Abstract
Heterozygosity for the CCR5 Δ32 allele is associated with delayed progression to AIDS in human immunodeficiency virus type 1 (HIV-1) infection. Here we describe an unusual HIV-1 isolate from blood of an asymptomatic individual who was heterozygous for the CCR5 Δ32 allele and had reduced levels of CCR5 expression. The primary virus used CCR5, CXCR4, and an unusually broad range of alternative coreceptors to enter transfected cells. However, only CXCR4 and CCR5 were used to enter primary T cells and monocyte-derived macrophages, respectively. Full-length Env clones had an unusually long V1/V2 region and rare amino acid variants in the V3 and C4 regions. Mutagenesis studies and structural models suggested Y308, D321, and to a lesser extent K442 and E444, contribute to the broad coreceptor usage of these Envs, whereas I317 is likely to be a compensatory change. Furthermore, database analysis suggests covariation can occur at positions 308/317 and 308/321 in vivo. Y308 and D321 reduced dependence on the extracellular loop 2 (ECL2) region of CCR5, while these residues along with Y330, K442, and E444 enhanced dependence on the CCR5 N-terminus compared to clade B consensus residues at these positions. These results suggest that expanded coreceptor usage of HIV-1 can occur in some individuals without rapid progression to AIDS as a consequence of changes in the V3 region that reduce dependence on the ECL2 region of CCR5 by enhancing interactions with conserved structural elements in G-protein-coupled receptors.
Keywords: HIV-1, CCR5Δ32, Env, V3, CCR5
Introduction
Human immunodeficiency virus type 1 (HIV-1) enters cells via interaction of the viral envelope glycoproteins (Env) with CD4 and a coreceptor. Macrophage (M)-tropic HIV-1 viruses primarily use CCR5 (R5) as a coreceptor, whereas T-cell line-tropic HIV-1 viruses use CXCR4 (X4) (Berger, Murphy, and Farber, 1999; Doms and Trono, 2000; Moore et al., 2004). Dual-tropic viruses (R5X4) use both coreceptors. A subset of viruses can also use alternative coreceptors including CCR3, CCR2b, CCR8, Apj, Strl33 (BONZO/CXCR6), Gpr1, Gpr15 (BOB), CX3CR1, ChemR23, or RDC1 for entry (Berger, Murphy, and Farber, 1999; Doms and Trono, 2000; Moore et al., 2004). However, usage of coreceptors other than CCR5 and CXCR4 by primary viruses in vitro is rare (Zhang et al., 1998), and infection of primary cells occurs, with few exceptions, exclusively via CCR5 or CXCR4 (Cilliers et al., 2005; Moore et al., 2004). R5 strains predominate during primary infection and the asymptomatic phase, whereas expansion of viral coreceptor usage and emergence of X4 or R5X4 strains is frequently associated with rapid disease progression.
Delayed or slow HIV-1 disease progression can be defined by lack of development of an AIDS defining illness for at least 10 years after infection with a slowly declining CD4+ T-cell count. Viral genetic factors associated with slow progression or nonprogression include mutations in the HIV-1 gag, rev, vif, vpr, vpu, env and nef genes (Churchill et al., 2004; Churchill et al., 2006; Deacon et al., 1995; Kirchhoff et al., 1995; Michael et al., 1997; Shioda et al., 1997; Wang et al., 2000). Host genetic factors linked to a delay in the onset of AIDS and prolonged survival include the CCR5 Δ32 mutation, CCR2b-V64I polymorphism, and certain HLA haplotypes (Dean et al., 1996; Eugen-Olsen et al., 1997; Huang et al., 1996; Smith et al., 1997) (reviewed in (O'Brien and Moore, 2000; Roger, 1998))). The CCR5 Δ32 mutation, which results in a 32-nucleotide deletion, is common in Caucasians, with heterozygosity in 15 to 20% and homozygosity in 1%. Individuals homozygous for the CCR5 Δ32 allele are highly resistant to HIV-1 transmission (O'Brien and Moore, 2000), whereas heterozygotes are susceptible but typically have delayed CD4+ T-cell decline and prolonged survival compared to CCR5 wt/wt individuals (Dean et al., 1996; Eugen-Olsen et al., 1997; Huang et al., 1996; Michael et al., 1997). Among CCR5 Δ32/wt heterozygotes, there is large variation in levels of CCR5 expression (Cohen et al., 1997; de Roda Husman et al., 1999). Slow progression of HIV-1 disease has been correlated with reduced levels of CCR5 expression on CD4+ T-lymphocytes and monocytes compared to levels in CCR5 wt/wt individuals (Cohen et al., 1997; de Roda Husman et al., 1999). Nonetheless, there is considerable overlap between CCR5 expression levels in CCR5 Δ32/wt heterozygotes and individuals with the CCR5 wt/wt genotype (de Roda Husman et al., 1999).
In this study, we isolated and characterized HIV-1 from blood of an asymptomatic individual who was heterozygous for the CCR5 Δ32 allele and had reduced levels of CCR5 cell surface expression. In addition to using CCR5 and CXCR4, the virus has highly expanded utilization of alternative coreceptors that is broader than that of any previously described HIV-1 virus. Mutagenesis studies and structural models suggested Y308 and D321 in the V3 region of gp120, and to a lesser extent K442 and E444 in the C4 region, contribute to the broad coreceptor usage of Envs cloned from the viral isolate. Furthermore, studies using mutant CCR5 coreceptors indicated Y308, D321, Y330, K442, and E444 alter dependence on the N-terminal and extracellular loop 2 (ECL2) regions of CCR5. The results suggest that expanded coreceptor usage of HIV-1 can occur in some individuals without rapid progression to AIDS as a consequence of changes in the V3 region that enhance interactions with conserved structural elements in G-protein-coupled receptors (GPCRs).
Results
Clinical history and isolation of HIV-1
The subject is a homosexual male who was infected with HIV-1 via sexual contact and first tested seropositive for HIV-1 in May 1989. As of 2006, the subject remained asymptomatic with no AIDS defining illness. His antiretroviral therapy (ART), plasma HIV-1 RNA levels, and CD4 counts are summarized in Supplementary Table 3. The subject was seropositive for cytomegalovirus, hepatitis A, hepatitis C, and Toxoplasma gondii. Genetic analysis of CCR5 alleles by PCR demonstrated heterozygosity for the CCR5 Δ32 deletion (data not shown). Two-color FACS staining of peripheral blood mononuclear cells (PBMC) collected in October 2003 demonstrated that the mean percentage of CCR5+ cells in the CD4+ T-lymphocyte fraction was 0.9% (n=2, SD=0.08) as compared with 19.3% in healthy HIV-1-negative control subjects (n=7, SD=10.15). HIV-1 was isolated from PBMC collected in August 2000 by coculture with CD8-depleted donor PBMC as described (Gorry et al., 2001). Attempts to isolate HIV-1 from cryopreserved PBMC collected in October 1998 and March 1999 were unsuccessful.
Coreceptor usage
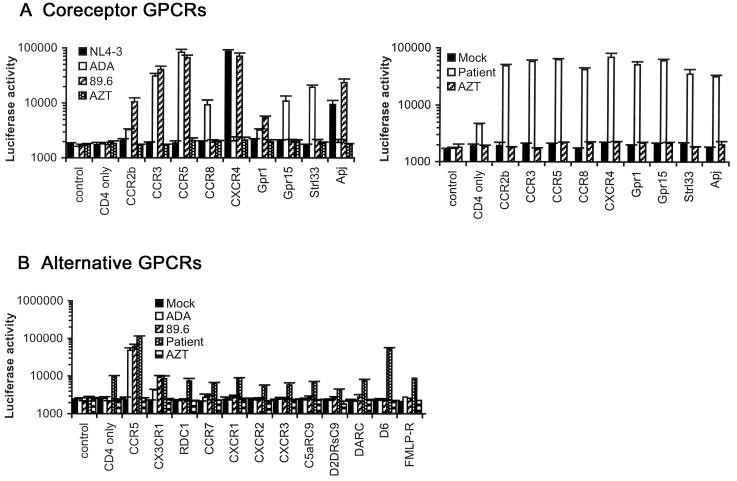
The ability of the primary virus isolate to utilize CCR5, CXCR4, or alternative coreceptors for virus entry was first determined in canine Cf2-Luc cells (Gorry et al., 2001) (Fig. 1A). The X4 NL4-3, R5 ADA, and R5X4 89.6 HIV-1 viruses were used as positive controls (Gorry et al., 2001; Gorry et al., 2002b). The primary virus used both CCR5 and CXCR4 for virus entry. Efficient usage of CCR2b, CCR3, CCR8, Gpr1, Gpr15, Strl33 and Apj was also demonstrated. Compared to 89.6, usage of CCR2b, Gpr1, and Apj by the primary virus was approximately 5-, 20- and 2-fold greater, respectively. Compared to ADA, usage of CCR8, Gpr15 and Strl33 by the primary virus was approximately 5-, 6- and 2-fold greater. The virus could also enter canine Cf2-Luc cells transfected with CD4 alone, albeit at low levels. Coreceptor usage in human U87 astrocytoma cells was similar to that in Cf2-Luc cells with the exception that virus entry was not detected in U87 cells transfected with CD4 alone (data not shown), suggesting that entry in Cf2-Luc cells expressing CD4 alone was mediated by an endogenous canine coreceptor. The promiscuous pattern of coreceptor usage resembles that of some SIV and HIV-2 virus strains (Edinger et al., 1998; Morner et al., 1999; Reeves et al., 1999; Reeves et al., 1997; Rucker et al., 1997), which can infect certain cell types lacking CD4. However, virus entry mediated by CCR3, CCR5, CXCR4, or the endogenous coreceptor expressed on Cf2-Luc cells was strictly CD4-dependent (data not shown). We next tested whether the primary virus could utilize other GPCRs as coreceptors. Virus entry was detected in Cf2-Luc cells coexpressing CD4 and the promiscuous CC-chemokine receptor D6 (Nibbs et al., 1997), but not in cells coexpressing CD4 and CX3CR1, Rdc1, CCR7, CXCR1, CXCR2, CXCR3, complement 5a anaphylatoxin receptor (C5aRC9), dopamine receptor 2-short form (D2DrsC9), duffy antigen/receptor for chemokines (DARC), or formyl-methionine-leucine-phenylalanine receptor (FMLP-R) (Fig. 1B). Cell surface expression of these GPCRs was readily detected (M. Farzan and H. Choe, unpublished observations). Preincubation with zidovudine (AZT) abolished infection (data not shown). The broad and efficient usage of alternative coreceptors by the patient virus was unique by comparison to over 50 other primary HIV-1 viruses analyzed in similar assays ((Gorry et al., 2001; Gorry et al., 2002b; Gray et al., 2005; Lawson et al., 2004; Solomon et al., 2005) and data not shown). Thus, the patient virus demonstrates unusually broad and efficient usage of alternative coreceptors compared to other HIV-1 strains.
Figure 1. Coreceptor usage.
(A) Cf2-Luc cells were transfected with pcDNA3-CD4 alone or cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33 or Apj and infected with equivalent amounts of each control HIV-1 virus (left panel) or patient-derived virus (right panel). Control cells were transfected with pcDNA3 plasmid only. Mock-infected cells were treated with culture medium. Cell lysates were prepared at 48 h post-infection and assayed for luciferase activity. (B) Cf2-Luc cells transfected with pcDNA3-CD4 alone or cotransfected with pcDNA3-CD4 and plasmids expressing CCR5, CX3CR1, RDC1, CCR7, CXCR1, CXCR2, CXCR3, C5aRC9, D2DRsC9, DARC, D6 or FMLP-R were infected with equivalent amounts of each HIV-1 virus. HIV-1 entry was measured as above. Data are represented as means from duplicate infections. Error bars represent standard deviations. Similar results were obtained in two independent experiments.
Replication kinetics
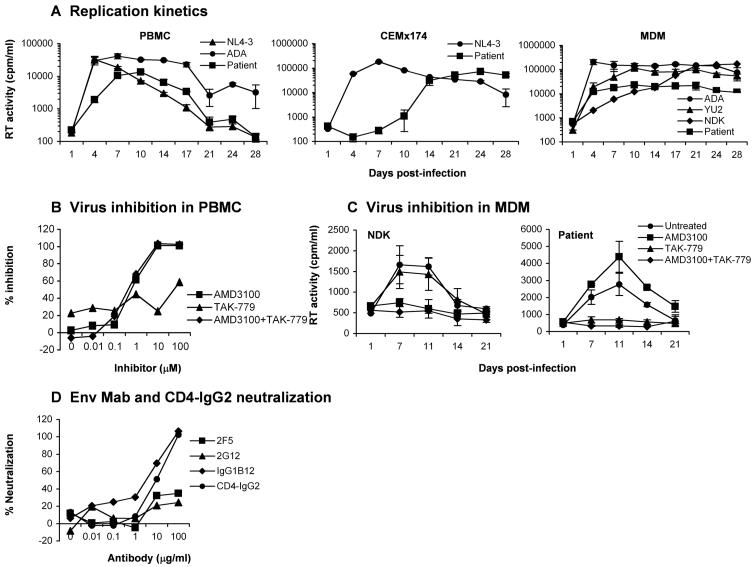
We examined the capacity of the primary virus to replicate in PBMC, CEMx174 cells, and monocyte-derived macrophages (MDM) (Fig. 2A). The ADA and NL4-3 viruses were used as positive controls for replication in PBMC; NL4-3 was used as a positive control for replication in CEMx174 cells; ADA, the R5 YU2 strain, and the X4 NDK strain (Ancuta et al., 2001; Gorry et al., 2001) were used as positive controls for replication in MDM. In PBMC, the primary virus replicated to low levels compared to NL4-3 and ADA, reaching peak levels of replication at day 10 post-infection compared to days 4 and 7 for NL4-3 and ADA, respectively. In CEMx174 cells, the primary virus reached peak levels of virus replication at day 24 post-infection compared to day 7 for NL4-3. In MDM, the primary virus replicated at low levels compared to ADA, YU2 and NDK. These results, together with experiments showing that the same primary virus stock had similar infectivity on Cf2-Luc cells coexpressing CD4 and CCR5 or CXCR4 compared to the control viruses (Fig. 1), suggest that the primary virus has reduced replication capacity in PBMC, CEMx174, and MDM compared to several laboratory and primary HIV-1 strains.
Figure 2. Replication kinetics and sensitivity to coreceptor inhibitors and antibody neutralization.
(A) PBMC, CEMx174 cells and MDM were infected with equivalent amounts of each HIV-1 virus, as described in Materials and Methods, and cultured for 28 days. Virus production in culture supernatants was measured by RT assays. (B) PBMC were treated with concentrations of CCR5 (TAK-779) or CXCR4 inhibitors (AMD3100), or both inhibitors increasing 10-fold from 0.01 to 100 μM, and infected with the patient virus in the presence of each concentration of inhibitor. Virus production in culture supernatants at day 14 post-infection was measured by quantitation of soluble HIV-1 p24 Ag. Production of p24 Ag was calculated as a percentage of the amount produced in the absence of inhibitors, and then expressed as the percentage of inhibition relative to cultures containing no inhibitor. (C) MDM were treated with TAK-779 (100 nM), AMD3100 (1.2 μM) or both for 1 h prior to infection. Untreated cells contained no inhibitor. Cells were infected with equivalent amounts of HIV-1 NDK or the patient isolate and cultured for 21 days in the presence of each inhibitor. HIV-1 production in culture supernatants was measured by RT assays. (D) Virus stocks were treated with concentrations of MAbs or CD4-IgG2 increasing 10-fold from 0.01 to 100 μM for 30 min, and used to infect PBMC. Virus production in culture supernatants and calculation of percent neutralization was determined as per (B). Values shown are means from duplicate infections. Error bars represent standard deviations (A, C). Results are representative of two independent experiments using cells obtained from different donors.
Sensitivity to CCR5 and CXCR4 inhibitors
The efficient usage of alternative coreceptors in transfected cells raised the possibility that infection of primary CD4+ cells by the primary virus might be mediated by alternative coreceptors. Therefore, sensitivity to small molecule inhibitors of CCR5 or CXCR4 (TAK-779 and AMD3100, respectively) was tested in PBMC and MDM. We first determined sensitivity to AMD3100 and TAK-779 in PBMC at a range of concentrations previously shown to be effective at inhibiting infection by other primary HIV-1 viruses (Gorry et al., 2002a). AMD3100 or AMD3100 and TAK-779 in combination abolished infection of PBMC at day 14 post-infection, whereas treatment with TAK-779 alone had only a modest inhibitory effect (Fig. 2B). Similar results were obtained at days 7 and 10 post-infection (data not shown). The 50% inhibitory concentration (IC50) and IC90 of AMD3100 at day 14 post-infection was 0.65 μM and 5.5 μM, respectively, whereas the IC50 and IC90 of TAK-779 at day 14 was 58 μM and >100 μM, respectively. PBMC from a donor homozygous for the CCR5 Δ32 allele supported replication of the primary virus, which was completely abolished by AMD3100 (data not shown). These results demonstrate that the virus primarily uses CXCR4 for infection of PBMC. We then determined sensitivity to AMD3100 and TAK-779 in MDM at concentrations previously shown to completely inhibit infection of MDM or microglia (Gorry et al., 2001; Simmons et al., 1998) (Fig. 2C). AMD3100 abolished infection of MDM by NDK, but had no inhibitory effect on the primary virus. TAK-779 abolished infection of MDM by the primary virus, but had no inhibitory effect on NDK. AMD3100 and TAK-779 in combination abolished infection of MDM by either virus. These results demonstrate exclusive use of CCR5 for infection of MDM. Thus, despite broad coreceptor utilization in transfected cells, infection of PBMC and MDM was mediated only by CXCR4 and CCR5, respectively.
We previously showed that primary human adult astrocytes transduced with CD4 via an adenovirus vector and primary brain microvascular endothelial cells (BMVECs) that express low levels of CD4 can support infection by a subset of HIV-1, HIV-2 and SIV isolates in a CCR5- and CXCR4-independent manner (Willey et al., 2003). To further investigate whether the primary virus can use alternative coreceptors for virus entry in primary human cells, we infected these cells with the primary virus, as previously described (Willey et al., 2003). No evidence of infection of CD4-expressing astrocytes or BMVECs by the primary virus was observed, whereas both cell types supported productive virus replication by the R5X4 HIV-1 viruses GUN-1v and HAN-2, the HIV-2 strain TER, and the SIV strain 17Efr (data not shown). These studies provide further evidence that infection of primary cells by the virus is unlikely to be mediated by alternative coreceptors.
Sensitivity to antibody neutralization
We next measured the sensitivity of the primary virus to neutralization by Env monoclonal antibodies (MAbs) and CD4-IgG2, as previously described (Trkola et al., 1995) (Fig. 2D). The neutralizing reagents were human MAbs 2F5 (Muster et al., 1994; Trkola et al., 1995), 2G12 (Trkola et al., 1995; Trkola et al., 1996), IgG1b12 (Burton et al., 1991; Burton et al., 1994), and the tetrameric CD4-IgG2 molecule (Allaway et al., 1995). The virus was moderately sensitive to neutralization by IgG1b12 and CD4-IgG2, but showed a high level of resistance to neutralization by 2F5 and 2G12, similar to that of 2 other primary viruses studied in parallel (Fig. 2D and data not shown). Thus, the primary virus is relatively resistant to neutralization by Env MAbs and CD4-IgG2, similar to the majority of primary HIV-1 viruses.
Neutralization of heterologous viruses by the subject's plasma
We determined the ability of the subject's plasma (obtained in September, 2001; see Supplementary Table 3) to inhibit infection of PBMC by 6 clade B and 2 clade A HIV-1 viruses as compared with HIVIG and MAbs IgG1b12, 2F5, and 2G12, as previously described (Mascola et al., 2002). The IC50s and IC90s are summarized in Table 1. The subject's plasma and HIVIG neutralized infection by 6 diverse clade B and 1 of 2 clade A HIV-1 viruses at plasma dilutions increasing 5-fold from 1:5 to 1:625, or HIVIG concentrations increasing 10-fold from 10 μg/ml to 10 mg/ml (Mascola et al., 2002). In contrast, the MAbs IgG1b12, 2F5 and 2G12, at concentrations increasing from 0.05 μg/ml to 50 μg/ml, neutralized infection by a subset of the viruses tested. Compared to plasma and sera from other HIV-1-infected individuals, the potency and breadth of the cross-neutralizing activity of the patient's plasma was in the top 10% of HIV-1-positive plasma/sera (J. Mascola, unpublished data). Together, these studies suggest that the subject's plasma has high levels of cross-neutralizing antibodies.
Table 1.
Virus neutralization studies
| Plasma or Ab |
Clade B HIV-1 |
Clade A HIV-1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SF162 | BaL | 89.6 | dBR07 | aBL01 | 6101 | UG031 | RW020 | ||
| Patienta | IC50 | 255 | 220 | 261 | 553 | 90 | 105 | 187 | 32 |
| IC90 | 42 | 31 | 64 | 256 | 18 | 27 | 10 | 7 | |
| b12b | IC50 | 0.4 | 1.1 | 0.3 | 0.6 | 12 | >50 | >50 | >50 |
| IC90 | 2.7 | 12 | 5.5 | 22 | >50 | >50 | >50 | >50 | |
| 2F5 | IC50 | 2.9 | 1.1 | 0.1 | 1.1 | 3.1 | 50 | 9 | 0.3 |
| IC90 | >50 | >50 | 2 | >50 | >50 | >50 | >50 | 4 | |
| 2G12 | IC50 | 17 | 0.1 | >50 | >50 | >50 | 29 | >50 | 0.6 |
| IC90 | >50 | 10 | >50 | >50 | >50 | >50 | >50 | 5 | |
| HIVIG | IC50 | 40 | 320 | 13 | 140 | 685 | 850 | 5420 | 2004 |
| IC90 | 680 | 6590 | 430 | 2610 | >10000 | 8700 | >10000 | 6050 | |
IC50 and IC90 values of patient plasma neutralization are expressed as the reciprocal of plasma dilution required to inhibit 50 or 90%, respectively, of viral infection of CD8-depleted PBMC. The patient plasma was obtained in September, 2001 (see Supplementary Table 3).
IC50 and IC90 values of Env MAbs and HIVIG are expressed as the concentration (μg/ml) required to inhibit 50 or 90%, respectively, of viral infection of CD8-depleted PBMC.
Characterization of full-length gp160 Env clones
The gp160 coding region of HIV-1 Env was cloned into the pCR3.1-Uni expression vector. Full-length, functional Env clones were identified by Western blot analysis of gp120/gp160 in transfected 293T cells and by fusion assays. Four Env clones that express distinct gp160 and gp120 proteins detected by Western blot analysis of transfected 293T cells (Fig. 4A) were sequenced (Fig. 3). Env gp160 amino acid sequences were uninterrupted in Env clones 6, 16 and 30, but contained a premature truncation of 4 amino acids in the gp41 cytoplasmic region in clone 12. The net charge of the V3 variable loops was +5 in all 4 Env clones. The Env clones contained an asparagine-rich insertion of 10 amino acids at positions 131 to 140 in the V1 variable region, which is unique among 207 clade B Envs screened in the Los Alamos database. Two similar asparagine-rich insertions of 11 and 6 amino acids were identified at positions 192 to 202 and 208 to 213, respectively, in the V2 variable region. These insertions result in a net gain of one potential N-linked glycosylation site in V1, and 3 potential N-linked glycosylation sites in V2. The total number of potential N-linked glycosylation sites in gp120 was 20 to 21 compared to 25 in the clade B consensus sequence.
Figure 4. Expression and function of full-length Env clones in cell-cell fusion and infection assays.
(A) 293T cells were cotransfected with 15 μg pCR3.1 Env-expressing plasmid (Envs 6, 12, 16 and 30) or pSVIII Env-expressing plasmid (ADA, HXB2 and 89.6 Envs) and 2 μg pLTR-Tat. At 72 h post-transfection, cell lysates were analyzed by Western blotting using rabbit anti-gp120. The positions of gp160 and gp120 are indicated on the right. (B) 293T effector cells transfected with Env and Tat as above were mixed with Cf2-Luc cells transfected with pcDNA3-CD4 only or cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33, Apj or D6 and incubated at 37°C for 12 h. Control 293T cells were transfected with ΔKS Env. Mock-transfected Cf2-Luc cells were transfected with pcDNA3 only. Cell lysates were then prepared and assayed for luciferase activity. (C) HIV-1 luciferase reporter viruses pseudotyped with each Env were generated and used to infect Cf2th cells transfected with pcDNA3-CD4 only or cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33, Apj or D6. Control virus was produced by pseudotyping with a non-functional Env (ΔKS Env). Cell lysates were prepared at 60 h post-infection and assayed for luciferase activity. Data are represented as means from duplicate wells in one experiment. Error bars represent standard deviations. Results are representative of two independent experiments, each performed in duplicate.
Figure 3. Env amino acid sequences.
Amino acid sequences were obtained from gp160 env genes cloned from genomic DNA of PBMC infected with the primary virus as described in Materials and Methods. Amino acid alignments of Envs from the patient virus are compared to the clade B consensus sequence. Dots indicate residues identical to the clade B consensus and dashes indicate gaps.
To determine whether the Env clones are representative of the predominant HIV-1 variants in the viral quasispecies, fusion and single round entry assays were performed (Fig. 4B, C). 293T cells expressing each Env (Fig. 4A) were fused with Cf2-Luc cells cotransfected with CD4 and a coreceptor as described (Gorry et al., 2002a) (Fig. 4B). 293T cells expressing a non-functional Env or expressing the ADA, HXB2 or 89.6 Env were used as negative and positive controls, respectively. The primary Env clones functioned in fusion assays with Cf2-Luc cells coexpressing CD4 and CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33, Apj or D6. A low level of fusion was also observed with Cf2-Luc cells expressing CD4 only. We performed single round entry assays in Cf2th cells expressing CD4 only or coexpressing CD4 and each of the coreceptors (Fig. 4C). HIV-1 pseudotyped with Env clones 6 and 30 entered cells expressing CD4 alone, or coexpressing CD4 and CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33, Apj or D6. In contrast, HIV-1 pseudotyped with Env clones 12 and 16 entered cells coexpressing CD4 and CCR2b, CCR3, CCR5, CXCR4, Gpr15, Strl33 or Apj, but not cells expressing CD4 alone or coexpressing CD4 and CCR8, Gpr1 or D6. The levels of virus entry mediated by Env clones 12 and 16 were lower than those mediated by Env clones 6 and 30. Together, these results demonstrate broad coreceptor usage by 4 Env clones in fusion and single round entry assays, similar to the pattern of coreceptor usage of the primary virus isolate (Fig. 1).
Analysis of Env determinants contributing to broad coreceptor usage
To investigate mechanisms underlying the broad coreceptor usage of these primary Envs, we analysed sequences of the V3 region, which modulates coreceptor usage (reviewed in (Hartley et al., 2005)), and the conserved coreceptor binding site on the Env core (Rizzuto et al., 1998) to identify amino acid variants that might influence Env-GPCR interactions. In the V3 region, patient Envs had Y308, a rare amino acid variant (1.5% of Clade B Envs, n=23,470) at a position implicated in modulating resistance to neutralizing antibodies (Zhang et al., 2002) and a small molecule CCR5 inhibitor (Kuhmann et al., 2004); I317, an amino acid that contributed to the ability of JR-CSF to use an N-terminal deletion mutant of CCR5 (Platt et al., 2001); D321 (or E321) and Y330, amino acid changes that affect the overall charge of V3 and thus may alter interactions with the tyrosine-sulfated N-terminal region of GPCRs; and T332, a change that results in the loss of a potential N-linked glycosylation site relative to the Clade B consensus (Fig. 5A). In the conserved coreceptor binding site on the Env core, patient Envs had K442 and E444 in the β22 strand near the base of V3, amino acid variants that could potentially affect electrostatic interactions at the Env-GPCR interface.
Figure 5. Analysis of Env determinants contributing to broad coreceptor usage.
(A) V3 and C4 amino acid sequences of the Clade B consensus and patient Env clones were aligned with Clustal X. Dots represent amino acids identical to the Clade B consensus sequence. Residues important for CCR5 binding (Rizzuto et al., 1998) in the C4 region are indicated. Amino acid variants analyzed in mutagenesis studies are highlighted in gray. (B) HIV-1 luciferase reporter viruses pseudotyped with each Env were generated and used to infect Cf2th cells cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR5, CXCR4, Gpr1, Gpr15, Strl33, or D6. Control virus was produced by pseudotyping with ΔKS Env. Cell lysates were prepared at 60 h post-infection and assayed for luciferase activity. Data were normalized to parental Env-30 activity and are represented as means from two to three independent experiments, each performed in duplicate. Error bars represent standard error of the mean. *, p<0.05; **, p<0.01, student's t test. inset, 293T cells were cotransfected with 15 μg Env-expressing plasmids and 2 μg pLTR-Tat. At 72 h post-transfection, cell lysates were analyzed by Western blotting using goat anti-gp120. The positions of gp160 and gp120 are indicated on the right.
To investigate the contribution of these amino acid variants to broad coreceptor usage, we used site-directed mutagenesis to change each of these amino acids to the Clade B consensus residue in patient Env-30. The parental and mutant Envs were expressed at similar levels as determined by Western blotting (Fig. 5B, inset). The ability of Envs to use CCR5, CXCR4, and alternate coreceptors Gpr1, Gpr15, Strl33, and D6 was analyzed in cell-to-cell fusion and single round virus entry assays (Fig. 5B and data not shown). The Y308H mutation resulted in an increased ability to use CCR5, and to a lesser extent CXCR4, but reduced the capacity of Env-30 to use Gpr1, Gpr15, Strl33, and D6 (p=0.001, 0.02, 0.02, and 0.04, respectively; student's t test). Additionally, a D321G mutation in Env-30 resulted in an increased utilization of CCR5, CXCR4, and D6 (p=0.002, 0.049, and 0.006, respectively), but decreased utilization of Gpr1 and Gpr15 (p=0.006 and 0.048, respectively). In contrast, a I317F mutation drastically reduced the capacity of Env-30 to use all coreceptors tested (p<0.01). A K442Q mutation resulted in a significant decrease in utilization of Gpr1 and Strl33 (p=0.001 and 0.01, respectively) and a minor decrease in utilization of D6 that did not reach statistical significance, while a E444R change reduced utilization of CXCR4 and Gpr15 (p<0.01). A Y330H mutation reduced the ability of Env-30 to use Gpr1 (p=0.04), but enhanced utilization of Gpr15 (p=0.01) and had no significant effect on utilization of Strl33 and D6. A T332N mutation had no significant effect on entry efficiencies and patterns of coreceptor usage compared to the parental Env-30 (Fig. 5B). These results suggest that Y308, D321, and to a lesser extent K442 and E444, contribute to enhanced gp120 interactions with alternate coreceptors, and raise the possibility that I317 may be a compensatory change in patient Env-30.
Molecular modeling of V3 determinants
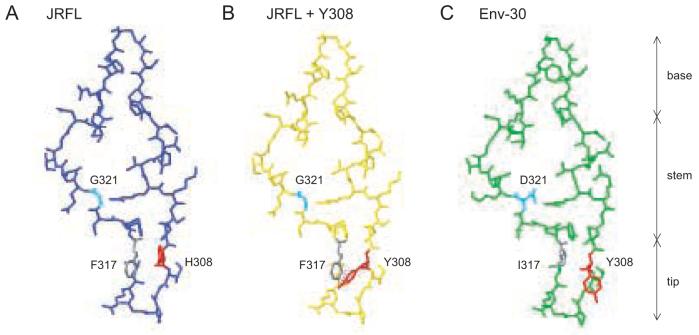
To elucidate a mechanism for enhanced gp120-coreceptor interactions in Env-30, we used Swiss PDB viewer to model the amino acids at positions 308, 317, and 321 on the JRFL gp120-CD4-X5 CD4i Ab crystal structure (2B4C) (Huang et al., 2005). The Clade B consensus amino acids at positions 308 and 317 in the V3 tip region are His and Phe, respectively (Fig. 6A). If the His at 308 is changed to Tyr, there is an increased potential for steric clashing when the aromatic ring of Phe is present at position 317 (Fig. 6B), whereas the smaller side chain of Ile at this position can accommodate the Tyr at 308 (Fig. 6C). This finding suggests that the loss of Env function in the I317F mutant may be due to the lack of a compensatory change at position 308. Consistent with this prediction, when Y308 is present in Clade B Envs (n=360), there is a decreased frequency of amino acids with bulky aromatic side chains such as Phe and Trp, but an increased frequency of amino acids with small hydrophobic chains such as Ile and Val at position 317 compared to Clade B Envs (n=23,470) (Table 2), suggesting that covariation may occur at positions 308/317 in vivo. In Envs with Y308 (n=360 Env sequences from 77 patients), I317 appeared in 23.9% (n=86 Env sequences from 11 patients) compared to 4.1% (n=959) of Clade B Envs in the database (n=23,470) (Table 2). At position 321, Gly may increase the flexibility of the V3 stem, allowing increased gp120 binding to GPCRs. An Asp at this position may decrease the flexibility of the stem, but may also enhance the ability of Env-30 to interact with some GPCRs that contain positive charges in the N-terminus, such as Gpr1 and Gpr15. In Envs with Y308, there is an increased frequency of charged amino acids at position 321 compared to Clade B Envs, raising the possibility that covariation may also occur at 308/321 (Table 2).
Figure 6. Structural modeling of Env V3 determinants contributing to broad coreceptor usage.
Swiss PDB viewer was used to model amino acid changes at positions 308, 317, and 321 on the V3 region of the JRFL gp120-CD4-X5 CD4i Ab crystal structure (2B4C) (Huang et al., 2005). JRFL V3 has a single amino acid change relative to the clade B consensus (N301Q). Red, position 308. Gray, position 317. Cyan, position 321. (A) V3 region of JRFL with H308, F317, and G321. (B) A Y308 change in JRFL results in steric clashing with F317 (dotted lines). (C) Changes Q301N, K305R, S306G, Y308H, F317I, G321D, E322K, I324V, Q328K, H330Y, and N332T were introduced in JRFL to produce a model of the V3 region of Env-30. Positions Y308, I317, and D321 are indicated.
Table 2.
Frequency and co-variation of unusual amino acid variants in vivo.
| Clade B Envs (n=23470)a | Envs with Y308 (n=360) | |||||
|---|---|---|---|---|---|---|
| Position | Amino Acid | Frequency (%)b | n | Frequency (%) | n | Significancec |
| 317 | F | 78.6 | 18446 | 23.6 | 85 | ** |
| W | 6.4 | 1500 | 0.6 | 2 | ** | |
| L | 5.1 | 1194 | 5.0 | 18 | ||
| I | 4.1 | 959 | 23.9 | 86d | ** | |
| V | 2.7 | 644 | 45.3 | 163 | ** | |
| Y | 1.8 | 411 | 0.0 | 0 | ** | |
| other | 1.3 | 316e | 1.7 | 6f | ||
| 321 | G | 79.8 | 18740 | 36.7 | 132 | ** |
| E | 4.1 | 970 | 19.7 | 71 | ** | |
| D | 2.8 | 667 | 7.5 | 27 | ** | |
| K | 2.0 | 460 | 26.9 | 97 | ** | |
| R | 1.7 | 389 | 8.1 | 29 | ** | |
| other | 9.6 | 2244g | 1.1 | 4h | ||
V3 sequence alignment was obtained from the Los Alamos database in October 2005. Sequence data for Envs with Y308 is from 77 patients.
Frequency was calculated as [(n/total number of sequences)×100].
Differences between groups were significant as calculated by Fisher's exact test.
p<0.005.
Envs with Y308/I317 are from 11 patients
Other indicates amino acids M, S, K, R, C, Q, H, P, T, G, A, D, and E (in descending order of frequency), or could not be determined.
Other indicates amino acids M, S, K, E, and P (in descending order of frequency).
Other indicates amino acids T, A, Q, N, S, I, and V (in descending order of frequency), a gap in the alignment, or could not be determined.
Other indicates amino acid Q or could not be determined.
Analysis of Env interactions with attenuated CCR5 coreceptors
GPCRs are 7-transmembrane receptors with an extracellular, post-translationally modified acidic N-terminal region and three extracellular loops (ECLs). The N-terminal region and ECL2 contain the major determinants for gp120 binding and coreceptor activity (Dragic et al., 1998; Farzan et al., 1998; Farzan et al., 1999; Kuhmann et al., 1997; Lee et al., 1999; Olson et al., 1999; Rabut et al., 1998; Rucker et al., 1996). The N-terminus of CCR5 has Tyr sulfation modifications at several sites that are important for mediating HIV entry (Farzan et al., 1999). To determine whether amino acid variants in Env-30 influence interactions with the N-terminus or ECL2 regions of GPCRs, Cf2 cells expressing CD4 and either wild-type CCR5 (CCR5(wt)), CCR5(Y14N) which has a point mutation in the N-terminal region that changes a Tyr sulfation site important for mediating HIV-1 entry to an N-linked glycosylation site (Farzan et al., 1999; Kuhmann et al., 2000), CCR5(Δ18) which contains a deletion of the first 18 amino acids of the N-terminal region, or CCR5(G163R) which contains a point mutation in the transmembrane region associated with a conformational change in ECL2 that is detrimental to HIV-1 entry (Platt et al., 2001; Siciliano et al., 1999), were infected with viruses expressing wild-type or mutant Envs. Primary, macrophage-tropic R5 viral isolates use CCR5(Y14N) and CCR5(G163R) at low efficiencies (1.5% and 10-20% compared to CCR5(wt), respectively) (Kuhmann et al., 2000; Platt et al., 2001) and are unable to use CCR5(Δ18)(Platt et al., 2005). In contrast to primary R5 Envs, Env-30 used CCR5(wt) and CCR5(G163R) with similar efficiencies (Fig. 7). Y308H and D321G had reduced utilization of CCR5(G163R) relative to CCR5(wt) compared to Env-30 (Fig. 7), suggesting that amino acid variants at these positions in patient Env-30 reduce dependence on the ECL2 region of CCR5. The Y308H, Y330H, K442Q, and E444R mutants used CCR5(Y14N) for entry more efficiently compared to Env-30 (Fig. 7). None of the Envs could use CCR5(Δ18) efficiently, consistent with the critical role of the N-terminal region of GPCRs in binding and entry (Dragic et al., 1998; Farzan et al., 1998; Farzan et al., 1999; Kuhmann et al., 1997; Rabut et al., 1998; Rucker et al., 1996). However, the Y308H, D321G, T332N, and E444R mutations resulted in an increase in the ability of Env-30 to use CCR5(Δ18), albeit at very low efficiencies. Cell surface expression levels of CD4 and CCR5 on transfected Cf2th cells were similar based on staining with CD4-FITC (RPA-T4) and CCR5-PE (2D7) and FACS analysis (data not shown). Together, these results suggest that Y308 and D321 reduce dependence on the ECL2 region of CCR5, while these residues along with Y330, K442, and E444 increase dependence on the CCR5 N-terminus compared to clade B consensus residues at these positions.
Figure 7. Env interactions with attenuated CCR5 coreceptors.

HIV-1 luciferase reporter viruses pseudotyped with each Env were generated and used to infect Cf2th cells cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR5(Y14N), CCR5(Δ18), or CCR5(G163R). Control virus was produced by pseudotyping with (ΔKS Env. Cell lysates were prepared at 60 h post-infection and assayed for luciferase activity. Results for each mutant CCR5 were normalized to levels of luciferase activity in cells expressing CCR5(wt) for each Env and are represented as means from two independent experiments, each performed in duplicate. Error bars represent standard deviations.
Discussion
In this study, we isolated and characterized an unusual HIV-1 strain from blood of an asymptomatic individual who was heterozygous for the CCR5 Δ32 allele and had reduced levels of CCR5 expression. The primary virus is dual-tropic and exhibits unusually broad and efficient utilization of alternative coreceptors. The repertoire of alternative coreceptors used is greater than that of any previously described HIV-1 virus, and more closely resembles that of some SIV and HIV-2 viruses than that of HIV-1 viruses (Morner et al., 1999; Rucker et al., 1997; Siciliano et al., 1999). However, in contrast to some SIV and HIV-2 strains that can infect cells in the absence of CD4 (Reeves et al., 1999; Reeves et al., 1997), CCR5- and CXCR4-mediated entry by the primary virus is strictly CD4-dependent. Previous studies suggest that expanded use of alternative coreceptors is associated with rapid HIV-1 disease progression (Bjorndal et al., 1997; Connor et al., 1997). However, the virus we described here was isolated from an individual who was asymptomatic for 18 years and had slow disease progression, suggesting that expanded tropism of HIV-1 can occur in some individuals who do not have rapid disease progression.
Despite efficient usage of many alternative coreceptors in transfected cells, the virus exclusively used CXCR4 or CCR5 to enter primary T cells and monocyte-derived macrophages, respectively. A similar pattern of coreceptor preference for infection of these primary cells was described for the R5X4 89.6 isolate. CD4-expressing astrocytes and BMVECS were not infected by the primary virus, whereas both cell types supported infection by other unusual HIV-1 and HIV-2 strains. Thus, infection of primary cells was mediated only by CXCR4 and CCR5. In addition to using CCR2b, CCR3, CCR5, CCR8, CXCR4, Gpr1, Gpr15, Strl33 and Apj, the patient virus also utilized the promiscuous CC-chemokine receptor D6 (Nibbs et al., 1997). Only two previous studies demonstrated HIV-1 entry mediated via D6 (Choe et al., 1998; Neil et al., 2005). D6 is expressed on lymphatic endothelial cells, but not on peripheral blood cells or vascular endothelial cells (Nibbs et al., 2001). In addition to entry mediated by D6, entry mediated by an endogenous canine coreceptor was demonstrated, albeit at very low levels. This property was not observed for over 50 other primary HIV-1 isolates tested in similar assays (data not shown). The identity of the endogenous canine coreceptor is unknown. However, entry via this coreceptor was not inhibited by AMD3100, suggesting it most likely is not CXCR4 (data not shown).
CCR5 cell surface expression levels were very low on the study subject's PBMC compared to other CCR5 Δ32 heterozygotes (Cohen et al., 1997; de Roda Husman et al., 1999). Previous studies demonstrated an increased frequency of R5X4 viruses in CCR5 Δ32 heterozygotes compared to CCR5 wt/wt individuals (Lathey et al., 2001; Zhang et al., 1998). We speculate that structural features that enhance interaction of the primary virus Env described herein with conserved structural elements in GPCRs may have resulted from adaptive evolution in the setting of very low levels of CCR5 cell surface expression. However, validation of this hypothesis would require longitudinal analysis of Env sequences in the patient's plasma and their functional characterization. The primary virus did not show enhanced sensitivity to antibody neutralization, but the subject's plasma had potent neutralizing activity against a wide variety of HIV-1 strains. This finding raises the possibility that the Env from the subject's virus may express epitopes that are immunogenic and neutralization functional, and may be responsible for at least some of the cross-reactive neutralizing activity of his plasma. A better understanding of these epitopes and their relationship to expanded coreceptor usage may provide insights relevant for improving HIV-1 immunogenicity and the elicitation of anti-HIV-1 neutralizing antibodies.
The broad usage of alternate coreceptors suggests that the Env of this primary virus has unique structural features that enhance interaction with conserved structural elements in GPCRs. In the V3 region, patient Envs had the unusual amino acid variants Y308 (present in 1.5% of Clade B Envs, n=23,470), I317 (4.1%), and D321 (2.8%). Y308H and D321G mutations increased the capacity of Env-30 to use CCR5 and to a lesser extent CXCR4, but decreased utilization of alternate coreceptors, and an I317F mutation drastically decreased the capacity of the Env to use any coreceptor. Structural modeling suggests that a Y308 change in the JRFL crystal structure results in steric clashing with F317 (the clade B consensus residue), whereas the smaller side chain of Ile at this position is accommodated. Thus, I317 is likely to be a compensatory change for Y308. D321 may enhance electrostatic interactions with the positively charged N-terminus of CCR5 and other GPCRs. These observations, together with evidence for covariation of amino acids at positions 308/317 and 308/321 in vivo based on database analysis, suggests that Y308, I317, and D321 may act cooperatively to enhance Env-GPCR interactions. Env-30 used CCR5(G163R), a mutant CCR5 which does not affect entry of SIVmac251 but reduces entry of primary macrophage tropic isolates such as ADA and SF162 (Kuhmann et al., 1997), for entry at levels comparable to those mediated by CCR5(wt). This finding is consistent with the promiscuous coreceptor usage of the patient isolate and Env-30, which resembles that of SIV or HIV-2 {Edinger, 1998 #89; Morner et al., 1999; Reeves et al., 1999; Reeves et al., 1997; Rucker et al., 1997). In addition to enhancing utilization of alternative coreceptors, Y308 and D321 reduced the dependence of Env-30 on the ECL2 region of CCR5, while introducing the clade B consensus residues H308, H330, K442, and E444 resulted in reduced dependence on the CCR5 N-terminus. These findings are consistent with a proposed model in which the tip of V3 interacts with ECL2 of GPCRs, while the more conserved stem and base of V3 interact with the N-terminal region (Huang et al., 2005; Xiang et al., 2005). K442 and E444 in the C4 region may work in concert with the changes in V3 by facilitating interactions with the highly acidic N-terminal region of CCR5 and other GPCRs. Changes in other regions of Env, such as those in the V1/V2 region that cooperatively modulate coreceptor usage with V3 (Nabatov et al., 2004; Pastore et al., 2006; Sullivan et al., 1993), may further enhance Env-coreceptor interactions. Based on our results, we propose that changes in the V3 region of Env-30 cooperatively enhance interactions with conserved structural elements in GPCRs, while changes in the base of V3 and the C4 region may enhance electrostatic interactions with the N-terminus of GPCRs.
Analysis of patient Env sequences revealed a unique insertion of 10 amino acids in the V1 region, and two asparagine-rich insertions of 11 and 6 amino acids in the V2 region. Similar extensions in the V2 region have been associated with slow HIV-1 progression or nonprogression (Shioda et al., 1997; Wang et al., 2000). The V1 insertion in this subject is unusual compared to other clade B Envs, and the V1/V2 region is longer than that of other Envs in the database. Changes in Env that reposition the V1/V2 loops can increase exposure of the coreceptor binding site (Kolchinsky et al., 2001). These findings raise the possibility that the unusually long V1/V2 region may reposition the V1/V2 loops, resulting in a conformation that enhances interaction of gp120 with conserved elements in GPCRs. Conserved elements in GPCRs important for HIV-1 entry include the tyrosine-rich sulfated regions in the N-terminus, which are important for CCR5-mediated HIV-1 entry (Farzan et al., 1999). GPCRs that mediate entry by the primary virus have many tyrosine residues within the N-terminus that are probably sulfated in close proximity, similar to the pattern of sulfated tyrosine residues in CCR5. Thus, the tyrosine-rich region in the N-terminus is likely to be one of the conserved elements in GPCRs involved in HIV-1 binding and virus entry.
The subject's slow disease progression despite harboring a dual-tropic HIV-1 strain with highly expanded coreceptor usage may reflect additional viral and/or host factors that can influence HIV-1 progression. The primary virus had attenuated replication kinetics in PBMC, CEMx174, and MDM compared to several laboratory strains (Fig. 3A) and other primary viruses (Gorry et al., 2001). However, we did not determine whether the virus studied here was one of the majority sequences present in the patient. Mutations in the nef gene can cause viral attenuation and reduced pathogenicity of HIV-1 strains (Deacon et al., 1995; Kirchhoff et al., 1995). However, we found no deletions or other obvious defects in nef genes cloned from the primary virus, which were fully functional for CD4 and MHC-I downregulation (K. Agopian and D. Gabuzda, unpublished data). Further studies are required to determine whether the primary virus contains any mutations in other viral genes that might be associated with slow disease progression, and whether it is a major variant in the patient viral quasispecies in vivo.
In summary, we describe an unusual dual-tropic HIV-1 strain with highly expanded coreceptor usage isolated from an asymptomatic individual who was heterozygous for the CCR5 Δ32 allele and had low CCR5 cell surface expression. In this subject, highly expanded coreceptor usage of HIV-1 occurred without progression to AIDS, suggesting that R5X4 HIV-1 strains with broadened coreceptor usage can be harbored by some individuals without rapid disease progression. Our results suggest that changes in the V3 and C4 regions, possibly a consequence of adaptive evolution in the setting of very low levels of CCR5 expression and viral escape from neutralizing antibodies, act cooperatively to broaden coreceptor usage by enhancing interactions with conserved structural elements in GPCRs. These results lead to a better understanding of HIV Env-GPCR interactions and provide insights that may facilitate development of vaccines and therapeutics that target virus entry.
Materials and Methods
Cells
Peripheral blood mononuclear cells were purified from blood of healthy HIV-1-negative donors by Ficoll-Hypaque density gradient centrifugation, stimulated with 2 μg/ml phytohemagglutinin (PHA) for 3 days and cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 20 units/ml interleukin-2 (IL-2; Boehringer Mannheim, Germany). CD8+ T-cells were depleted by magnetic separation with anti-CD8-conjugated magnetic beads (Miltenyi Biotech, Auburn, CA). Monocyte-derived macrophages were purified from PBMC by plastic adherence and cultured for 5 days in RPMI 1640 medium supplemented with 10% (v/v) human AB+ serum and 12.5 ng/ml M-CSF. Cf2-Luc cells (Etemad-Moghadam et al., 2000), derived from the Cf2th canine thymocyte cell line (Choe et al., 1996), stably express the luciferase gene under the control of the HIV-1 LTR. Cf2-Luc cells were cultured in DMEM medium supplemented with 10% (v/v) FBS, and 0.7 mg/ml G418. Cf2th cells were cultured in the same medium without G418. CEMx174 cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS.
Isolation of HIV-1
HIV-1 was isolated from patient PBMC by coculture with CD8-depleted donor PBMC as described previously (Gorry et al., 2001). Briefly, 2 × 106 patient cells were added to 5 × 106 CD8-depleted PBMC from a normal uninfected donor, incubated at 37°C for 1 h, and then cultured in 10 ml growth media containing 20 U/ml IL-2. Fifty percent media changes were performed twice weekly. Five million fresh PHA-activated, CD8-depleted PBMC from a different donor were added at every second media change. Supernatants were tested for reverse transcriptase (RT) activity using [3H]dTTP incorporation. Supernatants testing positive for RT were filtered through 0.45 μm filters and stored at −80°C.
Coreceptor usage
To determine coreceptor usage by the patient HIV-1 isolate, Cf2-Luc cells were co-transfected with 10 μg of plasmid pcDNA3-CD4 and 20 μg of plasmid pcDNA3 containing CCR2b, CCR3, CCR5, CCR8, CXCR4, CX3CR1, Gpr1, Gpr15, Strl33, Apj, CX3CR1, Rdc1, CCR7, CXCR1, CXCR2, CXCR3, C5aRC9, D2DrsC9, DARC, D6 or FMLP-R using the calcium phosphate method, and infected 48 h later by incubation with 10,000 3H cpm RT units of HIV-1 in the presence of 2 μg/ml polybrene as described previously (Gorry et al., 2001). Mock-infected cells were treated with culture medium. Cf2-Luc cells mock-transfected or transfected with pcDNA3-CD4 alone were used as negative controls. After overnight infection, virus was removed and the cells were cultured for an additional 48 h prior to lysis in 200 μl cell lysis buffer. Cell lysates were cleared by centrifugation, and assayed for luciferase activity according to the manufacturers' protocol (Promega).
HIV-1 replication kinetics
Five million PHA-activated PBMC were infected by incubation with 50,000 3H cpm RT units of virus supernatant in a volume of 2 ml for 3 h at 37°C. Virus was then removed and PBMC were washed 3 times with PBS and cultured in media containing 20 U/ml IL-2 for 28 days. Monocyte-derived macrophages were isolated from PBMC by plastic adherence and allowed to mature for 5 days prior to seeding in six-well tissue culture plates at approximately 90% confluence. Virus equivalent to 50,000 3H cpm RT units in a volume of 2 ml was allowed to adsorb to the cell monolayers for 3 h at 37°C. Virus was then removed and cells were rinsed 3 times with PBS prior to addition of 2 ml culture medium. Five million CEMx174 cells were infected by incubation with 50,000 3H cpm RT units of virus supernatant in a volume of 2 ml for 3 h at 37°C. Virus was then removed and cells were washed 3 times with PBS and cultured for 28 days. Fifty percent media changes were performed twice weekly and supernatants were tested for HIV-1 by RT assays.
Virus inhibition studies
The effects of the coreceptor inhibitors TAK-779 (Baba et al., 1999) and AMD-3100 (Donzella et al., 1998; Schols et al., 1997) on virus replication in PBMC were assayed as described elsewhere (Gorry et al., 2002a; Trkola et al., 1998). Briefly, PBMC were incubated for 30 min with a range of concentrations of each inhibitor (0.01 to 100 μM) prior to infection with the patient virus isolate. Virus replication was measured by production of HIV-1 p24 antigen in culture supernatants for 14 days (Trkola et al., 1995). The production of p24 antigen in the presence of an inhibitor was expressed as a percentage of the amount produced in control cultures containing no inhibitor. For virus inhibition studies in MDM, cells were pre-incubated with 100 nM TAK-779 or 1.2 μM AMD3100 for 1 h prior to infection with HIV-1 isolates containing the same concentration of inhibitor, as described previously (Gorry et al., 2001). Infected cells were cultured for 21 days in the presence of each inhibitor. Fifty percent media changes were performed weekly and supernatants were tested for HIV-1 by RT assays.
Neutralization assays
Human monoclonal antibodies (MAb) against HIV-1 gp120 (IgG1b12 and 2G12) and gp41 (2F5), the tetrameric CD4-immunoglobulin (CD4-IgG2) molecule, and purified polyclonal anti-HIV immunoglobulin (HIVIG) have been described previously (Allaway et al., 1995; Burton et al., 1991; Burton et al., 1994; Mascola et al., 2002; Muster et al., 1994; Trkola et al., 1995; Trkola et al., 1996). Neutralization of replication of the patient virus isolate in PBMC was assessed as described previously (Trkola et al., 1995). Briefly, virus was incubated for 30 min with a range of concentrations of each Mab or CD4-Ig2 (0.01 to 100 μg/ml) prior to infection. Virus replication and calculation of percent neutralization was measured as described above. Neutralization of heterologous viruses by the subject's plasma was assessed as described by Mascola et. al. (Mascola et al., 2002).
PCR amplification, HIV-1 Env cloning and sequence analysis
Genomic DNA was extracted from PBMC infected with the patient virus isolate using the DNeasy DNA extraction kit (Qiagen). Full length Env genes were amplified from genomic DNA with RTth XL polymerase and nested primers using hot start AmpliWax PCR Gem 50 (Applied Biosystems), as described previously (Gorry et al., 2002a). Env PCR-product DNA was gel purified and cloned into pCR3.1-Uni (Invitrogen). Functional full-length Env clones were identified by Western blot analysis of gp120/gp160 in transfected 293T cells and by fusion assays. Env clones were sequenced using a model 3100 Genetic Analyzer (Applied Biosystems). Y308H, I317F, D321G, Y330H, T332N, K442Q, and E444R mutant Env plasmids were created by PCR-based mutagenesis and changes were verified by DNA sequencing.
Western blot analysis
For analysis of Env expression, 293T cells were transfected with 15 μg of different pCR3.1 Env clones, or 15 μg pSVIII plasmid expressing ADA, HXB2 or 89.6 Env plus 2 μg pLTR-Tat plasmid. At 72 h after transfection, cells were rinsed twice in PBS and resuspended in 400 μl of ice cold lysis buffer for 20 min, followed by centrifugation at 15,300 × g for 10 min to remove cellular debris. Cell lysates were separated in 8.5% SDS-PAGE gels, and analyzed by Western blotting using rabbit anti-gp120 polyclonal antisera (American Biotechnologies Inc.) or goat anti-gp120 polyclonal antisera (NIH AIDS Research and Reference Reagent program). Env proteins were visualized using horseradish peroxidase-conjugated anti-rabbit or anti-goat immunoglobulin G antibodies and enhanced chemiluminescence (Perkin Elmer).
Fusion assays
293T cells (1×105) cotransfected with 15 μg Env-expressing plasmid and 2 μg pLTR-Tat were mixed with Cf2-Luc cells (1×106) that had been cotransfected with 10 μg pcDNA3-CD4 and 20 μg pcDNA3 expressing an alternative coreceptor as indicated, then incubated at 37°C in 0.75 ml culture medium. Mock-transfected Cf2-Luc cells were transfected with pLTR-Tat only. Control 293T cells were co-transfected with pLTR-Tat and a non-functional Env (pSVIII-ΔKS Env). Twelve hours later, cells were harvested and assayed for luciferase activity as described above
Single round entry assays
An Env complementation assay was used to quantitate HIV-1 entry as described (Choe et al., 1996). Briefly, recombinant HIV-1 luciferase reporter viruses were generated by cotransfection of 293T cells by the calcium phosphate method with 16 μg of pNL4-3env-Luc, which contains an HIV-1 provirus with a deletion in the env gene and a replacement of the nef gene with a luciferase gene, and 6 μg of pCR3.1Env or pSVIIIEnv plasmid. Cf2th cells intended for use as target cells were cotransfected with 10 μg pcDNA3-CD4 and 20 μg pcDNA3 expressing an alternative coreceptor as indicated. Plasmids expressing CCR5 Y14N, CCR5 Δ18, and CCR5 G163R were kindly provided by D. Kabat. Approximately 48 h after transfection, these cells were infected by incubation with 20,000 3H cpm RT units of recombinant luciferase reporter viruses. Reporter viruses pseudotyped with a non-functional Env (pSVIII-ΔKSenv) were used as negative controls. Sixty hours later, cells were harvested and assayed for luciferase activity.
Nucleotide sequence accession numbers
Nucleotide sequences were submitted to GenBank (accession numbers AY624304 through AY624307).
Supplementary Material
Acknowledgements
We are indebted to the subject who provided blood samples and details of his clinical history for this study. We thank J. Sodroski, P. Kwong, D. McPhee, N. Saksena, and J. Wang for helpful discussions, A. Dunne for preparation of clinical samples, and L. Gray for assistance with figures. We are also grateful to J. Sodroski for providing Cf2th and Cf2-Luc cells, J. Sodroski, R. Doms, S. Peiper and D. Kabat for coreceptor plasmids, H. Choe for D2DRsC9 and DARC plasmids, D. Montefiore for HIV-1 6101, and the NIH AIDS Research and Reference Reagent program, Division of AIDS, NIAID, NIH, for primary HIV-1 isolates (contributed by H. Gendelman, J. Levy, S. Gartner, and the UNAIDS Network for HIV Isolation and Characterization, DAIDS, NIAID), TAK-779, AMD3100 (DAIDS, NIAID), antibodies used in neutralization assays (contributed by D. Burton, P. Parren, and H. Katinger), and goat anti-sera used in Western blotting.
This work was supported by NIH NS37277 to D.G. and AI41420 to J.P.M. Core facilities were supported by Center for AIDS Research grants and the DFCI/Harvard Center for Cancer Research grant. S.M.C. was supported by the Australian National Center in HIV Virology Research and by an Australian National Health and Medical Research Council (NHMRC) Principal Research Fellowship. P.R.G. was supported in part by NHMRC 251520 and NIH R21 AI054207. P.C. was supported by NIH MH64408 and amfAR 02802-30-RG. A.M. was supported in part by a NSF fellowship. P.R.G. is a recipient of an NHMRC R. Douglas Wright Biomedical Career Development Award. D.G., J.P.M., and P.C. are Elizabeth Glaser Scientists who were supported by the Pediatric AIDS Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allaway GP, Davis-Bruno KL, Beaudry GA, Garcia EB, Wong EL, Ryder AM, Hasel KW, Gauduin MC, Koup RA, McDougal JS, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 1995;11(5):533–9. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- Ancuta P, Bakri Y, Chomont N, Hocini H, Gabuzda D, Haeffner-Cavaillon N. Opposite effects of IL-10 on the ability of dendritic cells and macrophages to replicate primary CXCR4-dependent HIV-1 strains. J Immunol. 2001;166(6):4244–53. doi: 10.4049/jimmunol.166.6.4244. [DOI] [PubMed] [Google Scholar]
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Procedings of the National Academy of Sciences USA. 1999;96(10):5698–703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. Journal of Virology. 1997;71(10):7478–87. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A. 1991;88(22):10134–7. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf ME, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. Journal of Virology. 1998;72(7):6113–8. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Churchill M, Sterjovski J, Gray L, Cowley D, Chatfield C, Learmont J, Sullivan JS, Crowe SM, Mills J, Brew BJ, Wesselingh SL, McPhee DA, Gorry PR. Longitudinal analysis of nef/long terminal repeat-deleted HIV-1 in blood and cerebrospinal fluid of a long-term survivor who developed HIV-associated dementia. J Infect Dis. 2004;190(12):2181–6. doi: 10.1086/425585. [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL, Cooke IR, Deacon NJ, Gorry PR. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol. 2006;80(2):1047–52. doi: 10.1128/JVI.80.2.1047-1052.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilliers T, Willey S, Sullivan WM, Patience T, Pugach P, Coetzer M, Papathanasopoulos M, Moore JP, Trkola A, Clapham P, Morris L. Use of alternate coreceptors on primary cells by two HIV-1 isolates. Virology. 2005 doi: 10.1016/j.virol.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Cohen OJ, Vaccarezza M, Lam GK, Baird BF, Wildt K, Murphy PM, Zimmerman PA, Nutman TB, Fox CH, Hoover S, Adelsberger J, Baseler M, Arthos J, Davey RT, Jr., Dewar RL, Metcalf J, Schwartzentruber DJ, Orenstein JM, Buchbinder S, Saah AJ, Detels R, Phair J, Rinaldo C, Margolick JB, Pantaleo G, Fauci AS. Heterozygosity for a defective gene for CC chemokine receptor 5 is not the sole determinant for the immunologic and virologic phenotype of HIV-infected long-term nonprogressors. J Clin Invest. 1997;100(6):1581–9. doi: 10.1172/JCI119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1--infected individuals. Journal of Experimental Medicine. 1997;185(4):621–8. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roda Husman AM, van Rij RP, Blaak H, Broersen S, Schuitemaker H. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J Infect Dis. 1999;180(4):1106–15. doi: 10.1086/314987. [DOI] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270(5238):988–91. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien SJ. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Doms RW, Trono D. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 2000;14(21):2677–88. doi: 10.1101/gad.833300. [DOI] [PubMed] [Google Scholar]
- Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, Moore JP. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4(1):72–7. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, Sakmar TP, Moore JP, Maddon PJ. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72(1):279–85. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Hoffman TL, Sharron M, Lee B, Yi Y, Choe W, Kolson DL, Mitrovic B, Zhou Y, Faulds D, Collman RG, Hesselgesser J, Horuk R, Doms RW. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. Journal of Virology. 1998;72(10):7934–40. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74(9):4433–40. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugen-Olsen J, Iversen AK, Garred P, Koppelhus U, Pedersen C, Benfield TL, Sorensen AM, Katzenstein T, Dickmeiss E, Gerstoft J, Skinhoj P, Svejgaard A, Nielsen JO, Hofmann B. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. Aids. 1997;11(3):305–10. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72(2):1160–4. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–76. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75(21):10073–89. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002a;76(12):6277–92. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Zhang C, Wu S, Kunstman K, Trachtenberg E, Phair J, Wolinsky S, Gabuzda D. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5 Delta 32 allele. Lancet. 2002b;359(9320):1832–4. doi: 10.1016/S0140-6736(02)08681-6. [DOI] [PubMed] [Google Scholar]
- Gray L, Sterjovski J, Churchill M, Ellery P, Nasr N, Lewin SR, Crowe SM, Wesselingh S, Cunningham AL, Gorry PR. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology. 2005;337:384–398. doi: 10.1016/j.virol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21(2):171–89. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-Containing HIV-1 gp120 Core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2(11):1240–3. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332(4):228–32. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75(7):3435–43. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71(11):8642–56. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74(15):7005–15. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78(6):2790–807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathey JL, Tierney C, Chang SY, D'Aquila RT, Bettendorf DM, Alexander HC, Santini CD, Hughes AM, Barroga CF, Spector SA, Landes JE, Hammer SM, Katzenstein DA. Associations of CCR5, CCR2, and stromal cell-derived factor 1 genotypes with human immunodeficiency virus disease progression in patients receiving nucleoside therapy. J Infect Dis. 2001;184(11):1402–11. doi: 10.1086/324427. [DOI] [PubMed] [Google Scholar]
- Lawson VA, Silburn KA, Gorry PR, Paukovic G, Purcell DF, Greenway AL, McPhee DA. Apoptosis induced in synchronized human immunodeficiency virus type 1-infected primary peripheral blood mononuclear cells is detected after the peak of CD4+ T-lymphocyte loss and is dependent on the tropism of the gp120 envelope glycoprotein. Virology. 2004;327(1):70–82. doi: 10.1016/j.virol.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Lee B, Sharron M, Blanpain C, Doranz BJ, Vakili J, Setoh P, Berg E, Liu G, Guy HR, Durell SR, Parmentier M, Chang CN, Price K, Tsang M, Doms RW. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274(14):9617–26. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Louder MK, Winter C, Prabhakara R, De Rosa SC, Douek DC, Hill BJ, Gabuzda D, Roederer M. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J Virol. 2002;76(10):4810–21. doi: 10.1128/JVI.76.10.4810-4821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael NL, Chang G, Louie LG, Mascola JR, Dondero D, Birx DL, Sheppard HW. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3(3):338–40. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors--central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20(1):111–26. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73(3):2343–9. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68(6):4031–4. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatov AA, Pollakis G, Linnemann T, Kliphius A, Chalaby MI, Paxton WA. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J Virol. 2004;78(1):524–30. doi: 10.1128/JVI.78.1.524-530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Aasa-Chapman MM, Clapham PR, Nibbs RJ, McKnight A, Weiss RA. The promiscuous CC chemokine receptor D6 is a functional coreceptor for primary isolates of human immunodeficiency virus type 1 (HIV-1) and HIV-2 on astrocytes. J Virol. 2005;79(15):9618–24. doi: 10.1128/JVI.79.15.9618-9624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbs RJ, Kriehuber E, Ponath PD, Parent D, Qin S, Campbell JD, Henderson A, Kerjaschki D, Maurer D, Graham GJ, Rot A. The beta-chemokine receptor D6 is expressed by lymphatic endothelium and a subset of vascular tumors. Am J Pathol. 2001;158(3):867–77. doi: 10.1016/s0002-9440(10)64035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbs RJ, Wylie SM, Yang J, Landau NR, Graham GJ. Cloning and characterization of a novel promiscuous human beta-chemokine receptor D6. J Biol Chem. 1997;272(51):32078–83. doi: 10.1074/jbc.272.51.32078. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- Olson WC, Rabut GE, Nagashima KA, Tran DN, Anselma DJ, Monard SP, Segal JP, Thompson DA, Kajumo F, Guo Y, Moore JP, Maddon PJ, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73(5):4145–55. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Nedellec R, Ramos A, Pontow S, Ratner L, Mosier DE. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J Virol. 2006;80(2):750–8. doi: 10.1128/JVI.80.2.750-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Kuhmann SE, Rose PP, Kabat D. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J Virol. 2001;75(24):12266–78. doi: 10.1128/JVI.75.24.12266-12278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Shea DM, Rose PP, Kabat D. Variants of human immunodeficiency virus type 1 that efficiently use CCR5 lacking the tyrosine-sulfated amino terminus have adaptive mutations in gp120, including loss of a functional N-glycan. J Virol. 2005;79(7):4357–68. doi: 10.1128/JVI.79.7.4357-4368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut GE, Konner JA, Kajumo F, Moore JP, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):3464–8. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira JM, Moniz-Pereira J, Clapham PR. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73(9):7795–804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, McKnight A, Potempa S, Simmons G, Gray PW, Power CA, Wells T, Weiss RA, Talbot SJ. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231(1):130–4. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Roger M. Influence of host genes on HIV-1 disease progression. Faseb J. 1998;12(9):625–32. doi: 10.1096/fasebj.12.9.625. [DOI] [PubMed] [Google Scholar]
- Rucker J, Edinger AL, Sharron M, Samson M, Lee B, Berson JF, Yi Y, Margulies B, Collman RG, Doranz BJ, Parmentier M, Doms RW. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. Journal of Virology. 1997;71(12):8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker J, Samson M, Doranz BJ, Libert F, Berson JF, Yi Y, Smyth RJ, Collman RG, Broder CC, Vassart G, Doms RW, Parmentier M. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87(3):437–46. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- Schols D, Struyf S, Van Damme J, Este JA, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186(8):1383–8. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan MK, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71(7):4871–81. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274(4):1905–13. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, McKnight A, Dejucq N, Hibbitts S, Power CA, Aarons E, Schols D, De Clercq E, Proudfoot AE, Clapham PR. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72(10):8453–7. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, Goedert JJ, O'Brien TR, Jacobson LP, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner MW, O'Brien SJ. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277(5328):959–65. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- Solomon A, Lane N, Wightman F, Gorry PR, Lewin SR. Enhanced replicative capacity and pathogenicity of HIV-1 isolated from individuals infected with drug-resistant virus and declining CD4+ T-cell counts. J Acquir Immune Defic Syndr. 2005;40(2):140–8. doi: 10.1097/01.qai.0000173460.75322.93. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67(6):3674–9. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Paxton WA, Monard SP, Hoxie JA, Siani MA, Thompson DA, Wu L, Mackay CR, Horuk R, Moore JP. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol. 1998;72(1):396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69(11):6609–17. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Spira TJ, Owen S, Lal RB, Saksena NK. HIV-1 strains from a cohort of American subjects reveal the presence of a V2 region extension unique to slow progressors and non-progressors. Aids. 2000;14(3):213–23. doi: 10.1097/00002030-200002180-00002. [DOI] [PubMed] [Google Scholar]
- Willey SJ, Reeves JD, Hudson R, Miyake K, Dejucq N, Schols D, De Clercq E, Bell J, McKnight A, Clapham PR. Identification of a subset of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus strains able to exploit an alternative coreceptor on untransformed human brain and lymphoid cells. J Virol. 2003;77(11):6138–52. doi: 10.1128/JVI.77.11.6138-6152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Farzan M, Si Z, Madani N, Wang L, Rosenberg E, Robinson J, Sodroski J. Functional mimicry of a human immunodeficiency virus type 1 coreceptor by a neutralizing monoclonal antibody. J Virol. 2005;79(10):6068–77. doi: 10.1128/JVI.79.10.6068-6077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman KJ, Brown RC, Phair JP, Neumann AU, Ho DD, Wolinsky SM. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72(11):9307–12. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV., Jr. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol. 2002;76(2):644–55. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.