflower mosaic virus 35S promoter region of a luciferase expression construct, pJD312, was replaced with a region of MuDR fragment. The 5′ to 3′ positions of the MuDR fragment fused to the expression cassette are indicated in brackets. pJB12 (pJD312 without the Cauliflower mosaic virus 35S promoter) was used as a negative control, and pMB5 (TIRB:Adh1 intron:LUC) was used as a positive control.
Luciferase expression was normalized using GUS activity derived from pJB4 as described in Methods.
Characterization of Heritable Genomic Deletions
In addition to somatic deletions, numerous heritable MuDR deletions have been reported (Hsia and Schnable, 1996; Lisch et al., 1999); however, the frequency of such deletions has not been quantified. We crossed active Mutator plants to and by standard maize anthocyanin tester lines lacking MuDR. Twelve families of progeny were examined for MuDR copy number and for the presence of deletions in MuDR elements (Table 3). SstI-digested genomic DNA was probed with a mixture of mudrA and mudrB fragments to identify deleted MuDR and then reprobed with the intergenic region DNA fragment to determine which of the already identified deleted elements were missing this specific region. Altogether, we found 20 newly arisen ΔMuDR elements plus 22 parental deletion derivatives segregating in the progeny. Some newly arisen derivatives found in several progeny plants represented a tassel sector in a parental plant that allowed transmission to multiple individuals.
Table 3.
Survey for Deleted MuDR (ΔMuDR) Elements in Active Mutator Plants
| F1 Family a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | T55 | T57 | T62 | T63 | T64 | T56 | T65 | T69 | SK26 | SK30 | SB03 | SB36 | Total |
| Number of ΔMuDRb | 4/6 c | 2/3 | 1/2 | 4/6 | 2/3 | 0/0 | 1/1 | 2/3 | 2/5 | 0/3 | 0/4 | 2/6 | 20/42 |
| Deletion of IGR d | 1/2 e | 0/1 | 1/2 | 1/2 | 1/2 | 0/0 | 0/0 | 1/1 | 1/2 | 0/2 | 0/4 | 2/3 | 8/21 |
| Plants carrying ΔMuDR | 5/6 f | 12/12 | 4/4 | 6/6 | 6/6 | 0/6 | 1/6 | 5/10 | 12/12 | 11/11 | 11/11 | 4/16 | 77/106 |
Mutator plants were crossed to/by anthocyanin tester lines to produce F1 progeny ears; the resulting F1 plants were screened for the presence of ΔMuDR elements using DNA blot hybridization.
Genomic DNA was prepared from parents and sibling plants of each F1 family. SstI-digested genomic DNA was probed with a mixture of mudrA (A) and mudrB (B) fragments (see Figure 1A). Bands smaller than the 4.7-kb intact MuDR were scored as deleted MuDR.
Number of new ΔMuDR/total number of ΔMuDR detected in the family. The total ΔMuDR count includes new (found in only one individual) and parental (segregating or present in all members of the family) deletions. Segregating parental bands of the identical size were considered to represent one deletion event that occurred in a previous generation.
Blots used for the detection of ΔMuDR were reprobed with an intergenic region (IGR) DNA fragment (see Figure 1A). ΔMuDR bands that failed to hybridize were scored as deletions of the intergenic region. Intergenic region deletions of identical size were counted as one event.
Number of new ΔMuDR elements that lack the intergenic region/total number of ΔMuDR in the family that lacks the intergenic region.
Number of plants carrying a ΔMuDR/number of total plants examined.
Strikingly, 21 of the 42 analyzed deletions (50%) and 8 of 20 new deletions (40%) removed the 3′ untranslated region (UTR) of mudrA and/or mudrB containing the intergenic region, as determined by the failure to detect cross-hybridization with the 250-bp intergenic region probe. The deletion frequency was exceptionally high; considering just the singular deletions, events occurred in 18.9% of the plants in one generation (20 deletions in 106 plants). Most plants carry 5 to 20 copies of MuDR elements (copy number reconstruction data not shown); thus, these internal deletions occur at a frequency of 9.4 × 10−3 to 3.8 × 10−2 per element; 20 new deletions arose from an estimated 530 to 2120 copies of MuDR elements in 106 plants. Because very short deletions could have been overlooked, we consider our data to be minimal estimates of ΔMuDR production.
Structure and Transcript Types of Two Heritable ΔMuDR Elements
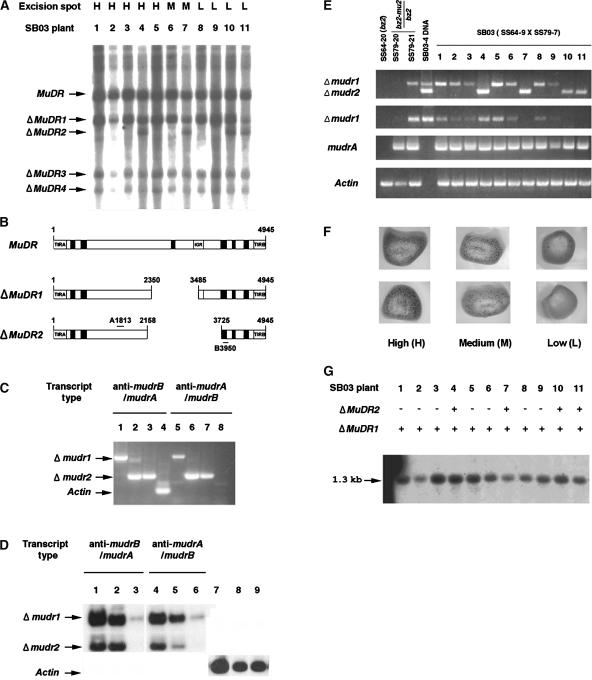
To evaluate the impact of antisense messages transcribed from germinally transmittable ΔMuDR derivatives, we characterized two specific ΔMuDR derivatives and their transcripts. DNA gel blot analysis of 11 sibling Mutator plants, SB03-1 to SB03-11, showed that this family has four ΔMuDR derivatives: three deletions (ΔMuDR1, -3, and -4) were transmitted to all siblings, and ΔMuDR2 was segregating in the family (Figure 4A). Based on an analysis of progenitors, we determined that ΔMuDR1 was generated during the growth of the great grandparent of the SB03 plants and became homozygous after self-pollination, whereas ΔMuDR2 was produced in a grandparent (data not shown). Using PCR, we amplified and sequenced regions surrounding the deletion junctions of ΔMuDR1 and ΔMuDR2. Figure 4B shows simplified schemes. ΔMuDR1 has an intact mudrB gene but lacks 1136 bp (positions 2350 to 3485). Consequently, a segment spanning the 3′ coding region and the 3′ UTR of mudrA as well as most of the intergenic region are missing. ΔMuDR2 is missing 1568 bp (positions 2158 to 3725), including part of mudrB, all of the intergenic region, and ∼40% of mudrA. The 1.0 kb of ΔMuDR1 and 0.57 kb of ΔMuDR2 sequence examined were perfectly collinear with MuDR except for these deletions.
Figure 4.
Molecular and Structural Analyses of Two Germinally Deleted MuDR Elements and Their Transcripts.
(A) Detection of heritable deleted MuDR derivatives in sibling plants grown from SB03. Genomic DNA was prepared from these plants, digested with SstI, and blotted and probed with the A and B probe described in Figure 1B. Four deleted elements (ΔMuDR1 to ΔMuDR4) were found and are indicated with arrows. H, high; M, medium; L, low.
(B) Schemes of the intact MuDR and two ΔMuDR elements. Primers 3 and 12 shown in Figure 1A were used to amplify portions of ΔMuDR1 and ΔMuDR2. The amplified fragments were sequenced to confirm the structures.
(C) Directional RT-PCR cloning of antisense mudrB/mudrA (anti-mudrB/mudrA) and antisense mudrA/mudrB (anti-mudrA/mudrB) from total RNA of plants SB03-1 (lanes 1 and 5), SB03-4 (lanes 2 and 6), and SB03-11 (lanes 3 and 7). Single-stranded cDNA was obtained using either primer 12 (lanes 1, 2, and 3) or primer 3 (lanes 5, 6, and 7). After removal of template RNA and inactivation of the reverse transcriptase, cDNA was amplified using primer 3 or primer 12, as appropriate. Control amplification of sense actin transcripts (lane 4), but not antisense transcripts (lane 8), confirmed that the RT-PCR was directional in this experiment. Data show RT-PCR products fractionated on a 1% agarose gel and stained with ethidium bromide.
(D) Amplification of the Δmudr transcripts from several RNA sources. Total RNA was extracted from plant SB03-4 and fractionated into poly(A+) and poly(A−) RNA as described in Methods. Directional RT-PCR was performed using total RNA (lanes 1, 4, and 7), poly(A−) RNA (lanes 2, 5, and 8), or poly(A+) RNA (lanes 3, 6, and 9) as described in (C). The amplified RT-PCR products were fractionated on an agarose gel and detected using the A and B DNA probes after blotting on a nylon membrane. Sense actin transcripts were amplified as a control.
(E) Transcripts of ΔMuDR1 and ΔMuDR2 and expression levels of sense mudrA transcripts. Transcripts were amplified from total RNA of SB03 plants, from two plants in the family of the male parent (SS79-20 and SS79-21), and from a plant in the family of the female parent (SS64-20). Primers 3 and 12 were used for the amplification of both Δmudr1 and Δmudr2 transcripts (top gel). RT-PCR with primers 4 and 12 produces only Δmudr1 transcripts (second gel). Sense mudrA transcripts were amplified using primers 5 and 9 (third gel). Note that these primers can amplify two known mudrA transcripts, because intron 3 is spliced in only ∼80% of the transcripts (Hershberger et al., 1995); in this experiment, most transcripts appear to be spliced, because the smaller 490-bp product is visualized prominently by ethidium bromide staining. The Actin gel (bottom) indicates that RT-PCR amplification was performed as a control to determine if equal amounts of RNA were used in the experiment. Portions of the ΔMuDR1 and ΔMuDR2 elements were amplified by PCR from genomic DNA of plant SB03-4 to use for a size comparison with the RT-PCR products (top two gels only; lane 4). The PCR amplification was for 31 cycles.
(F) Representative kernels of the SB03 ear (SS64-3 × SS79-7) were classified as high, medium, or low spotted. Mu excision from a mutable bz2-mu2 allele restores a functional Bz2 gene, and this is a visual indicator of somatic Mutator activity, which varied from high (>150 spots on a 2 mm × 2 mm aleurone area of the kernels) to medium (10 to 150 spots) to low (<10 spots).
(G) Analysis of the methylation status at HinfI sites within the MURA binding sites in the TIRs of Mu1 and Mu2 elements. Genomic DNA extracted from SB03 individuals was digested with HinfI, and the resulting DNA gel blot was probed with a Mu1/Mu2-specific probe (Raizada and Walbot, 2000). In this family, there were no Mu2 elements; consequently, only the 1.3-kb fragment generated from Mu1 was detected.
Directional RT-PCR was performed to distinguish transcripts initiating in TIRA from those initiating in TIRB of ΔMuDR1 and ΔMuDR2. Based on its intact mudrB structure, we expected chimeric sense mudrA/antisense mudrB and mudrB transcripts from ΔMuDR1. However, both mudrA/antisense mudrB and mudrB/antisense mudrA transcripts were amplified from this deletion derivative (Figure 4C, lanes 1, 2, 5, and 6). Similarly, Lisch et al. (1999) found that deletion of just 174 bp from the 3′ end of mudrA resulted in the termination failure of mudrB. Therefore, it appears that a DNA structure in the 3′ UTR of mudrA or in the intergenic region is important for the successful termination of mudrB transcription. ΔMuDR2 also produced both antisense mudrA and antisense mudrB read-through transcripts, as expected, because both genes lack a 3′ UTR and the intergenic region is missing (Figure 4C, lanes 2, 3, 6, and 7). As determined by sequencing, the amplified RT-PCR fragments were shown to be identical to the corresponding deletion derivative (data not shown). No novel splicing events were observed in either ΔMuDR1 or ΔMuDR2 transcripts. Interestingly, the majority of chimeric transcripts from both deletion derivatives lacked polyadenylation (Figure 4D, lanes 2 and 5), suggesting that the chimeric transcripts may be retained in the nucleus. As a control, the majority of actin transcripts were polyadenylated, suggesting that they are cytoplasmic mRNA (Figure 4D, lane 9).
Epigenetic Regulation of ΔMuDR Transcript Abundance and Lack of Measurable Impact on mudrA Transcript Level and Mu1 TIR Methylation
With confirmation that heritable deletion derivatives are transcriptionally active, we next examined the relative abundance of sense and chimeric antisense transcripts. RT-PCR was used to simultaneously amplify antisense mudrA and antisense mudrB transcripts (Figure 4E, top two gels) of ΔMuDR1 and/or ΔMuDR2. From the same RNA preparation, the sense mudrA transcripts of an intact MuDR also were amplified (Figure 4E, third gel from the top). Interestingly, the expression of either Δmudr1 and/or Δmudr2 antisense transcripts did not significantly reduce the relative abundance of mudrA in 9 of 11 plants examined. Additionally, Δmudr1 transcripts in a sibling of the male parent of the SB03 family (SS79-21) did not reduce mudrA abundance compared with that in the sibling plant SS79-20, which did not inherit this deletion (or any other detectable deletion derivative). Stochastic epigenetic silencing of Mutator activity in SB03-9 plant is the most likely explanation for the decrease of both mudrA and Δmudr1 transcript abundance in those plants (Bennetzen et al., 1993).
Second, transcripts from ΔMuDR1 and ΔMuDR 2 did not affect the methylation status of Mu TIRs. SB03 sibling plants were grown from kernels with different frequencies of Mu excision scored in the aleurone (Figure 4F). Decreased spotting frequency is an indicator of the incipient loss of Mutator activity, which is accompanied by methylation of the TIRs of Mu elements (Chandler and Walbot, 1986). HinfI cleaves unmethylated sites within the TIRs of Mu1 and Mu2 elements, resulting in 1.3- or 1.6-kb fragments, respectively. Because the HinfI sites are within the MURA transposase binding site defined in vitro (Benito and Walbot, 1997), lack of methylation is a molecular phenotype indicative of a transcriptionally and functionally active MuDR. We found that all SB03 plants retained unmethylated TIRs in the Mu1 elements (Figure 4G), indicating that MuDR is active.
Third, ΔMuDR1, the older deletion derivative present in all SB03 plants, had low transcript levels in plants that also inherited ΔMuDR2 (SB03-7, -10, and -11), with the exception of plant SB03-4. Δmudr1 transcripts were detected easily in all plants that inherited only ΔMuDR1. Therefore, it appears that some deletion derivatives affect the transcript abundance of other deletion transcripts without influencing MuDR transcripts.
Generation and Characterization of Transgenic Plants Expressing Antisense mudrA or Antisense mudrB Transcripts
Nearly all somatic excision events of engineered Mu1 (RescueMu) elements occur during or after the last cell division (Raizada et al., 2001c), similar to the very late excision observed in mutable alleles with MuDR and nonautonomous Mu insertions. We hypothesize that this late excision timing ensures that most internal deletions within MuDR also occur late in development. If this is true, then antisense transcripts produced so late in development may not influence MuDR activities in the soma, because MuDR transcripts and protein products already are present. Furthermore, the vast majority of somatic deletions are nontransmissible: only those ΔMuDR arising in lineages that enter meiosis can be transmitted to the next generation. Transcript analysis of the two transmissible deleted MuDR, ΔMuDR1 and ΔMuDR2, showed that the majority of these messages were not polyadenylated properly (Figure 4D). By contrast, substantial amounts of polyadenylated antisense messages were detected easily from a seedling leaf of a Mutator plant (Figure 1C, lane 4).
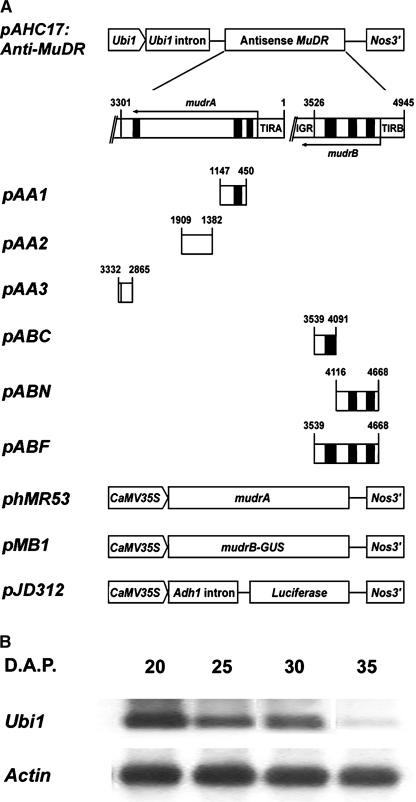
Transgenic plants constitutively expressing antisense mudrA or antisense mudrB were used to further analyze the impact of antisense RNA expression on the abundance of sense MuDR messages and Mutator somatic excision activities. First, the effectiveness of transiently expressed antisense transcripts, transcribed from different regions of the MuDR element, was tested in reducing corresponding sense transcripts in maize Black Mexican Sweet protoplasts (see Figure 5A for schemes of the fusion constructs and Table 4 for the transient analysis data). This study identified the most effective region of antisense mudrA (exon 4 and the 3′ UTR) and antisense mudrB (full-length antisense mudrB including the 3′ UTR and more than half of the 5′ UTR). We then generated transgenic maize expressing antisense mudrA (pAA3; pAHC17:Anti-mudrA) or antisense mudrB (pABF; pAHC17:Anti-mudrB) under the control of the maize Ubiquitin1 (Ubi1) promoter.
Figure 5.
Promoter Constructs Used in Transient Gene Expression Analysis and Ubi1 Expression in Developing Aleurone Layers.
(A) Schemes of constructs used in the experiments testing the effectiveness of antisense MuDR fragments in reducing corresponding sense transcript. PCR-amplified MuDR fragments were cloned into the 3′ region of pAHC17 to make a transcriptional fusion with the DNA segment in an antisense orientation with respect to the ubiquitin promoter. Numbers on each PCR fragment represent the positions in MuDR. Drawings are not to scale. CaMV, Cauliflower mosaic virus; Nos, nopaline synthase.
(B) Ubi1 expression in the aleurone layers of developing kernels. Total RNAs were prepared from aleurone layers of developing kernels at different days after pollination (D.A.P.). The RNAs were fractionated on a 1.2% agarose gel and probed with a Ubi1-specific oligonucleotide.
Table 4.
Transient Analysis of the Effectiveness of Antisense MuDR Fragments in Suppression of the Corresponding Sense Gene Expression in Maize Black Mexican Sweet Protoplasts
| Reporter Expression b
|
||
|---|---|---|
| Construct a | Comparison with phMR53 (%) | Comparison with pMB1 (%) |
| Antisense mudrA | ||
| phMR53 only | 100 ± 3.9 | |
| phMR53 + pABF [3539 to 4668] | 98 ± 5.8 | |
| phMR53 + pAA1 [1147 to 450] | 90 ± 8.1 | |
| phMR53 + pAA2 [1909 to 1382] | 94 ± 6.3 | |
| phMR53 + pAA3 [3332 to 2865] | 78 ± 4.7 | |
| Antisense mudrB | ||
| pMB1 only | 100 ± 4.5 | |
| pMB1 + pAA3 [3332 to 2865] | 98 ± 5.4 | |
| pMB1 + pABF [3539 to 4668] | 70 ± 6.4 | |
| pMB1 + pABN [4116 to 4668] | 96 ± 6.5 | |
| pMB1 + pABC [3539 to 4091] | 106 ± 8.4 | |
Schemes of the constructs used in this experiment are shown in Figure 5A. The 5′ to 3′ positions of the antisense MuDR fragment in the Ubi1-driven expression vector are indicated in brackets.
Expression data were normalized using luciferase expression derived from pJD312.
Previous work established that a maize Ubi1 promoter is highly active in most maize tissues, including roots, developing anthers, and pollen (Raizada et al., 2001a). We confirmed that Ubi1 transcripts also are abundant in aleurone cells through 30 days after pollination (Figure 5B), when mitotic proliferation has finished and when most Mu1 excisions have occurred (Levy and Walbot, 1990; Raizada and Walbot, 2000). Table 5 summarizes the genetic analysis and transgene expression of five independent antisense transgenic lines for each construct. In nearly all T0 plants and their T1 progeny, there was an excellent correspondence of herbicide resistance, which was used as the selection marker, and transgene expression: most herbicide-resistant T0 and T1 seedling plants expressed the corresponding transgene (41 of 43 plants tested). Furthermore, we detected no expression of the antisense transgene in plants sensitive to Basta (data not shown). Exceptionally, two plants derived from the Anti-mudrB-8 and -10 insertion events conferred resistance to the herbicide, but expression of the transgene could not be detected. Collectively, these results indicate that both the Bar gene and the antisense mudrA or antisense mudrB gene construct were transcriptionally active initially and had inserted at a linked locus. In general, transgene expression, determined by sensitivity to the Basta treatment, was relatively well maintained through several generations for plants carrying Anti-mudrA (moderate to good maintenance in Anti-mudrA-3, -4, -5, and -11 and poor maintenance in Anti-mudrA-12). By contrast, Anti-mudrB transgene expression was lost frequently (poor maintenance in Anti-mudrB-2, -8, and -10 and moderate to poor maintenance in Anti-mudrB-1 and -3). The biological significance of the frequent silencing of Anti-mudrB expression has yet to be examined.
Table 5.
Characterization of the Transgenic Lines Used in This Study
| Cosegregation of the Transgene with Herbicide Resistance a
|
Transgene Segregation
|
||||
|---|---|---|---|---|---|
| Genetic Locus | T0 Plant | T1 Seedling | T1 Plant | T2 Plant | Transgene/Herbicide Stability (Progression from T0 to T2) |
| Anti-mudrA-3 | 3/3 b, medium c | 3/3 d | 11 (40/120) e,f | N.D. g | Moderate to poor h |
| Anti-mudrA-4 | 3/3, weak | 1/1 | 8 (34/75) | 4 (24/54) e,f | Moderate to good |
| Anti-mudrA-5 | 3/3, weak | 2/2 | 4 (29/59) | 4 (24/54) | Good |
| Anti-mudrA-11 | 3/3, medium | 1/1 | 8 (49/83) | 1 (6/12) | Moderate to good |
| Anti-mudrA-12 | 3/3, medium | 1/1 | 8 (28/73) | 3 (1/41) | Poor |
| Anti-mudrB-1 | 3/3, medium | 2/2 | 3 (30/39) | 3 (12/35) | Moderate to poor |
| Anti-mudrB-2 | 3/3, weak | 1/1 | 6 (29/83) | 2 (2/28) | Poor |
| Anti-mudrB-3 | 3/3, weak | 2/2 | 8 (43/65) | 4 (20/45) | Moderate |
| Anti-mudrB-8 | 2/3, weak | N.D. | 9 (15/88) | N.D. | Poor |
| Anti-mudrB-10 | 2/3, strong | N.D. | 9 (4/79) | 2 (0/35) | Poor |
Cosegregation (linkage) of transgene expression and herbicide resistance was evaluated by RNA gel blot analysis and a Basta sensitivity test, respectively, for the indicated samples.
Number of plants expressing the transgene in the eighth leaf/number of Basta-resistant plants.
Relative level of transgene expression determined by an RNA gel blot analysis. Assigning weak expression as 1.0, medium corresponds to fourfold to eightfold higher and strong corresponds to more than eight times higher expression than the weak level.
Number of plants expressing the transgene in the fifth leaf/number of Basta-resistant plants.
Number of ear families tested in each transgenic line (number of Basta-resistant plants/total plants tested). Data are the sum of results obtained from all families of that line. Transgenes were transmitted through the egg.
Transgene expression was determined in families of plants by examining resistance to Basta applied to the tenth leaf.
N.D., not determined.
Families were ranked by the ratio of T1 or T2 plants expressing the transgene to those lacking it. Maintenance of ranges of the ratio from the T0 to the T2 generation was scored as good (1 ± 0.2), medium (1 ± 0.4), or poor (<0.5). A ratio <0.5 predicts a high probability of transgene silencing in the next generation.
Antisense MuDR Transgene Expression Does Not Affect mudrA and mudrB Transcript Levels or Mutator Somatic Excision Activity
We selected progeny from two independently transformed Anti-mudrA lines (HSA9 carrying Anti-mudrA-4 and HSA22 carrying Anti-mudrA-5) and two Anti-mudrB lines (HSA3 carrying Anti-mudrB-1 and HSA32 carrying Anti-mudrB-3) for detailed analysis. These transgenic lines maintained good expression of both the Bar gene and the transgene, with some examples of silencing in lines with Anti-mudrB-1 (Table 5). Nonetheless, HSA3 of the Anti-mudrB-1 transgenic line showed good expression of both genes.
From a program of genetic crosses, we found that antisense transgenes did not influence somatic excision frequency (Tables 6 and 7). For example, Bz2 T0 plants carrying Anti-mudrA-4 or -5 were crossed with bz2 testers. T1 Basta-resistant (Bz2/bz2) plants grown from purple kernels then were crossed by a bz2-mu2 Mutator male plant to produce HSA9 (Anti-mudrA-4) and HSA22 (Anti-mudrA-5) ear families. Because the Mutator parent was homozygous for the bz2-mu2 reporter allele, all T2 progeny ears should segregate 1:1 for purple:spotted bronze kernels, and half of each color class should contain the antisense transgene. At this stage, 40 to 60% of the kernels were spotted, indicating that there was no impact of the antisense construct on somatic excision activity. As is typical of Mu-induced mutable alleles, most excision sectors consisted of 1 to 16 cells in transgenic and nontransgenic plants, indicative of similar excision timing.
Table 6.
Effect of Antisense Transgene Expression on Somatic Excisions of bz2-mu2
| Number of Progeny Ears with Spotted Kernels within the Indicated Percentage a
|
||||||
|---|---|---|---|---|---|---|
| Genetic Locus (Female Parent) | 10 to 20 | 21 to 29 | 30 to 39 | 40 to 60 | >60 (%) | Total Ears |
| Anti-mudrA-4(−) b | 4 | 8 | 56 | 4 | 72 | |
| Anti-mudrA-4(+) b | 8 | 4 | 53 | 3 | 68 | |
| Anti-mudrA-5(−) | 4 | 19 | 61 | 4 | 88 | |
| Anti-mudrA-5(+) | 12 | 60 | 4 | 76 | ||
| Anti-mudrB-1(−) | 8 | 64 | 72 | |||
| Anti-mudrB-1(+) | 19 | 19 | ||||
| Anti-mudrB-3(−) | 7 | 36 | 64 | 12 | 119 | |
| Anti-mudrB-3(+) | 12 | 36 | 48 | |||
T2 Mutator parents were tested for transgene expression as described in footnote b and were crossed by bz2 testers to generate progeny ears. The percentage of spotted kernels in the resulting ears was calculated, and each ear was placed into a category as indicated.
Transgene expression was determined in families of plants by examining resistance to Basta applied to the tenth leaf. (−), sensitive; (+), resistant.
Table 7.
Effect of Antisense mudrA Expression on the Frequency of Mu1 Somatic Excision from bz2-mu2
| Excision Frequency
|
|||
|---|---|---|---|
| Sample | High (>150 a) | Medium (10 to 150) | Low (<10) |
| Anti-mudrA (−) | 65.0 ± 14.0 b | 30.2 ± 11.4 | 4.0 ± 3.2 |
| Anti-mudrA (+) | 66.4 ± 9.4 | 27.8 ± 8.1 | 5.8 ± 2.8 |
T3 progeny ears described in Table 6 were sampled from transgenic plants carrying Anti-mudrA-4 or Anti-mudrA-5.
The number of excision spots in a 2 mm × 2 mm area was counted for each kernel using a Nikon stereo zoom microscope. Most excision sectors consist of 1 to 16 cells, and there was no significant difference in the excision timing depending on the expression of the transgene (data not shown).
Spotted kernels were classified by their excision frequency as high, medium, and low. Numbers represent the percentage of kernels on each ear that belong to the indicated frequency category ± sd. Data were taken from counting of 1235 spotted kernels/8 ear families that do not express the transgene and of 1085 spotted kernels/7 ears families that express the antisense mudrA transgene.
We tested for antisense transgene effects in the subsequent generation. A population of plants was grown from T2 spotted kernels, categorized for Basta resistance, and crossed again by bz2 tester. We expected that individuals with a bz2-mu2/bz2 Anti-mudrA/− genotype would produce 25% spotted progeny kernels if the antisense transgene completely suppressed Mutator activity, whereas control plants lacking the transgene would produce 50% spotted kernel progeny. On a per ear basis, as shown in Table 6 for the lines examined with or without an antisense transgene, most ears fell into the 40 to 60% category, which is a frequency normally found in standard Mutator lines [78% of the progeny in the Anti-mudrA-4(+) family and 79% in the Anti-mudrA-5(+) family], and these were very similar to the sister lines lacking the transgene. On the other hand, 18% (12 of 68 ears) of Anti-mudrA-4(+) and 16% (12 of 76 ears) of Anti-mudrA-5(+) progeny ears produced T3 ears with <40% spotted kernels. Control families that did not contain the antisense mudrA transgene also gave similar results (17% in a line sister to Anti-mudrA-4 and 26% in a line sister to Anti-mudrA-5 progeny ears). Therefore, stochastic silencing is the best explanation for ears with <40% spotted kernels in the transgenic Anti-mudrA-4(+) and Anti-mudrA-5(+) lines.
On a per kernel basis, the expression of antisense mudrA also did not affect the frequency of Mu1 somatic excisions (Table 7). In the control that did not carry Anti-mudrA, 65% of spotted kernels were classified as high spotted, 30.2% were classified as medium spotted, and 4.8% were classified as low spotted. Expression of an antisense mudrA transgene did not alter this distribution.
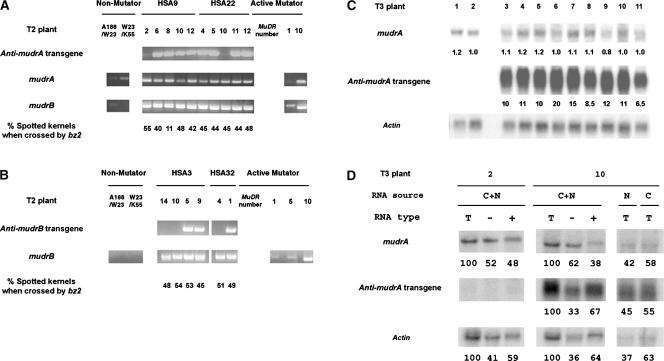
RT-PCR was used to determine whether expression of an antisense transgene reduced the abundance of the corresponding sense transcripts in T2 plants. mudrA and mudrB transcript levels are proportional to MuDR copy number in active Mutator lines (Rudenko and Walbot, 2001). Increasing amounts of mudrA and mudrB amplification from Mutator plants carrying higher MuDR copies (Figures 6A and 6B) support our idea that the RT-PCR in this experiment can be used to compare transcript levels between plants of one family. As a check for equal RNA samples, similar amounts of mudrB from plants were amplified from members of the HSA9 and HSA22 families in which an antisense mudrA locus is segregating (Figure 6A). Figures 6A and 6B show gene expression in representative T2 plants and the spotting phenotype of their resulting T3 ears for the Anti-mudrA and Anti-mudrB lines, respectively. Neither antisense mudrA nor antisense mudrB suppressed sense gene expression significantly compared with sibling plants without the transgene. Plants expressing antisense mudrA (HSA9-6, -8, -10, and -12 as well as HSA22-4, -5, -11, and -12) contained similar levels of mudrA transcripts compared with two control plants, HSA9-2 and HSA22-10, that lacked the transgene. Moreover, the frequency of excision per ear was similar in plants with the Anti-mudrA transgene and in the control plants. There were 40% spotted kernels in the ear of HSA9-6 × bz2 tester and 48% in the ear of HSA22-12 × bz2 compared with the control crosses, which had 55% in HSA9-2 × bz2 and 45% in HSA22-10 × bz2 (Figure 6A). Similarly, antisense mudrB transgenes had no quantitative impact on the expression of mudrB or the frequency of somatic excision in the T2 or T3 generation (Figure 6B, Table 6).
Figure 6.
Molecular and Genetic Analyses of the Impact of Antisense Transgene Expression on mudrA and mudrB Transcript Abundance and on Somatic Excision Activity.
(A) and (B) Two transgenic families for each Anti-mudrA (A) or Anti-mudrB (B) were examined. These are T2 families generated by crossing to a bz2-mu2 active Mutator line in a previous generation. RNA was extracted from transgenic plants and from individuals of two non-Mutator plants, the HiII hybrid (A188 × B73) used to make transgenic plants and W23 × K55 (bz2 tester), and from active Mutator plants with varying numbers of MuDR elements (1, 5, or 10 copies). RT-PCR was performed to amplify antisense mudrA transcripts (using a primer residing in the nopaline synthase [Nos] 3′ region and primer 9), antisense mudrB transcripts (using the Nos 3′ primer and primer 8), endogenous mudrA transcripts (primers 5 and 9), and mudrB transcripts (primers 8 and 13). Two different splicing products were amplified for mudrB, as expected. Previously, data from RNase protection assays were used to estimate that mudrB retaining only the last intron (corresponding to the lower band in the bottom gel in [A]) represented 75% of total mudrB transcripts and that the transcript containing both introns 2 and 3 (corresponding to the upper band in [A]) represented 20% of the total RNA extracted from leaf samples of a high-copy MuDR line (Hershberger et al., 1995). PCR was performed for 31 cycles, and the amplified fragments were visualized by staining with ethidium bromide. Each transgenic individual (bz2/bz2-mu2) was crossed to the W23 × K55 bz2 tester to allow the scoring of somatic excision spots in the progeny.
(C) Expression of antisense transgene and endogenous mudrA in T3 sibling plants. Total RNAs were prepared from seedlings of spotted sibling kernels of a T3 progeny ear (HSA22-12 × bz2). An RNA gel blot was probed with the Anti-A DNA fragment to detect the expression of the genes. The abundance of messages was quantified using a Kodak image analysis system as described in the Methods section, and they are normalized to the expression of actin. The numbers below each blot represent the relative abundance of messages compared with that of mudrA in plant 2. Plants 1 and 2 are control plants that do not carry the Anti-mudrA transgene.
(D) Polyadenylation and localization of transgenic anti-mudrA messages. Whole-plant total RNA (C+N), nuclear total RNA (N), or cytosolic total RNA (C) was isolated from leaves of plant 2 or plant 10 described in (C). The whole-plant total RNA (T) was fractionated into poly(A−) RNA (−) or poly(A+) RNA (+), and the RNA preparations were separated on an agarose gel and then blotted on a nylon membrane. The abundance of messages probed with Anti-A DNA fragments was quantified as described in (C). The numbers below each blot represent the percentages. For whole-plant RNA preparations (C+N), total RNA from the preparation was designated as 100%. The abundance of messages derived from the cytoplasmic or nuclear preparation is expressed as a fraction of the sum from both sources. Fifteen micrograms of total RNA or equivalent amounts of poly(A−) RNA and poly(A+) RNA or 10 μg of cytosolic RNA or the cell-volume equivalent amount of nuclear RNA (1 μg) were used for the RNA gel blot analysis (Alonso-Caplen et al., 1992).
As a second check of RNA abundance, we examined the expression of Anti-mudrA and mudrA on an RNA gel blot (Figure 6C). Using an Anti-A fragment (Figure 1A) as a probe, we detected both endogenous mudrA (∼2.8 kb) and Anti-mudrA (∼600 bp) transcripts simultaneously on the same blot and compared the molar ratios of the transcripts. The T3 plants expressed antisense mudrA at 6.5 times (plant 11) to 20 times (plant 6) higher levels than endogenous mudrA. This massive expression of antisense RNA, however, did not affect the abundance of the sense transcripts significantly compared with that in sibling plants lacking the antisense transgene. All transgenic plants still expressed 80 to 120% of mudrA compared with a sibling control plant that does not carry the transgene (Figure 6C, plant 2). Similar results were found for antisense mudrB T3 plants (data not shown). About two-thirds of the transgene Anti-mudrA transcripts were polyadenylated, and slightly more than half of the total transgene transcripts were localized in the cytosol (Figure 6D, second gel from the top). Polyadenylation of the endogenous mudrA transcripts in the transgenic plants was less efficient compared with that of other transcripts, such as actin. By quantification of RNA gel blot intensities, we found a 10- to 12-fold molar excess of anti-mudrA transcripts compared with endogenous mudrA in both the cytoplasmic and nuclear fractions (Figure 6D, right gels); however, the abundance of total mudrA transcripts was unaffected (Figure 6C). We conclude that neither the abundance of mudrA and mudrB transcripts nor the somatic excision frequency or its timing is sensitive to the presence of a massive amount of antisense transcripts for these gene products in the nucleus and the cytosol of active Mutator plants.
DISCUSSION
Antisense transcripts can arise from active antisense promoters (Spicer and Sonenshein, 1992), from read-through transcription initiated from neighboring genes (Acheson, 1984), from transcription driven by an RNA-dependent RNA polymerase using small complementary RNAs or an inverted repeat transgene array as a primer (Cogoni and Macino, 1999), or from a gene transcription unit in trans (Lee et al., 1993). In this study, we demonstrate that chimeric sense/antisense transcripts of MuDR originate mainly from deletion derivatives of MuDR and not from read-through in an intact MuDR element. Antisense RNA structures and the absence of strong promoters within the mudrA or mudrB coding region suggest that transcription probably initiates normally at +162 in TIRB and at +168 in TIRA or the nearby second initiation site at +252.
Internal deletions that preferentially remove the 3′ UTR of mudrA and/or mudrB, including the intergenic region, either eliminate termination or polyadenylation signals or in some way prevent their use. As a consequence of frequent MuDR deletions and convergent transcription of the sense genes, these defective MuDR elements are prone to produce abundant chimeric sense/antisense transcripts that accumulate in active Mutator lines. Those antisense RNAs whose sequences are perfectly complementary to the endogenous sense RNA could interfere with sense RNA stability and translation or induce post-transcriptional gene silencing. We showed here, however, that the sense transcript abundances and Mutator activities are not obviously regulated by naturally occurring antisense RNAs. This lack of suppression also is evident in transgenic plants expressing massive amounts of antisense mudr transcripts, even when both sense and antisense transcripts are present in the same subcellular compartment. The antisense-expressing construct was proven to reduce the abundance of the corresponding sense transcripts in non-Mutator Black Mexican Sweet protoplasts over an ∼24-h incubation (Table 4), but in intact plants there was no suppression effect; presumably, different conditions exist in situ in a Mutator plant carrying active MuDR elements than in the protoplasts of a non-Mutator line. Alternatively, the ratio of sense to antisense transcripts could differ in the electroporated protoplasts compared with intact plants containing an integrated transgene.
Structural Analysis of Antisense MuDR Transcripts
Only active Mutator plants produce antisense transcripts detectable by RNA gel blot hybridization analysis (Hershberger et al., 1995; Joanin et al., 1997). The origin of only one antisense transcript has been documented to date: a heritable deletion derivative (d202) missing 174 bp at the 3′ end of the mudrA gene encodes chimeric sense/antisense transcripts for both mudrA and mudrB (Lisch et al., 1999). However, this single transcript type does not account for the RNase protection results, indicating that many portions of mudrA and mudrB RNA are present in antisense form (Hershberger et al., 1995). In a multicopy MuDR plant, we identified diverse antisense transcripts and recovered 13 different chimeric transcripts from a small leaf disc of a single plant. These transcripts lacked 7 to 2909 bp of internal MuDR sequence, and none of the missing sequences occurred at points that match the consensus features of maize intron borders. We propose that diverse somatic deletion derivatives arising during organ development are a major source of somatic antisense transcripts. Because we used RT-PCR primers residing within either mudrA or mudrB, MuDR elements deleted in the intergenic region may have been recovered preferentially. Thus, the antisense hybrid transcripts analyzed here could represent only a subset of antisense MuDR transcript diversity.
Germinal Deletions within MuDR
Plants with heritable ΔMuDR have provided fruitful genetic material to determine the roles of mudrA and mudrB in Mutator activities (Lisch et al., 1999). Despite their utility, the frequency of MuDR deletions has not been quantified. Analyzing families in which the immediate Mutator progenitors had several copies of MuDR, we found that 18.9% of progeny contained an independent, newly arising deletion derivative. Per element, the mutation frequency is >10−2, many orders of magnitude greater than deletions within standard maize genes. In most cases, the ΔMuDR appeared to be multicopy, indicating that amplification, most likely by transposition, happened late in the life cycle of the previous generation. Considering only new deletions, 40% of the deletion events removed the intergenic region. This short 225-bp intergenic region and the neighboring sequences containing the 3′ UTRs of mudrA and mudrB were deleted preferentially. The intergenic region is composed of five distinct sets of direct repeats (11 to 27 bp in length) and three copies of one inverted repeat (12 bp in length) that can be modeled to fold into extended intrastrand duplexes (Hershberger et al., 1995). Removal of the intergenic region could affect the successful termination of MuDR transcription by changing DNA or RNA topology. The absence of the normal 3′ UTRs may result in a majority of the chimeric sense/antisense RNAs lacking polyadenylation.
Inability of Antisense MuDR Transcripts to Suppress MuDR Regulatory Elements and Mutator Activities
Although germinally transmitted ΔMuDR elements are transcriptionally active and produce chimeric sense/antisense transcripts, they have no measurable impact on mudrA transcript levels or Mu1 TIR methylation. As determined by in situ hybridization, antisense mudrA transcripts colocalized with sense mudrA and mudrB transcripts in many tissues of active Mutator plants, and the sense and antisense transcripts may have been paired in vivo (Joanin et al., 1997).
As with documented cases of quelling in fungi, RNA interference in animals, and cosuppression in transgenic plants (reviewed by Matzke et al., 2001; Vance and Vaucheret, 2001), structural properties of ΔMuDR transcripts would be predicted to lead to RNA-based epigenetic gene silencing (Walbot and Rudenko, 2002). Ectopically expressed antisense RNAs are thought to pair with sense transcripts and feed into the double-stranded RNA–induced degradation pathway (Stam et al., 2000; Van Houdt et al., 2000; Serio et al., 2001), and in such cases, antisense transcripts can effectively silence homologous genes. Furthermore, the 3′ UTRs of mudrA and mudrB transcripts share repeat motifs that could fold into double-stranded structures (Hershberger et al., 1995). In recent studies, such intrastrand duplexes have been shown to be much more efficient than antisense RNAs in inducing homologous host gene silencing (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Wesley et al., 2001).
Therefore, it is particularly interesting that sense transcripts and Mutator activities were not suppressed by either endogenous or transgene-encoded antisense mudrA and antisense mudrB RNA. In some previous studies assessing the impact of antisense transgenes, there was a lack of correlation between antisense and sense transcript abundance. For example, in antisense chs petunia transformants, Blokland et al. (1994) found no direct correlation between the presence of antisense RNA and chs suppression. A high level of antisense expression did not necessarily result in suppression of the corresponding endogenous gene, although chs silencing was the common outcome. Previously, in situ RNA analysis demonstrated that both sense and antisense MuDR transcripts were present in the same tissues and in the same cells (as in the endothelium, where both transcript types were present in all cells) (Joanin et al., 1997). In the present study, both sense and transgene-encoded antisense messages were detected in the nucleus and cytoplasm by RNA gel blot analysis, but we do not know whether the molar ratio of sense to antisense transcripts is similar in all cells. It is also unknown whether transcripts localized in the same compartment interact to form double-stranded RNA.
It is possible that antisense RNA has additional roles or fates beyond the formation of double-stranded RNA with sense transcripts. In that regard, we were interested in the observation that most ΔMuDR1 and ΔMuDR2 transcripts lack polyadenylation, a post-transcriptional modification required for efficient transport of mRNAs into the cytoplasm (Huang and Carmichael, 1996). If most ΔMuDR RNA is compartmentalized inside the nucleus, contact with properly processed mudrA and mudrB transcripts in the cytosol would be reduced, which might prevent chimeric sense/antisense transcripts from blocking the translation of the sense transcripts or triggering sense transcript degradation in that same subcellular location. A significant fraction of nuclear MuDR transcripts also were poly(A−) RNA, particularly in lines undergoing silencing (Rudenko et al., 2003); hence, antisense poly(A−) transcripts might act in that cellular compartment to disrupt sense transcript maturation. Molecular and genetic analysis using antisense transgenic plants, however, demonstrated that substantial antisense transcript accumulates in both the nucleus and the cytoplasm and that the antisense transcripts are present in greater than ∼10-fold molar excess over the corresponding sense transcripts. Therefore, the tolerance of Mutator activities cannot be explained by the compartmentalization of the antisense transcripts.
Studies of antisense regulation in animals suggest two other mechanisms that might operate in MuDR regulation. X chromosome inactivation in mouse is initiated by an accumulation of Xist RNA; however, expression of the neighboring Tsix gene blocks this accumulation in a cis-limited manner. Tsix transcription starts 15 kb downstream of Xist and continues across the Xist locus, resulting in antisense RNA complementary to the Xist sense messages; the antisense transcripts effectively suppress Xist on that X chromosome but not on other ones (Lee et al., 1999; Stavropoulos et al., 2001). Such cis regulation seems unlikely to be effective for MuDR, because messages transcribed from deleted elements would suppress only the sense transcripts encoded from the same deleted MuDR element. Transcripts originating from the dispersed MuDR elements in multicopy lines would be unaffected, although all copies of MuDR cease transcription during Mutator silencing (Hershberger et al., 1995). Alternatively, cosuppression of Drosophila I transposon activity by I-containing sense or antisense transgenes requires at least several and up to 10 generations to be established (Jensen et al., 1999). Thus, we cannot exclude the possibility that antisense repression of MuDR transposon activities requires many generations to establish the silenced state in these highly active Mutator plants.
Escape of MuDR/Mu from Maize Regulatory Mechanisms
DNA transposons are parasitic and mutagenic agents (Martienssen, 1998). They are hypothesized to increase the host's genetic diversity through transposition while ensuring their own propagation (Schwarz-Sommer et al., 1985). However, uncontrolled transposition, resulting in chromosomal instability or an excess of lethal alleles, could compromise host viability and thus end the relationship of each transposon with its host. Natural selection pressure should drive both partners to limit transposition but ensure transposon propagation, resulting in a spectrum of regulatory mechanisms. Two defensive features in maize plants limit these harmful effects of uncontrolled MuDR/Mu propagation: transposition is restricted to terminally differentiated cells, and coordinated epigenetic Mutator silencing occurs to quench the activities of multiple MuDR elements.
On the other hand, there are at least three examples of MuDR strategies by which the transposable element may evade several host defensive mechanisms. First, a subset of hMuDR elements escapes transcriptional downregulation during Mutator silencing (Rudenko and Walbot, 2001). Second, 21- to 26-nucleotide MuDR- or hMuDR-specific small RNAs are found in both non-Mutator and active or silencing Mutator plants, but their expression is not correlated with the epigenetic silencing of the element (Rudenko et al., 2003). This is particularly interesting because it is well known that transgenes expressing double-stranded RNA induce post-transcriptional gene silencing when coding sequences are used and induce transcriptional gene silencing when promoter sequences are used; small RNAs are thought to play a major role in both types of silencing phenomena (Sijen et al., 2001). Third, antisense transcripts resulting from the inevitable generation of frequent internal deletions do not affect MuDR transcript abundance and Mutator activities. In other words, MuDR is not suppressed by the homology-dependent gene silencing machinery of the host, at least that component of the mechanism triggered by antisense transcripts. Additionally, ΔMuDR elements are more likely to be silenced by chimeric transcripts than is an active MuDR. The deleted elements are coordinately silenced independently of transcription by intact MuDR elements. By contrast, active MuDR elements do not repress their own expression; rather, they enhance their own transcription and/or mRNA abundance (Raizada et al., 2001b).
We show here that antisense MuDR transcripts are not dominant-negative regulators of Mutator activities, but much remains to be learned before we fully understand the mechanism by which MuDR elements escape from antisense-mediated regulation. Detailed analysis of Mutator plants, such as investigation of whether specific endogenous or transgene-encoding antisense RNA types increase small RNA molecules in the plants or determination of whether a threshold level of the small molecules is needed to induce antisense-mediated negative regulation, may reveal important clues for understanding antisense-tolerant mechanisms of the MuDR elements.
METHODS
Plant Materials
All maize (Zea mays) Mutator stocks used were derived from lines originally supplied by D.S. Robertson (Iowa State University, Ames), including a line with the a1-mum2 allele. bz2 mutable alleles were recovered in our laboratory. Mutator lines have been maintained by selfing and by outcrossing to bz2 or a1 tester lines, as appropriate. The active Mutator lines SK26, SK30, SB03, and SB36 have a high number of MuDR elements (10 to 25 copies) and have the bz2-mu2 mutable allele. They were crossed to a W23 bz2 tester line in the previous generation. M857, M858, and M874 are active Mutator lines carrying several unmethylated, 4.9-kb MuDR elements and very few Mu1, Mu2, or Mu3 elements; these lines were the gift of V.L. Chandler. Sibling plants of these lines were crossed to anthocyanin tester plants to produce progeny lines: M857 to bz2 (T56 and T65), M858 to bz2 (T55, T63, and T64), M858 to c2 (T57), M858 to r-g tester (T62), and M874 to bz1 (T69).
RNA Preparation, Reverse Transcription PCR Amplification, Subcloning, and Sequence Analysis of PCR Products
Total plant RNA and nuclear or cytoplasmic RNA preparations were isolated from 10 to 20 g of leaf tissue as described previously (Rudenko et al., 2003) using either RNeasy Maxi kits (Qiagen, Valencia, CA) or Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA was fractionated into poly(A+) and poly(A−) RNA after determination of RNA concentration using an Oligotex mRNA Midi Kit (Qiagen) and then kept in the same volume of RNA storage buffer (Ambion, Austin, TX) as the initial total RNA sample. RNA preparations for reverse transcription (RT) PCR analysis were treated further with RNase-free DNase I (Gibco BRL, Gaithersburg, MD). For RT-PCR amplification of the antisense mudrB transcripts shown in Figure 1C, sense-strand cDNA was reverse-transcribed (OneStep RT-PCR kit; Qiagen) at 50°C for 30 min using the strand-specific primer B3843 (5′-TTCGAAGAGCACTCTCGTGTC-3′). Reverse-transcribed cDNA was amplified after adding the opposite strand-specific primer, A1621 (5′-CTTGCATCTGCTACTGGTGTAGATGGCC-3′). To amplify antisense mudrA transcripts, A1621 was used as the RT primer and B3843 was added for PCR amplification. Control reactions were preincubated with DNase-free RNase (Boehringer Mannheim, Indianapolis, IN) according to the manufacturer's protocol.
For the retrieval of polyadenylated antisense transcripts, amplification of the 3′ cDNA ends was performed from total RNA using a 5′/3′ rapid amplification of cDNA ends kit (Boehringer Mannheim) according to the manufacturer's instructions. In brief, reverse transcription was performed at 45°C using an oligo(dT)-anchored primer (5′-GACCACGCGTATCGATGTCGATTTTTTTTTTTTTTTTT-3′). Antisense mudrA or antisense mudrB cDNA then was amplified using a PCR-anchor primer (5′-GACCACGCGTATCGATGTCGAC-3′) and strand-specific primers residing in different regions of the antisense strand of mudrA or mudrB. PCR fragments were gel-purified from the RT-PCR product and subcloned into pT-Adv; plasmids were transformed into Escherichia coli strain DH10B according to the supplier's protocol (Clontech, Palo Alto, CA). Double-strand cycle sequencing with Big-Dye terminators was performed at the Stanford University Protein and Nucleic Acid Biotechnology Facility.
The antisense cDNAs listed in Table 1 were amplified and cloned as described above but using the RT-PCR primer sets indicated in the same table. All RT-PCR procedures used OneStep RT-PCR (Qiagen) and were performed using DNase-treated RNA samples and the following primers: A493 (5′-TTCCCAACTCCCCCGATGTAG-3′), A1813 (5′-CCGTATGCTGAGAGAAGAGAATGC-3′), A2298 (5′-ATGACTAAACATGTCGTGAATGCAG-3′), A2713 (5′-TCTAGCTACAAGTGCCCTTTGAAT- GG-3′), A2907 (5′-TACAGCCAGACGTAGCAGAGG-3′), A2952 (5′-ATCCACAGCCAGAGACAGAACAATTGGG-3′), IGR3501 (5′-TATAACACACATGAATAACACTGAGCC-3′), A3276 (5′-GGGCTTGTTCTTAGCAGT- CTTACAACC-3′), IGR3469 (5′-CAAGCAATCTGTTTCTCTGAACTG- GGCG-3′), B3950 (5′-TCTAGATGATGATGAACTTGGTGATGGG-3′), B4334 (5′-CCATTGCCGTGGAGAAGGTTGAAGCAG-3′), B4473 (5′-ATT- GTCCACCGAGCAAAGTGG-3′), and NOS3′ (5′-CCGGCAACAGGATTC- AATCTTAAGAAAC-3′). The maize actin primers pMAcl 5′ and pMAcl 3′ (Raizada and Walbot, 2000) were added to the reactions as an internal RT-PCR and RNA quantity control. When RT-PCR products were analyzed using DNA gel blot analysis, 1 μL of RT-PCR product after 26 cycles of PCR amplification was separated by electrophoresis on a 1.0% agarose gel and visualized by standard DNA gel blot analysis using gene-specific probes as indicated in the legend of Figure 4D.
Genomic DNA Analysis
Plant genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. DNA aliquots of 5 μg were digested with SstI or HinfI, fractionated by electrophoresis on 0.8% agarose gels, transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech, Piscataway, NJ), and probed with α-32P-dCTP–labeled DNA fragments as indicated in the legends of Figures 1B, 4A, and 4G. Hybridization and washing of the membranes were performed as described previously (Warren and Hershberger, 1994).
RNA Gel Blot Analysis
Total RNA preparations were heat-denatured at 55°C for 15 min and fractionated on a 1.2% agarose gel containing formaldehyde. After transfer to a Hybond N+ membrane, the membrane was probed with α-32P-dCTP–labeled DNA probes and washed under the conditions described previously (Hershberger et al., 1995). When an oligonucleotide was used as a probe to detect the expression of Ubi1, a gene-specific oligonucleotide (5′-GCAGATACTTTGACAACC-3′) was end-labeled with α-32P-dCTP and hybridization and washing were performed as described by Christensen et al. (1992).
Hybridization Probes
Transposon-specific double-stranded DNA probes A, B, and IGR were generated by PCR amplification from the pMu9.6 MuDR clone (Raizada and Walbot, 2000) using gene-specific primers. The mudrA probe (A) corresponds to MuDR positions 464 to 3276, the mudrB probe (B) corresponds to positions 3501 to 4495, the intergenic region probe (IGR) corresponds to positions 3297 to 3548, and the antisense mudrA probe (Anti-A) corresponds to positions 2865 to 3332. The Mu1/Mu2-specific probe and an actin probe were prepared as described by Raizada and Walbot (2000). Primer sets used for the MuDR-specific PCR amplifications were as follows: A (5′-CAGTTTCAATTCGCTAGACTCCAACGG-3′ and 5′-CTGGGCTTGTTCTTAGCAGTCTTACAACC-3′); B (5′-TATAACACACATGAATAACACTGAGCC-3′ and 5′-TTCTCTGCTCCTGTGCGG- ATGGATTG-3′); IGR (5′-CAGTTTTAGTTGCCAGTTCGTTGCTTCC-3′ and 5′-TATCTGTTTTGTGTTATATTTTATCT-3′); and Anti-A (5′-TTGGATCCCTAAAGAGCTTCGGACTTCTTC-3′ and 5′-ATGGATCCGAACCTGGAAGCAACGAACTGGC-3′).
Plasmid Construction
Plasmids were constructed using routine DNA manipulation protocols (Sambrook et al., 1989) and purified from bacterial cultures using Plasmid Max Prep Kits (Qiagen). The structures of the plasmid constructs are diagrammed in Figures 3 and 5A. Detailed descriptions for pJB4, pJB12, pMB5 (Benito and Walbot, 1994), pJD312 (Luehrsen et al., 1992), pAHC17 (Christensen and Quail, 1996), pBARGUS (Fromm et al., 1990), phMR53 (Raizada and Walbot, 2000), and pMB1 (Ono et al., 2002) are available in the literature.
Antisense Promoter Construct
Regions of MuDR were amplified by PCR from pMu9.6 (Raizada and Walbot, 2000) using the PstI- and KpnI-adapted primer sets listed below. The enzyme-digested PCR fragments were subcloned into pJD312 digested with the same enzymes to replace the Cauliflower mosaic virus 35S promoter fragment of the luciferase expression plasmid with the MuDR fragment. The resulting constructs harbor a MuDR fragment followed by the first intron of the maize Adh1 gene inserted upstream of the luciferase coding region. The primer sets used for the PCR construction of individual plasmids were as follows: (1) pAP1 [4612 to 3307] (5′-CAGCTGCAGCGGCAATGCTGGACCGATTC-3′ and 5′-GTTGGTACCTTGCCAGTTCGTTGCTTCCC-3′); (2) pAP2 [4292 to 2143] 5′-CAGCTGCAGGATTATACAAACTCATACACTC-3′ and 5′-CGAGGTACCCTGTTT- CATCGTAGGCGAAGG-3′); (3) pBP1 [445 to 1900] (5′-AAGCTGCAGGATCCATGGACTTGACGCCC-3′ and 5′-TAAGGTACCCTGCTGGATACATGTGCTCT-3′); (4) pBP2 [2124 to 3540] (5′-CAGCTGCAGACAATGGAACTGTTTCATCG-3′ and 5′-TAAGGTACCTGTGTGTTATAT- GTTATCTG-3′); (5) pBP3 [1212 to 2871] (5′-CACCTGCAGTGGAAGGAGGAGGACTACTAC-3′ and 5′-TAAGGTACCGGGATCCAATTTTTT- GTGGTG-3′); and (6) pBP4 [3297 to 3443] (5′-GGGCTGCAGTCAGTTTTAGTTGCCAGTTCG-3′ and 5′-AATGGTACCCGACAATTGCAAGCA- ATCTG-3′).
Antisense RNA Expression Construct
Regions of mudrB or mudrA were amplified from pMu9.6 (Raizada and Walbot, 2000) by PCR using BamHI-adapted primer sets. The enzyme-digested PCR fragment was subcloned into pAHC17 digested with the same enzyme. Bacterial clones harboring fusion plasmids, which result in the expression of antisense mudrA or antisense mudrB fragments directed by the maize Ubi1 promoter, were selected. The primer sets used for the PCR construction of individual plasmid were as follows: (1) pABN [3539 to 4091] (5′-GACGGATCCATTAGTTCTTACAACCTC-3′ and 5′-CAAGGATCCCGGGTTTCTGGAACATGT-3′); (2) pABC [4116 to 4668] (5′-GTTGGATCCGAATGCAACAGTTTAG-3′ and 5′-CGCGGATCCTTG- CGGTCTCCTCTTCTC-3′); (3) pABF [3539 to 4668] (5′-GACGGATCC- ATTAGTTCTTACAACCTC-3′ and 5′-CGCGGATCCTTGCGGTCTCCT- CTTCTC-3′); (4) pAA1 [450 to 1147] (5′-TTTGGATCCATGGACTTG- ACGCCCAG-3′ and 5′-CATGGATCCTCCACGGGCAATCACCAC-3′); (5) pAA2 [1382 to 1909] (5′-GTAGGATCCTTGGGAGGAAAGCTTCCAG-3′ and 5′-GCTGGATCCATGTGCTCTGACCCAGCAT-3′); and (6) pAA3 [2865 to 3332] (5′-TTGGATCCCTAAAGAGCTTCGGACTTCTTC-3′ and 5′-ATGGATCCGAACCTGGAAGCAACGAACTGGC-3′).
The restriction site in each primer is underlined. The portion of MuDR amplified is indicated in brackets using the numbering system of Hershberger et al. (1991). The proper orientations of gene fragments in the final fusion constructs were confirmed by cycle sequencing at the Stanford University Protein and Nucleic Acid Biotechnology Facility.
Transient Gene Expression Analysis
Preparation of maize Black Mexican Sweet protoplasts and electroporation and transient expression assays were performed as described previously (Carle-Urioste et al., 1995). For the experiment testing antisense promoter activity, 20 μg of each MuDR fragment fusion construct (pAP1or pAP2 for antisense mudrA promoter activity and pBP1, pBP2, pBP3, or pBP4 for antisense mudrB promoter activity) was electroporated into 106 Black Mexican Sweet protoplasts together with 10 μg of β-glucuronidase (GUS)–expressing pJB4 plasmid. The transformed protoplasts then were collected after 24 h of incubation in growth medium, and the activity of GUS and luciferase in protoplast lysates was quantified as described previously (Carle-Urioste et al., 1995). GUS expression by pJB4 was used as an internal transformation control; luciferase expression was normalized using the GUS expression for the data presented in Table 2. pJB12 (pJD312 without the Cauliflower mosaic virus 35S promoter) was used as a negative control, and pMB5 (TIRB:Adh1 intron:LUC) was used as a positive control. Luciferase activity was expressed as the number of photons emitted using luciferin during a 10-s integrative counting period. GUS activity was measured as picomoles of methylumbelliferone converted from methylumbelliferyl-β-d-glucuronide per minute.
For the experiment evaluating the effectiveness of antisense MuDR fragments, the impact of transiently expressed antisense transcripts in reducing the abundance of the corresponding sense messages was examined either by RNA gel blot analysis (for antisense mudrA) or by measurement of the expression from a MURB-GUS fusion protein (for antisense mudrB) (Ono et al., 2002). In brief, 20 μg of each antisense construct (pAA1, pAA2, or pAA3 for antisense mudrA and pABN, pABC, or pABF for antisense mudrB) was electroporated into 106 Black Mexican Sweet protoplasts together with 10 μg of phMR53 and 10 μg of pJD312 (for the antisense mudrA constructs) or 20 μg of pMB1 and 10 μg of pJD312 (for the antisense mudrB constructs). Antisense mudrA-transformed protoplasts were collected after 24 h of incubation in growth medium, and total RNA was isolated using Trizol reagent. The expression of sense mudrA was examined by RNA gel blot analysis using the mudrA fragment as a probe (Figure 1A), and the abundance of the transcript was quantified using a Kodak Digital Science Electrophoresis Documentation and Analysis System 120 and its 1D Image Analysis Software (Eastman Kodak, New Haven, CT). Antisense mudrB–transformed protoplasts were collected as described above, and the ratios of luciferase to GUS activities in protoplast lysates were quantified as described above. Luciferase expression by pJD312 was used as an internal transformation control. The expression of mudrA transcript and MURB-GUS protein were normalized to luciferase units for the data presented in Table 4.
Transgenic Plants
The most effective antisense mudrA or antisense mudrB expression construct identified by the transient analysis, pAA3 (pAHC17:Anti-mudrA) or pABF (pAHC17:Anti-mudrB), along with an herbicide selection plasmid (pBARGUS), was cobombarded into embryogenic HiII calli using biolistic delivery at the Plant Transformation Facility at Iowa State University (www.agron.iastate.edu/ptf/web/mainframe.htm/). The transformed calli were checked by RNA gel blot analysis for transgene expression and tested for resistance to 3 mg/mL bialaphos, the active ingredient in Basta herbicide (Spencer et al., 1990). The doubly positive calli were regenerated into T0 plants. Plants and progeny from five different transgenic calli for each antisense construct were chosen for detailed study. Herbicide resistance was used to identify transgene-expressing plants in the field or greenhouse because Basta resistance perfectly cosegregated with transgene expression (Table 5). Herbicide-resistant plants were identified as described previously (Raizada and Walbot, 2000).
The mature T0 plants were crossed to or by bz2 or active Mutator lines with a high copy number of MuDR elements and homozygous for bz2-mu2 (Hershberger et al., 1995). Because the T0 plants were A1 Bz2 r-g/r-r, it was not possible to score somatic excision immediately in T1 progeny by crossing to these Mutator lines; all kernels were purple or mottled and Bz2/bz2-mu2. Somatic excision in T2 kernels was scored after herbicide-resistant T1 plants were crossed to/by bz2 (if they had been crossed to a Mutator line in the previous generation) or to/by an appropriate Mutator line (if they had been crossed to a non-Mutator tester in the previous generation) to generate a recessive background at bz2. In the T2 generation, half of the kernels on each ear were purple, and the other half of the kernels were scored for somatic excision of the segregating reporter allele. Progeny from spotted kernels in the T2 generation were tested further for Basta resistance and crossed again by bz2 tester to score the somatic excision on kernels. Phenotypic comparisons were made to highly active Mutator plants carrying the bz2-mu2 allele without an antisense transgene.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Virginia Walbot, walbot@stanford.edu.
Acknowledgments
We are especially grateful to Akemi Ono, who generously provided the genomic blots, and to Vicki L. Chandler (University of Arizona, Tucson), who provided lines M874, M865, M858, and M857 used in the experiment described in Table 3. We thank M. Raizada, G. Rudenko, S. Shaw, G. Nan, D. Goodman, M. Fitzgerald, and S. Shah for stimulating discussions, critical reading, and thoughtful comments on the manuscript. We appreciate T. DeHoog and M. Abreu for their field and laboratory assistance. This research was supported by a grant from the National Institutes of Health (GM49681).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014605.
References
- Acheson, N.H. (1984). Kinetics and efficiency of polyadenylation of late polyomavirus nuclear-RNA: Generation of oligomeric polyadenylated RNAs and their processing into messenger RNA. Mol. Cell. Biol. 4, 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Caplen, F.V., Nemeroff, M.E., Qiu, Y., and Krug, R.M. (1992). Nucleocytoplasmic transport: The influenza virus NS1 protein regulates the transport of spliced NS2 mRNA and its precursor NS1 mRNA. Genes Dev. 6, 255–267. [DOI] [PubMed] [Google Scholar]
- Benito, M.I., and Walbot, V. (1994). The terminal inverted repeat sequences of MuDR are functionally active promoters in maize cells. Maydica 39, 255–264. [Google Scholar]
- Benito, M.I., and Walbot, V. (1997). Characterization of the Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., Springer, P.S., Cresse, A.D., and Hendrickx, M. (1993). Specificity and regulation of the Mutator transposable element system in maize. Crit. Rev. Plant Sci. 12, 57–95. [Google Scholar]
- Bisseling, T. (1999). The role of plant peptides in intercellular signaling. Curr. Opin. Plant Biol. 2, 365–368. [DOI] [PubMed] [Google Scholar]
- Blokland, R., Geest, N., Mol, J.N.M., and Kooter, J.M. (1994). Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. 6, 861–877. [Google Scholar]
- Bourque, J.E. (1995). Antisense strategies for genetic manipulations in plants. Plant Sci. 105, 125–149. [Google Scholar]
- Brendel, V., Kleffe, J., Carle-Urioste, J.C., and Walbot, V. (1998). Prediction of splice sites in plant pre-mRNA from sequence properties. J. Mol. Biol. 276, 85–104. [DOI] [PubMed] [Google Scholar]
- Carle-Urioste, J.C., Marrs, K., Bodeau, J., and Walbot, V. (1995). Gene transfer to protoplasts: Transient gene expression analysis. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (New York: Springer-Verlag), pp. 106–111.
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., Sharrock, R.A., and Quail, P.H. (1992). Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689. [DOI] [PubMed] [Google Scholar]
- Chuang, C.-F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A., Benito, M.I., and Walbot, V. (1994). Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22, 2634–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm, M.E., Morrish, F., Armstrong, C., Williams, R., Thomas, J., and Klein, T.M. (1990). Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/Technology 8, 833–839. [DOI] [PubMed] [Google Scholar]
- Hershberger, R.J., Benito, M.-I., Hardeman, K.J., Warren, C., Chandler, V.L., and Walbot, V. (1995). Characterization of the major transcripts encoded by the regulator MuDR transposable element of maize. Genetics 140, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger, R.J., Warren, C.A., and Walbot, V. (1991). Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc. Natl. Acad. Sci. USA 88, 10198–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia, A.-P., and Schnable, P.S. (1996). DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon MuDR. Genetics 142, 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.Q., and Carmichael, G.G. (1996). Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., Gassama, M.-P., and Heidmann, T. (1999). Cosuppression of I transposon activity in Drosophila by I-containing sense and antisense transgenes. Genetics 153, 1767–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanin, P.J., Hershberger, R.J., Benito, M.J., and Walbot, V. (1997). Sense and antisense transcripts of the maize MuDR regulatory transposon localized by in situ hybridization. Plant Mol. Biol. 33, 23–36. [DOI] [PubMed] [Google Scholar]
- Knee, R., Li, A.W., and Murphy, P.R. (1997). Characterization and tissue-specific expression of the rat basic fibroblast growth factor antisense mRNA and protein. Proc. Natl. Acad. Sci. USA 94, 4943–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankenau, S., Corces, V.G., and Lankenau, D.H. (1994). The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol. Cell. Biol. 14, 1764–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.T., Davidow, L.S., and Warshawsly, D. (1999). Tsix, a gene antisense to Xist at the X-inactivation center. Nat. Genet. 21, 400–404. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Levy, A.A., and Walbot, V. (1990). Regulation of the timing of transposable element excision during maize development. Science 248, 1534–1537. [DOI] [PubMed] [Google Scholar]
- Lisch, D. (2002). Mutator transposons. Trends Plant Sci. 7, 498–504. [DOI] [PubMed] [Google Scholar]
- Lisch, D., Carey, C.C., Dorweiler, J.E., and Chandler, V.L. (2002). A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc. Natl. Acad. Sci. USA 99, 6130–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., Girard, L., Donlin, M., and Freeling, M. (1999). Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen, K.R., Dewet, J.R., and Walbot, V. (1992). Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 216, 397–414. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. (1998). Transposons, DNA methylation and gene control. Trends Genet. 14, 263–264. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., and Baron, A. (1994). Coordinate suppression of mutations caused by Robertson's Mutator transposons in maize. Genetics 136, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J.M., Pruss, G.J., and Vance, V.B. (2001). RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11, 221–227. [DOI] [PubMed] [Google Scholar]
- Ono, A., Kim, S.-H., and Walbot, V. (2002). Subcellular localization of MURA and MURB proteins encoded by the maize MuDR transposon. Plant Mol. Biol. 50, 599–611. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., Benito, M.-I., and Walbot, V. (2001. a). The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 25, 79–91. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., Brewer, K.V., and Walbot, V. (2001. b). A maize MuDR transposon promoter shows limited autoregulation. Mol. Gen. Genomics 265, 82–94. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., Nan, G.-L., and Walbot, V. (2001. c). Somatic and germinal mobility of the RescueMu transposon in transgenic maize. Plant Cell 13, 1587–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada, M.N., and Walbot, V. (2000). The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell 12, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko, G.N., Ono, A., and Walbot, V. (2003). Initiation of silencing of maize MuDR/Mu transposable elements. Plant J. 33, 1013–1025. [DOI] [PubMed] [Google Scholar]
- Rudenko, G.N., and Walbot, V. (2001). Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives. Plant Cell 13, 553–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwarz-Sommer, Z.A., Gierl, A., Cuypers, H., Peterson, P.A., and Saedler, H. (1985). Plant transposable elements generate the DNA sequence diversity needed in evolution. EMBO J. 4, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio, F.D., Schob, H., Iglesias, A., Tarina, C., Bouldoires, E., and Meins, F., Jr. (2001). Sense- and antisense-mediated gene silencing in tobacco is inhibited by the same viral suppressors and is associated with accumulation of small RNAs. Proc. Natl. Acad. Sci. USA 98, 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J.N.M., and Kooter, J.M. (2001). Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11, 436–440. [DOI] [PubMed] [Google Scholar]
- Simons, R.W., and Kleckner, N. (1988). Biological regulation by antisense RNA in prokaryotes. Annu. Rev. Genet. 22, 567–600. [DOI] [PubMed] [Google Scholar]
- Spencer, T.M., Gordon-Kamm, W.J., Daines, R.J., Start, W.G., and Lemaux, P.G. (1990). Bialaphos selection of stable transformants from maize cell culture. Theor. Appl. Genet. 2, 111–126. [DOI] [PubMed] [Google Scholar]
- Spicer, D.B., and Sonenshein, G.E. (1992). An antisense promoter of the murine c-myc gene is localized within intron 2. Mol. Cell. Biol. 12, 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Bruin, R., Blokland, R., Hoorn, R.A.L., and Mol, J.N.M. (2000). Distinct features of post-transcriptional gene silencing by antisense transgenes in single copy and inverted T-DNA repeat loci. Plant J. 21, 27–42. [DOI] [PubMed] [Google Scholar]
- Stavropoulos, N., Lu, N., and Lee, J.T. (2001). A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. USA 98, 10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terryn, N., and Rouze, P. (2000). The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci. 5, 394–396. [DOI] [PubMed] [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants: Defense and counterdefense. Science 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Van Houdt, H., Van Montagu, M., and Depicker, A. (2000). Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanisms. Mol. Gen. Genet. 263, 995–1002. [DOI] [PubMed] [Google Scholar]
- Walbot, V. (1986). Inheritance of Mutator activity in Zea mays as assayed by somatic instability of the bz2-mu1 allele. Genetics 114, 1293–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V. (1992). Strategies for mutagenesis and gene cloning using transposon tagging and T-DNA insertional mutagenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 49–82. [Google Scholar]
- Walbot, V., and Rudenko, G.N. (2002). MuDR/Mu transposable elements of maize. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A. Lambowitz, eds (Washington, DC: American Society of Microbiology), pp. 533–564.
- Warren, C.A., and Hershberger, R.J. (1994). Southern blots of maize genomic DNA. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 566–568.
- Waterhouse, P.M., Graham, M.W., and Wang, M.-B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.-B., and Finnegan, E.J. (2001). Role of short RNAs in gene silencing. Trends Plant Sci. 6, 297–301. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]