Abstract
Phosphorylation of histone H2A or H2AX is an early and sensitive marker of DNA damage in eukaryotic cells, although mutation of the conserved damage-dependent phosphorylation site is well tolerated. Here, we show that H2A phosphorylation is required for cell-cycle arrest in response to DNA damage at the G1/S transition in budding yeast. Furthermore, we show that the tandem BRCT domain of Rad9 interacts directly with phosphorylated H2A in vitro and that a rad9 point mutation that abolishes this interaction results in in vivo phenotypes that are similar to those caused by an H2A phosphorylation site mutation. Remarkably, similar checkpoint defects are also caused by a Rad9 Tudor domain mutation that impairs Rad9 chromatin association already in undamaged cells. These findings indicate that constitutive Tudor domain-mediated and damage-specific BRCT domain–phospho-H2A-dependent interactions of Rad9 with chromatin cooperate to establish G1 checkpoint arrest.
Keywords: H2A, DNA damage, Rad9, G1, yeast
Introduction
The eukaryotic DNA-damage response involves the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinase-dependent signal-transduction pathways that preserve genomic integrity (Shechter et al, 2004; Lavin et al, 2005). ATM/ATR phosphorylation of H2AX on serine 139 is required for retention of the check-point mediator proteins p53 binding protein 1 (53BP1) and mediator of DNA-damage checkpoint 1 (MDC1) on chromatin flanking DNA double-strand break (DSB) sites, and it acts to promote both checkpoint signalling and DSB repair (Stucki & Jackson, 2006). Similarly, the Saccharomyces cerevisiae ATM/ATR orthologues Tel1 and Mec1 also phosphorylate histone H2A (Downs et al, 2000) on serine 129 (S129) to recruit chromatin remodelling factors to sites of DSBs (Downs et al, 2004; Morrison et al, 2004; van Attikum et al, 2004) and promote efficient DSB repair (Downs et al, 2000; Redon et al, 2003).
Other histone modifications also participate in the DNA-damage response. For example, DSB-induced acetylation of lysine residues in histones H3 and H4 affects repair efficiency (Bird et al, 2002; Jazayeri et al, 2004; Masumoto et al, 2005), and histone lysine methylation is linked to the activation of checkpoint signalling (Huyen et al, 2004; Sanders et al, 2004; Wysocki et al, 2005; Botuyan et al, 2006). Human 53BP1, S. cerevisiae Rad9 and Schizosaccharomyces pombe Crb2 contain tandem Tudor domains (Tudor2 domains) that directly recognize methylated lysine residues (Huyen et al, 2004; Botuyan et al, 2006; Du et al, 2006). In budding yeast, mutation of either histone H3 lysine 79 (K79) or the Rad9 Tudor2 domain causes G1- and S-phase checkpoint defects but does not abolish the RAD9-dependent G2/M arrest (Wysocki et al, 2005). Therefore, we tested whether other known DNA damage-associated histone modifications might elicit cell-cycle phase-specific responses. We show that defective H2A S129 phosphorylation impairs checkpoint signalling in G1 but does not compromise the G2 checkpoint, and that phospho-H2A-dependent G1 checkpoint signalling involves the direct interaction between phosphorylated H2A and the tandem BRCT domain (BRCT2) of Rad9.
Results And Discussion
H2A phosphorylation is required for the G1 checkpoint
Although histone H2A phosphorylation promotes DNA repair in yeast, its role in checkpoint control has not been thoroughly explored (Downs et al, 2000; Redon et al, 2003). As both DSB-induced checkpoint signalling and DSB-induced H2A S129 phosphorylation in G1 cells depend on the MRX-Tel1 pathway (Grenon et al, 2001; Shroff et al, 2004), we used, for checkpoint analyses, an allele of H2A (hta-S129*) in which S129 is mutated to a STOP codon that removes the last four residues (Downs et al, 2000).
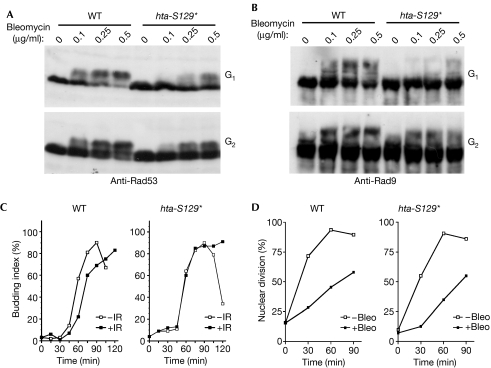
Notably, G1-arrested wild-type cells showed the characteristic phosphorylation-dependent Rad53 mobility shift in response to the radiomimetic drug bleomycin, whereas this Rad53 shift was markedly diminished in G1-arrested hta-S129* cells (Fig 1A, top panel). By contrast, the extent of Rad53 hyperphosphorylation following bleomycin treatment in G2/M-arrested hta-S129* cells was very similar to that in wild-type cells, with only a modest defect at very low doses (Fig 1A, bottom panel). Similarly, hta-S129* cells were severely defective in DNA-damage-induced hyperphosphorylation of Rad9, the key mediator of Rad53 activation (Sweeney et al, 2005); again this effect was more pronounced during G1 than in G2/M-arrested cells (Fig 1B). Consistent with this, wild-type cells delayed bud emergence—a marker of progression past START—following exposure to ionizing radiation during G1, whereas this response was completely absent in hta-S129* cells (Fig 1C). By contrast, hta-S129* cells were as proficient as wild-type cells in delaying the onset of anaphase following bleomycin treatment of G2/M-arrested cells (Fig 1D). On the basis of these findings, we conclude that the H2A C terminus, including the S129 phosphorylation site, is required for efficient damage-induced hyperphosphorylation of Rad9 and Rad53, and that this function is specifically crucial for the G1 checkpoint.
Figure 1.
Defective H2A phosphorylation abolishes the G1 checkpoint. (A) Analysis of Rad53 phosphorylation in response to bleomycin. G1-arrested (top panel) or G2-arrested (bottom panel) cells were treated with the indicated doses of bleomycin for 60 min before processing for immunoblot analysis. (B) Samples derived from cells treated as in (A) were probed with Rad9 antibodies. (C) G1 checkpoint assay. G1-arrested strains were irradiated with 100 Gy of IR or mock treated and then returned to fresh medium. Timed samples were scored for the percentage of budded cells. (D) G2 checkpoint assay. G2-arrested strains were treated with bleomycin and then returned to fresh medium. Timed samples were scored for nuclear division. Bleo, bleomycin; IR, ionizing radiation; WT, wild type.
Rad9:phospho-H2A binding controls the G1 checkpoint
The tandem BRCT (BRCT2) domain-dependent interaction of the human Rad9 homologue MDC1 with phosphorylated H2AX is required for the efficient localization of MDC1 to DSBs (Stucki et al, 2005). Therefore, we investigated whether the Rad9 BRCT2 domain could interact in a similar manner with phosphorylated H2A to mediate G1 checkpoint signalling. Phosphopeptide pull-down experiments showed that the purified Rad9 BRCT2 domain formed a relatively salt-stable interaction with a phosphorylated peptide comprising the C-terminal 13 residues of histone H2A surrounding S129, but not with an unphosphorylated control peptide (Fig 2A,B). By contrast, no phosphopeptide binding was observed for a BRCT2 mutant in which Lys 1088 was changed to methionine (Rad9-K1088M)—a mutation analogous to K1937M of MDC1 that abolishes H2AX phosphopeptide binding (supplementary Fig 1 online; Stucki et al, 2005). The BRCT2 domains of BRCA1 and MDC1 did not interact with this phosphopeptide (Fig 2C) and, unlike the phospho-H2A peptide itself, unrelated phospho-serine-containing peptides did not competitively inhibit binding of the Rad9 BRCT2 domain to beads bearing the phospho-H2A peptide (Fig 2D), underscoring the specificity of this interaction. These data therefore indicate that the Rad9 BRCT2 domain interacts specifically with phosphorylated H2A in a structurally analogous manner to the interaction between phosphorylated H2AX and MDC1.
Figure 2.
The Rad9 BRCT2 domain mediates a direct interaction with phosphorylated H2A. (A) In vitro peptide pull-down assay. Wild-type (Rad9 BRCT2) or mutant Rad9 BRCT2 ([K1088M]) domain was incubated with immobilized H2A peptides and bound proteins were visualized by Coomassie blue staining. (B) Phosphopeptide pulldown in the presence of increasing concentrations of NaCl (150, 300, 450 and 600 mM). (C) In vitro peptide pull-down assay. Indicated GST fusions were analysed as in (A). (D) Peptide competition assay. Rad9 GST–BRCT2 was incubated with competitor phosphopeptides (HSL: CFHPRRS(pSer)QGVL; hNKCC1: CEKLLRP(pSer)LAEL) and phospho-H2A binding was performed as in (A). GST, glutathione-S-transferase; H2A, unphosphorylated peptide; γH2A, phosphorylated peptide; MDC1, mediator of DNA-damage checkpoint 1.
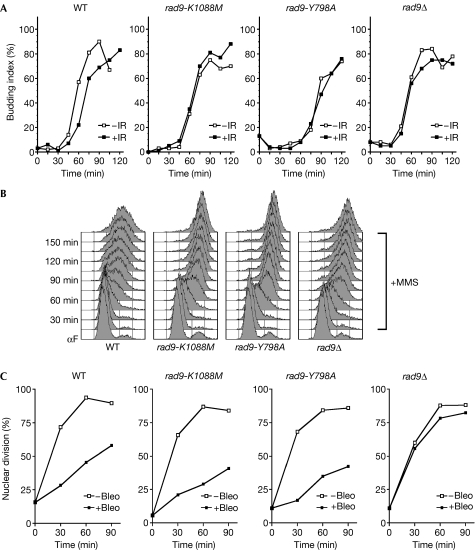
To investigate the functional importance of the phospho-dependent interaction between the Rad9 BRCT2 domain and H2A, we assessed the phenotypes of cells bearing the rad9-K1088M mutation. Similar to hta-S129* cells, G1-arrested rad9-K1088M cells were completely unable to delay bud emergence following ionizing radiation treatment (Fig 3A). We also analysed the role of the Rad9 BRCT2 domain in other Rad9-dependent checkpoints. Notably, wild-type cells slowed DNA replication in medium containing 0.02% methyl methane sulphonate (MMS), whereas rad9-K1088M cells rapidly completed S phase in the presence of MMS, similar to rad9Δ cells (Fig 3B), indicating that the BRCT2 domain is also involved in at least some aspects of the intra-S-phase checkpoint. As hta-S129A cells are reported to have an intact intra-S-phase checkpoint (Redon et al, 2003), this suggests that the Rad9 BRCT2 domain might have other targets in addition to phospho-S129 H2A. However—and again in a manner similar to the hta-S129* phenotype but in contrast to rad9Δ—the rad9-K1088M cells efficiently delayed anaphase following treatment with bleomycin during G2/M (Figs 1D,3C). This, in conjunction with the fact that Rad9-K1088M protein levels are similar to those of the wild-type protein (data not shown), indicates that rad9-K1088M is not a null allele. Consistent with the similar G1 checkpoint defect, rad9-K1088M cells also had G1-specific Rad9 and Rad53 phosphorylation defects similar to hta-S129* cells. Compared with wild-type cells, rad9-K1088M cells were almost completely defective for bleomycin- and ultraviolet-induced Rad9 hyperphosphorylation in G1 (Fig 4A, left panels; Fig 4B), but showed essentially normal levels of bleomycin-induced Rad9 hyperphosphorylation during G2/M (Fig 4A, right panels). Rad53 phosphorylation was also severely impaired in rad9-K1088M cells following ionizing radiation or ultraviolet irradiation of G1 cells (Fig 4C). Together with the phosphopeptide binding analyses, these results strongly indicate that the Rad9 BRCT2 domain mediates the phospho-S129 H2A signal to effect G1 checkpoint arrest.
Figure 3.
The Rad9 BRCT2 domain contributes to G1 checkpoint control. (A) G1 checkpoint assay. Indicated strains were treated as in Fig 1C and timed samples were scored for the percentage of budded cells. (B) Analysis of cell-cycle progression in the presence of methyl methane sulphonate (MMS). G1-arrested cells were released into medium containing 0.02% MMS. Timed samples were analysed for cell-cycle progression by flow cytometry. (C) G2 checkpoint assay. Indicated strains were treated as in Fig 1D and timed samples were scored for nuclear division. Bleo, bleomycin; IR, ionizing radiation; WT, wild type.
Figure 4.
The Rad9 BRCT2 domain is required for G1 checkpoint signalling. (A) Analysis of Rad9 phosphorylation in response to bleomycin. G1-arrested cells were treated as in Fig 1A and analysed by anti-Rad9 immunoblotting. (B) Analysis of Rad9 phosphorylation in response to ultraviolet-irradiation. G1-arrested strains were treated with 50 J/m2 ultraviolet radiation and then held in G1 for the indicated time period. (C) Analysis of Rad53 phosphorylation in response to IR and UV radiation. G1-arrested cells were irradiated with the indicated doses of X-ray or UV irradiation and then held in G1 for 30 min. αF, alpha factor; Bleo, bleomycin; IR, ionizing radiation; UV, ultraviolet; WT, wild type.
Rad9 Tudor2 and BRCT2 domains functionally cooperate
G1/S checkpoint defects similar to those identified here for BRCT2/H2A mutations were recently reported for substitution of the conserved tyrosine 798 (Y798) within the Tudor2 domain of Rad9 or loss of the Tudor2 domain ligand—H3 methyl-K79 (Wysocki et al, 2005). For direct comparison, we created an alanine substitution of RAD9 Y798 (rad9-Y798A) and a rad9-Y798A, K1088M double mutant. Strikingly, the phenotypes of rad9-K1088M cells and rad9-Y798A cells were virtually identical, with defective G1/S and intra-S-phase checkpoints but normal G2/M checkpoints (Fig 3), as well as impaired Rad9 and Rad53 hyperphosphorylation in response to DNA damage during G1 but not G2 (Fig 4A). Importantly, the phenotype of the rad9-Y798A, K1088M double mutant, with respect to Rad9 and Rad53 phosphorylation following DNA damage in G1, was similar to the two single-domain mutants (Fig 4A,C), indicating that the Tudor2 and BRCT2 domains function together to mediate efficient G1 checkpoint activation.
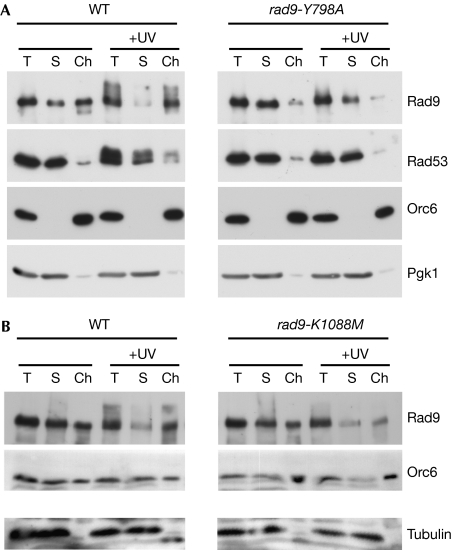
The similar checkpoint defects of Rad9 Tudor2 and BRCT2 domain mutations raised the possibility that both domains might be needed to target Rad9 to sites of damaged chromatin. To test this possibility, we treated G1-arrested spheroplasts with ultraviolet irradiation, and then separated extracts into soluble and chromatin-enriched fractions to analyse Rad9 association with chromatin (Fig 5). Surprisingly, we found that significant amounts of Rad9 were already associated with chromatin in extracts from undamaged G1 cells (Fig 5A). By contrast, Rad53 was found almost exclusively in the soluble fraction. After ultraviolet irradiation, wild-type Rad9 was depleted from the soluble pool and redistributed almost exclusively to the chromatin-enriched fraction (Fig 5A). Strikingly, mutation of the Tudor2 domain almost completely abolished Rad9 chromatin association in undamaged cells and, unlike the wild-type protein, Rad9-Y798A was not further recruited into chromatin (Fig 5A). This suggests that a constitutive chromatin association of Rad9 in undamaged cells is a prerequisite for its hyperphosphorylation and concomitant enrichment in chromatin following DNA damage. In contrast to the Tudor2 domain mutant, the basal chromatin association of the Rad9-K1088M BRCT2 mutant in undamaged cells was undiminished compared with the wild type, but it also failed to be further enriched in damaged chromatin or to be hyperphosphorylated after DNA damage (Fig 5B). The simplest explanation for these data is that the Rad9 Tudor2 domain mediates a constitutive association of Rad9 with chromatin during G1 phase in undamaged cells, as a prerequisite for its DNA damage-specific BRCT2-dependent enrichment in the vicinity of DNA damage. Although we cannot exclude the possibility that the observed distribution of Rad9 in otherwise undamaged G1 cells reflects a possible disruption of chromatin fibres that is recognized as DNA damage during extract preparation, we feel that this is unlikely and note that others did not observe a similar association of Rad9 in undamaged G2-arrested cells using an almost identical lysis procedure (Toh et al, 2006).
Figure 5.
The Rad9 Tudor2 and BRCT2 domains cooperate to establish association with damaged chromatin. (A) Analysis of total (T), soluble (S) and chromatin-enriched (Ch) fractions from extracts of G1-arrested wild-type and rad9-Y798A cells before and 30 min after treatment with ultraviolet irradiation. Blots were probed with antibodies as indicated. (B) Wild-type and rad9-K1088M cells were analysed as in (A). Orc2, origin recognition complex 2; Pgk1, phosphoglycerate kinase 1; UV, ultraviolet; WT, wild type.
Together, our results indicate that histone H2A phosphorylation is required for establishment of the G1 DNA-damage checkpoint and support a model in which this function involves a direct interaction between phosphorylated H2A and the Rad9 BRCT2 domain. We have also found that both the Tudor2 and BRCT2 domains of Rad9 are required for the G1 checkpoint response, and that Tudor2 domain-dependent chromatin localization of Rad9 in undamaged cells is crucial for the establishment of G1 checkpoint signalling. The nuclear redistribution of Rad9 in G1 cells after DNA damage is in marked contrast to that observed previously in G2 cells in which hypophosphorylated forms of Rad9 were retained only at later time points after ionizing radiation (Toh et al, 2006). This difference indicates that, in contrast to G1 cells, sustained chromatin binding is dispensable for correct checkpoint activation in G2, perhaps reflecting a requirement for a lower activation threshold.
Our results thereby confirm and extend the previously published finding that the Rad9 Tudor2 domain is required for the G1 checkpoint (Wysocki et al, 2005). While this manuscript was in preparation, our initial findings were independently confirmed by a report that H2A phosphorylation and H3 K79 methylation both control G1/S checkpoint arrest and Rad9 occupancy at DSBs in G1 (Javaheri et al, 2006). However, in contrast to their finding of a partial G1 arrest defect in hta-S129A strains—and more consistent with the findings reported in a previous study (Redon et al, 2003)—we have found that hta-S129* cells are completely G1 checkpoint-defective. The simplest explanation for this apparent discrepancy could be the higher dose of ionizing radiation used by Javaheri et al (2006).
On the basis of the available data, in particular the constitutive chromatin localization of Rad9 (Fig 5), we speculate that the Rad9 Tudor2 domain—and, by analogy, the equivalent domains of Rad9 counterparts in other organisms—might recognize multiple histone methylation marks to enable association with bulk chromatin. Indeed, in budding yeast, two distinct histone lysine methylases are required for the G1 checkpoint response (Giannattasio et al, 2005; Wysocki et al, 2005), and the Tudor2 domain of 53BP1 recognizes at least three histone methylation marks in vitro (Kim et al, 2006) and might recognize multiple ligands in vivo (Botuyan et al, 2006). Constitutive chromatin ‘scanning' could enhance the speed and efficiency of the DNA-damage response, particularly after low doses of DNA damage. H2A phosphorylation might then act to concentrate Rad9 near sites of damage through the BRCT2 domain and thus allow efficient Rad9 hyperphosphorylation and signalling. At present, it is unclear why G1 checkpoint activation requires two histone-binding modules that are largely dispensable in G2. As Rad9 is phosphorylated in normal post-G1 cells, and cyclin-dependent kinase (CDK) phosphorylation of the Rad9 orthologue Crb2 is required for the correct repair of ionizing radiation induced lesions (Caspari et al, 2002) and also for ionizing radiation induced focus formation in the absence of ligands for the Tudor2 and BRCT2 domains (Du et al, 2006), it is tempting to speculate that CDK activity controls both the choice of DNA-repair pathway and the mode of checkpoint activation. As Rad9 is also required for efficient DNA repair (Toh et al, 2006), such controls could act to regulate and coordinate checkpoint arrest and DNA repair mechanisms.
Methods
Yeast strain construction and checkpoint experiments. Strains were constructed using PCR-based allele replacement techniques (Erdeniz et al, 1997) and are listed in supplementary Table 1 online. Analysis of yeast checkpoint protein phosphorylation and cell-cycle progression was carried out as detailed in the supplementary information online.
GST fusion expression and purification. Sequences encoding residues 981–1,309 of Rad9 and residues 1,634–1,863 of BRCA1 encompassing the BRCT2 domains were cloned into the vector pGEX4T1 for bacterial expression and purified using glutathione-Sepharose (GE Healthcare, Rydalmere, Australia). Thrombin-cleaved BRCT2 domains were purified by cation-exchange chromatography, as detailed in the supplementary information online.
Peptide-binding assays. Synthetic biotinylated peptides (W. Mawby, University of Bristol, UK) were coupled to streptavidin dynabeads M-280 (Invitrogen, Mt Waverley, Australia) in TBS-T (Tris-buffered saline Tween-20) in excess, such that all available biotin-binding sites were blocked. The beads were then washed and incubated with purified BRCT2 domains (∼1–4 μg) in TBS-T for 90 min at 4°C, followed by extensive washing before analysis by SDS–polyacrylamide gel electrophoresis and Coomassie blue staining. For competition assays, BRCT2 domains were preincubated with 100 μg competitor peptides at 4°C for 2 h before addition to the immobilized phosphopeptide as above.
Chromatin fractionation. Chromatin fractionation was carried out, essentially as described previously (Donovan et al, 1997), on G1-arrested cells that were held in G1 for the entire procedure by inclusion of alpha-factor in all buffers. For ultraviolet irradiation, spheroplasts were washed and resuspended in PBS-S (PBS containing 1 M sorbitol), and treated with 30–100 J/m2 ultraviolet radiation (depending on the ultraviolet source) and then allowed to recover for 30 min at 30°C in YPD-S (YPD containing 0.7 M sorbitol, 25 mM Tris, pH 7.4) before washing and extract preparation. Effective fractionation was assessed using antibodies to either Orc6 (Weinreich et al, 1999) or Orc2 (Duncker et al, 2002).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Supplementary Fig 1
Acknowledgments
We thank N. Lowndes (Rad9, Rad53), B. Stillman (Orc6) and B. Duncker (Orc2) for antibodies, A. Traven for critical review of the manuscript and N. Tenis for technical support. A.H. and J.H. are supported by funding from the National Health and Medical Research Council, and the Clive and Vera Ramaciotti Foundation. Work in the S.P.J. laboratory was made possible by a Programme Grant from Cancer Research UK, and core infrastructure funding from Cancer Research UK and the Wellcome Trust.
References
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Murray JM, Carr AM (2002) Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev 16: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA 94: 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J (2004) Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16: 979–990 [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Shimada K, Tsai-Pflugfelder M, Pasero P, Gasser SM (2002) An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc Natl Acad Sci USA 99: 16087–16092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N, Mortensen UH, Rothstein R (1997) Cloning-free PCR-based allele replacement methods. Genome Res 7: 1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Grenon M, Gilbert C, Lowndes NF (2001) Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat Cell Biol 3: 844–847 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ (2006) Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci USA 103: 13771–13776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, McAinsh AD, Jackson SP (2004) Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci USA 101: 1644–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Birrell G, Chen P, Kozlov S, Scott S, Gueven N (2005) ATM signaling and genomic stability in response to DNA damage. Mutat Res 569: 123–132 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X (2004) INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775 [DOI] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004) Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3: 901–908 [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (2004) Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Jackson SP (2006) γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 5: 534–543 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF (2006) Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair 5: 693–703 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788 [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B (1999) The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci USA 96: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Fig 1