Abstract
The human C1 heterogeneous nuclear ribonucleoprotein particle protein (hnRNP protein) undergoes a cycle of phosphorylation–dephosphorylation in HeLa cell nuclear extracts that modulates the binding of this protein to pre-mRNA. We now report that hyperphosphorylation of the C1 hnRNP protein is mediated by a kinase activity in nuclear extracts that is RNA-dependent. Although the basal phosphorylation of the C1 hnRNP protein in nuclear extracts reflects a casein kinase II-type activity, its RNA-dependent hyperphosphorylation appears to be mediated by a different kinase. This is indicated by the unresponsiveness of the RNA-stimulated hyperphosphorylation to casein kinase II inhibitors, and the distinct glycerol gradient sedimentation profiles of the basal versus RNA-stimulated C1 hnRNP protein phosphorylation activities from nuclear extracts. RNA-dependent phosphorylation was observed both for a histidine-tagged recombinant human C1 hnRNP protein added to nuclear extracts and also for the endogenous C1 hnRNP protein. Additional results rule out protein kinase A, protein kinase C, calmodulin-dependent protein kinase II, and double-stranded RNA-activated protein kinase as the enzymes responsible for the RNA-dependent hyperphosphorylation of the C1 hnRNP protein. These results reveal the existence in nuclear extracts of an RNA-dependent protein kinase activity that hyperphosphorylates a known pre-mRNA binding protein, and define an additional element to be integrated into the current picture of how nuclear proteins are regulated by phosphorylation.
Approximately 20 characterized heterogeneous nuclear ribonucleoprotein particle proteins (hnRNP proteins) bind to pre-mRNA soon after its transcription and may play a role in the early posttranscriptional steps of pre-mRNA processing (1–3). We have recently reported that the C1 hnRNP protein, one of the most abundant hnRNP proteins, undergoes a cycle of phosphorylation–dephosphorylation in HeLa cell nuclear extracts which modulates its binding to pre-mRNA, and that the dephosphorylation step of this cycle is facilitated by the 3′ end of U6 small nuclear RNA (4, 5). In the course of this work we found that the phosphorylation level of the C1 hnRNP protein was greatly reduced in nuclear extracts in which endogenous nucleic acids had been previously eliminated by nuclease digestion (5). Pursuit of this finding has revealed the presence of a protein kinase activity in nuclear extracts that hyperphosphorylates the C1 hnRNP protein in an RNA-dependent manner.
MATERIALS AND METHODS
Nuclear extracts were prepared from exponentially growing HeLa cells as described (6). Nuclear extracts from a human T cell leukemia line (Jurkat) were obtained from Promega. Prior to use, nuclear extracts were incubated with micrococcal nuclease (Worthington) at 200 units/ml in the presence of 10 mM CaCl2 for 30 min at 30°C. The extracts were then brought to 4°C, EGTA was added to 75 mM, and the extracts were made 4.8 mM in MgCl2, 600 μM in ATP, and 30 mM in creatine phosphate. RNasin (Promega) was added to a final concentration of 660 units/ml. Recombinant C1 hnRNP protein was generated by introducing a full-length human C1 hnRNP protein cDNA clone (pHC12, ref. 7; provided by Gideon Dreyfuss, University of Pennsylvania) into the expression vector pET-14b (Novagen, Madison, WI). The resulting construct, PAF/C1, was expressed in Escherichia coli strain BL21[DE3]LysS and the histidine-tagged C1 hnRNP protein was purified on Ni2+-chelation resin (His-Bind; Novagen). The C1 protein was incubated in nuclear extract at concentrations of 100–200 μg/ml for 30 min at 30°C in the presence or absence of various RNAs as indicated. [γ-32P]ATP (NEN/DuPont; 3000 Ci/mmol, 400 μCi/ml final concentration; 1 Ci = 37 GBq) or [γ-32P]GTP (NEN/DuPont; 3000 Ci/mmol, 400 μCi/ml final concentration) was then added and the incubations continued for the times indicated. In all cases, at the end of the labeling period the reactions were incubated for an additional 30 min (30°C) with heparin (2 mg/ml), micrococcal nuclease (680 units/ml), and CaCl2 (2.5 mM). The samples were then cooled to 4°C and the C1 hnRNP protein was recovered by adding His-Bind resin and rocking the samples gently at 4°C for 2 hr followed by washing the resin twice in binding buffer (5 mM imidazole/0.5 M NaCl/20 mM Tris·HCl, pH 7.9), twice in wash buffer (60 mM imidazole/0.5 M NaCl/20 mM Tris·HCl, pH 7.9), and then once in elution buffer (1 M imidazole/0.5 M NaCl/20 mM Tris·HCl, pH 7.9). After collecting the resin by centrifugation, the supernatants were boiled in SDS sample buffer and subjected to electrophoresis in 14% polyacrylamide gels followed by autoradiography.
In some experiments the phosphorylation of endogenous C1 hnRNP protein in HeLa nuclear extracts was investigated by immunoselection with a monoclonal antibody, 4F4 (8), as described (4, 5).
RNA was prepared from HeLa nuclear extracts by two successive phenol/chloroform (1:1, vol/vol) extractions followed by adjustment of the final aqueous phase to 0.3 M sodium acetate and ethanol (67% vol/vol) precipitation. For preparation of HeLa cytoplasmic S100 RNA the postnuclear supernatant (6) was centrifuged in a Beckman 60Ti rotor at 38,000 rpm for 1 hr. The supernatant was removed and subjected to phenol/chloroform extraction and ethanol precipitation as above. RNA concentrations were determined by measurement of absorbance at 260 nm. Protein kinase inhibitors and their sources were 2,3-bisphosphoglycerate (Sigma), quercetin (3,3′,4′,5,7-pentahydroxylfavone·2H2O; Sigma), H-7 (1-[5-isoquinolinylsulfonyl]-3-methylpiperazine; Sigma), and staurosporine (Boehringer Mannheim).
RESULTS
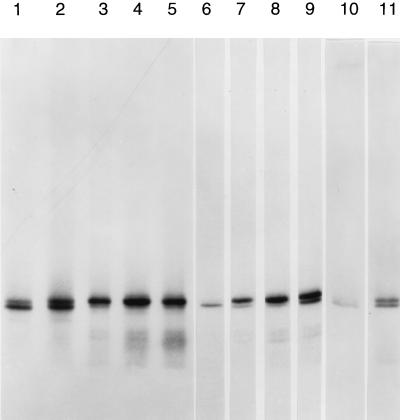
Incubation of human C1 hnRNP protein in micrococcal nuclease pretreated HeLa nuclear extract in the presence of [γ-32P]ATP resulted in the formation of two phosphorylated species, as shown in Fig. 1, lane 1. Addition of the Ser/Thr protein phosphatase 1/2A inhibitor okadaic acid resulted in the expected (4) increase in phosphorylation of both C1 hnRNP protein species (Fig. 1, lane 2). However, if the basal (no okadaic acid) reaction was supplemented by the addition of deproteinized nuclear extract RNA (final concentration, 150 μg/ml), the upper (more slowly migrating) of the two C1 hnRNP protein bands became more phosphorylated and the lower C1 hnRNP protein band became less phosphorylated (Fig. 1, lane 3, compare with lane 1). A similar RNA effect was observed when [γ-32P]GTP was used as the phosphate donor (data not shown; see also Fig. 3). The extent of the phosphorylation stimulation of the upper band was approximately proportional to the amount of RNA added (Fig. 1, lanes 6–9). The same stimulatory effect of nuclear RNA on C1 hnRNP protein phosphorylation was observed in nuclear extracts from another human cell line (Fig. 1, lanes 10 and 11), indicating that this may be a general phenomenon. Nuclear RNA also stimulated hyperphosphorylation of the C1 hnRNP protein in nuclear extracts that had not been previously treated with micrococcal nuclease (Fig. 2), indicating that the RNA effect is not a unique attribute of nuclease-digested extracts. Pretreatment of the RNA preparation with pancreatic DNase did not influence its capacity to stimulate C1 hnRNP protein phosphorylation, whereas pretreatment of the RNA preparation with micrococcal nuclease completely eliminated its capacity to stimulate phosphorylation (data not shown).
Figure 1.
Nuclear RNA stimulates C1 hnRNP protein phosphorylation. C1 hnRNP protein was incubated in nuclear extracts with [γ-32P]ATP, as detailed in Materials and Methods, and other additions as indicated. Lanes: 1, control (no other additions); 2, 1 μM okadaic acid; 3, 150 μg/ml nuclear extract RNA (no okadaic acid); 4, 150 μg/ml nuclear extract RNA plus 6 mM 2,3-bisphosphoglycerate; 5, 150 μg/μl nuclear extract RNA plus 12 mM 2,3-bisphosphoglycerate; 6, control (no other additions); 7, 50 μg/ml nuclear extract RNA; 8, 100 μg/ml nuclear extract RNA; 9, 200 μg/ml nuclear extract RNA; 10, C1 hnRNP protein incubated in micrococcal nuclease treated Jurkat nuclear extract as in lane 1, no added RNA; and 11, C1 hnRNP protein incubated as in lane 10 but with HeLa nuclear extract RNA, 75 μg/ml. Samples were subjected to gel electrophoresis in 14% polyacrylamide gels as described.
Figure 3.
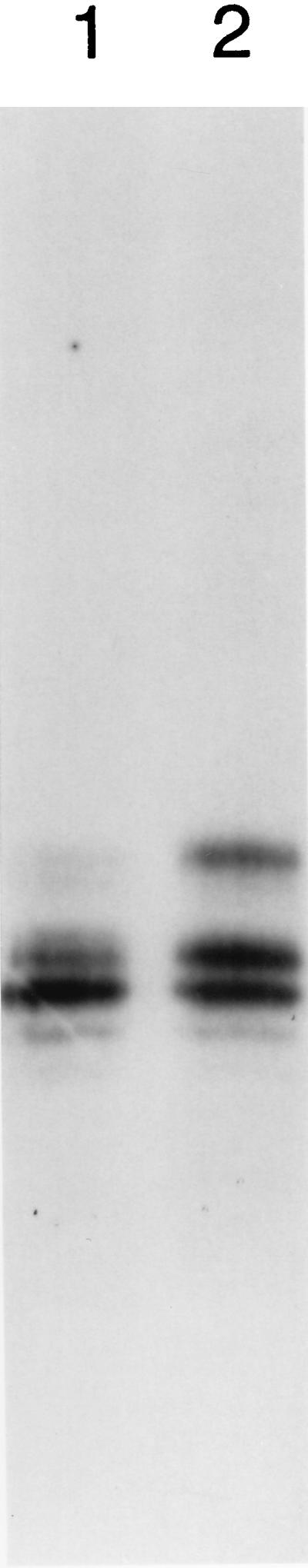
RNA also elicits hyperphosphorylation of endogenous C1 hnRNP protein in nuclear extract. HeLa nuclear extract (not pretreated with micrococcal nuclease) was incubated for 30 min (at 30°C) in the presence or absence of added nuclear extract RNA (final concentration of added RNA, 75 μg/ml) and then for an additional 60 min (at 30°C) with [γ-32P]GTP. The reactions were then processed as described followed by incubation with protein A-Sepharose coupled 4F4 monoclonal antibody for 2 hr at 4°C. After washing the resin, the bound C1 hnRNP protein was eluted (4, 5, 9, 10) and analyzed by electrophoresis and autoradiography. Lanes: 1, no added RNA; 2, added nuclear extract RNA.
Figure 2.

RNA stimulation of C1 hnRNP protein phosphorylation does not require prior micrococcal nuclease treatment of the nuclear extract. C1 hnRNP protein was incubated with [γ-32P]ATP as in Fig. 1, except that the HeLa nuclear extract was not pretreated with micrococcal nuclease. Lanes: 1, control (no other additions); 2, 1 μM okadaic acid; and 3, 150 μg/ml nuclear extract RNA (no okadaic acid).
The experiments shown in Figs. 1 and 2 involved recombinant human C1 hnRNP protein as the substrate. To investigate whether RNA also affects the phosphorylation of endogenous C1 hnRNP protein, nuclear extract (not treated with micrococcal nuclease) was incubated with [γ-32P]GTP in the presence or absence of added nuclear extract RNA, and the endogenous C1 hnRNP protein was immunoselected with a specific monoclonal antibody (4, 5). The pattern of phosphorylated endogenous C1 hnRNP proteins in the absence of added RNA (Fig. 3, lane 1) was somewhat more complex than that of the recombinant protein, probably due to the presence of endogenous phosphorylated C2 hnRNP protein which also is selected by this antibody (8), and agrees with the phosphorylation pattern for endogenous C1 and C2 hnRNP proteins we have previously reported (4, 5). The presence of added nuclear RNA (Fig. 3, lane 2) stimulated phosphorylation not only of the upper band seen without added RNA, but also caused a pronounced phosphorylation of an even more slowly migrating band, which is probably the aforementioned C2 hnRNP protein. Thus, a stimulatory effect of RNA on phosphorylation is seen both for recombinant and endogenous C1 hnRNP protein.
Previous work by us and others had indicated that casein kinase II (CKII) (or a CKII-like activity) is the major phosphorylating enzyme for the C1 hnRNP protein in isolated HeLa nuclei or nuclear extracts (4, 11, 12). However, as shown in Fig. 1, lanes 4 and 5, the RNA-stimulated hyperphosphorylation of the upper C1 hnRNP protein band was not blocked by 2,3-bisphosphoglycerate, an inhibitor of CKII. Moreover, RNA-stimulated phosphorylation of the upper C1 hnRNP protein band also occurred in the presence of another CKII inhibitor, quercetin (Table 1). In addition, when the recombinant C1 hnRNP protein was incubated in buffer together with purified CKII, there was no effect of RNA on phosphorylation (Fig. 4). (This experiment also rules out the possibility that RNA stimulates an autophosphorylation activity of the C1 hnRNP protein itself.) Additional results (Table 1) indicated that the RNA-stimulated activity is not protein kinase A, protein kinase C, calmodulin-dependent protein kinase II (16), or the double-stranded RNA-activated protein kinase, PKR (17). Thus, all the results indicate that the RNA-dependent phosphorylation of the C1 hnRNP protein is mediated by a kinase activity distinct from those previously described.
Table 1.
The C1 hnRNP protein hyperphosphorylation activity is distinct from several known kinases
| Inhibitor/activator* | Effect on RNA-dependent C1 hnRNP protein phosphorylation† |
|---|---|
| 2,3-Bisphosphoglycerate (CKII↓) | 0 |
| Quercetin (CKII↓) | 0 |
| H-7 (PKA↓) | 0 |
| Staurosporine (PKC↓)‡ | 0 |
| EGTA (CaM-PKII↓) | 0 |
| Poly(I)·poly(C) (PKR↑)§ | 0 |
Activity was assayed as in Fig. 1. Nuclear RNA was present at 100 μg/ml unless otherwise noted. 2,3-Bisphosphoglycerate was used at 6 and 12 mM; quercetin was used at 50 and 100 μM; H-7 was used at 10 μM; staurosporine was used at 2 and 5 nM; EGTA was used at 50 mM; and poly(I)·poly(C) was used at 100 μg/ml.
In parentheses are indicated the known inhibitory or stimulatory effects on various kinases. CKII, casein kinase II; PKA, protein kinase A; PKC, protein kinase C; CaM-PKII, calmodulin-dependent protein kinase II; PKR, double-stranded RNA-activated protein kinase.
Zero denotes no effect.
Staurosporine was initially thought to be a selective inhibitor of PKC (13, 14). Subsequent work (ref. 15, and references cited therein) has suggested that it is a preferential inhibitor of PKC, PKA, and certain of the cyclin-dependent kinases. Because staurosporine did not block the RNA-dependent phosphorylation of C1 hnRNP protein in the present investigation, the aforementioned caveats as to selectivity of action would, if anything, exclude even additional kinases.
Poly(I)·poly(C) was used in place of nuclear RNA.
Figure 4.
CKII-mediated phosphorylation of C1 hnRNP protein is not stimulated by RNA. C1 hnRNP protein was incubated for 2 hr at 30°C in the same buffer as used in Fig. 1 (but no nuclear extract) in the presence or absence of human CKII (Boehringer Mannheim) at a final concentration of 15 milliunits/ml, and in the presence or absence of HeLa nuclear extract RNA, at a final concentration of 75 μg/ml. Lanes: 1, C1 hnRNP protein and purified CKII; 2, C1 hnRNP protein, no CKII; 3, C1 hnRNP protein and CKII plus nuclear extract RNA; 4, C1 hnRNP protein plus nuclear extract RNA. Results similar to those shown in the figure were obtained when the incubation period of C1 hnRNP protein with CKII was 30 min or 1 hr. In addition to showing that the phosphorylation of C1 hnRNP protein by CKII is not stimulated by RNA (lanes 1 vs. 3), this experiment also indicates that the C1 hnRNP protein does not undergo autophosphorylation under these experimental conditions, either in the presence or absence of RNA (lanes 2 and 4).
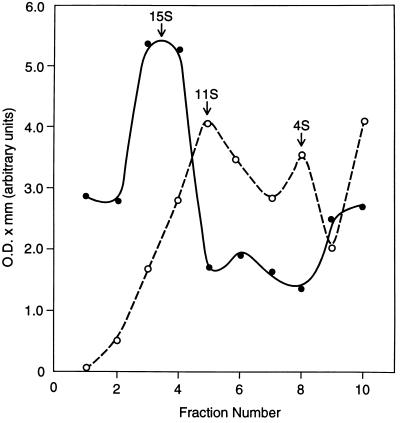
To determine whether the RNA-stimulated C1 hnRNP protein phosphorylation activity is physically distinct from this protein’s basal phosphorylation activity, nuclease-treated nuclear extract was centrifuged in glycerol gradients, followed by assay of C1 hnRNP protein phosphorylation in each fraction in the absence or presence of nuclear RNA. As shown in Fig. 5, the basal C1 hnRNP protein phosphorylation activity resided predominantly in a 15S peak, whereas the RNA-stimulated C1 hnRNP protein phosphorylation activity displayed a different sedimentation profile, with a bimodal distribution of activity at 11S and 4S. The RNA-stimulated C1 hnRNP protein phosphorylation activity is thus physically distinguishable from this protein’s basal phosphorylation activity, adding further evidence that it is a distinctive kinase.
Figure 5.
Glycerol gradient centrifugation resolves the RNA-stimulated kinase activity from the basal C1 hnRNP protein phosphorylation activity. Micrococcal nuclease-treated HeLa nuclear extract was layered on 10–30% (vol/vol) glycerol gradients and centrifuged for 18 hr at 40,000 rpm (Beckman SW41 rotor) at 4°C. The gradients were fractionated, and each fraction was divided into two equal portions. One was assayed for C1 hnRNP protein phosphorylation without exogenous RNA, and the other was assayed for C1 hnRNP protein phosphorylation in the presence of added nuclear RNA (final concentration, 125 μg/ml). The proteins were displayed by electrophoresis and autoradiograms exposed in the linear range of the film’s dpm vs. silver grain exposure curve were subjected to quantitative densitometry. The amounts of 32P in the C1 hnRNP protein bands were summed and plotted as a function of gradient position. •, No added RNA; ○, with nuclear RNA. The S values indicated by the arrows were estimated by interpolation of the gradient positions of apoferritin (18S) and alcohol dehydrogenase (8S) standards (18).
DISCUSSION
Phosphorylation of the C1 hnRNP protein is a key feature of its RNA binding activity (4). Interestingly, the phosphorylation of this protein is cell cycle-regulated (19), raising the possibility that its phosphorylation-dependent RNA binding activity may be functionally linked to regulated pre-mRNA metabolism during the G1, S, and G2 phases of the cell cycle. From the point of departure based on our observation that C1 hnRNP protein phosphorylation in HeLa cell nuclear extracts is reduced by prior nuclease treatment of the extract (5), we have gone on in the present investigation to uncover evidence for an RNA-dependent phosphorylation activity acting on both endogenous and exogenous (recombinant) C1 hnRNP protein in nuclear extracts. Several lines of evidence indicate that this activity is distinct from CKII, the enzyme that mediates basal phosphorylation of the C1 hnRNP protein, and additional results rule out several other known kinases: protein kinase A, protein kinase C, double-stranded protein kinase II, and calmodulin-dependent protein kinase II. The latter enzyme is known to phosphorylate the C1 hnRNP protein under some conditions (20).
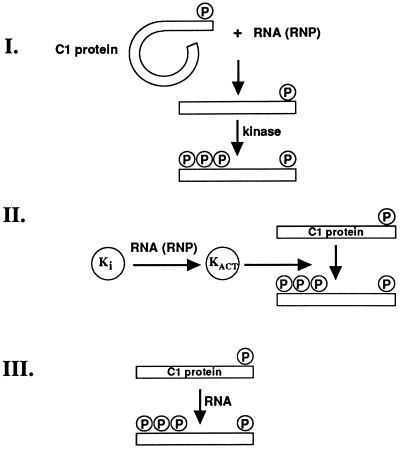
RNA-dependent phosphorylation of the C1 hnRNP protein was observed upon addition of HeLa nuclear extract RNA and HeLa cytoplasmic S100 RNA, but not with poly(A), yeast transfer RNA, double-stranded (linearized pGEM-1 plasmid) DNA, or single-stranded (phage M13) DNA. The RNA-dependent phosphorylation of the C1 hnRNP protein may reflect a specific activating RNA sequence, like the distinct DNA sequences that specifically activate the kinase that phosphorylates the SP1 transcription factor (21, 22). Alternatively, the C1 hnRNP protein’s phosphorylation may be activated by a broad class of RNA sharing common sequence or structural elements. For example, recent studies have revealed unanticipated features of the RNA elements that activate the double-stranded RNA-activated protein kinase, PKR (23, 24). In the present experiments, the RNA-dependent phosphorylation of the C1 hnRNP protein could reflect an RNA-mediated conformational event in the substrate protein which causes it, in turn, to be recognized by this particular kinase, an interaction of RNA with the kinase itself, or both (Fig. 6). It is of interest to note that the Tat-1 and Tat-2 proteins encoded by HIV-1 and HIV-2, respectively, interact both with a specific viral RNA element, transactivation response element (TAR), and a cellular protein kinase in a ternary RNA–protein kinase–protein substrate complex (28). In our experiments it is possible that a RNP complex, formed upon addition of RNA, is the actual hyperphosphorylation inducing entity. A (much) more speculative possibility is that RNA actually participates in the chemical step of the phosphorylation reaction (Fig. 6III). The labile energy available in the β-γ phosphodiester bond of ATP (25, 29) can be used by a polynucleotide kinase ribozyme (26), opening the theoretical possibility of (laboratory or natural) ribozymes that catalyze group transfer of the γ-phosphate of ATP to other substrates, e.g., amino acids in proteins. Ribozymes that catalytically operate on amino acids have been reported (27).
Figure 6.
Mechanistic possibilities for RNA-stimulation of C1 hnRNP protein hyperphosphorylation. In all three cases, the reaction pathway starts with the C1 hnRNP protein in its basal state of phosphorylation, indicated schematically by the presence of a single phosphoamino acid. (In reality, the basal state of C1 hnRNP protein phosphorylation in HeLa nuclear extracts consists of two phosphorylated serine residues, viz. Ser-107 and Ser-247; P. Dwen and T.P., unpublished results.) (I) The stimulating RNA (or an RNA-protein complex assembled from it, designated “RNP”) binds to the C1 hnRNP protein and changes its conformation into one conducive to hyperphosphorylation. (II) The effect of the RNA (or RNP) is to activate a latent kinase (KI). (III) The RNA itself mobilizes the labile energy in the β-γ phosphodiester bond of ATP (25) to catalyze, as a ribozyme, phosphate group transfer to the C1 hnRNP protein. Although III is very speculative, an ATP-hydrolytic ribozyme, viz. polynucleotide kinase, has been identified in RNA sequence space (26) and other ATP-binding RNAs have been observed, as have RNAs that act catalytically on amino acids (e.g., ref. 27).
Our previous findings that hyperphosphorylation of the C1 hnRNP protein negatively modulates its interaction with pre-mRNA (4) suggests that phosphorylation may actively release C1 hnRNP protein from pre-mRNA. The facts that the C1 protein is the first hnRNP protein to bind nascent pre-mRNA (30), and that this protein binds in a cooperative mode (31), suggest that active removal of C1 hnRNP protein would be a likely early step prior to assembly of the spliceosome. An RNA-dependent kinase could function to release bound C1 hnRNP protein from the pre-mRNA, which would be expected to destabilize other bound hnRNP proteins as well (30, 31). The fact that only a small fraction of the C hnRNP proteins are in the hyperphosphorylated state in vivo (19) is compatible with the idea that this represents the non-RNA bound fraction, because the great majority of C1 hnRNP protein in the nucleus is bound to pre-mRNA. That the C1 hnRNP protein is hyperphosphorylated via an RNA-dependent mechanism, possibly linked to its active removal from pre-mRNA as a requisite step prior to functional spliceosome assembly (4), draws attention to the plausibility of co-evolution of the splicing machinery and RNA-regulated protein modification enzymes as eukaryotes advanced.
Acknowledgments
We thank Sandra Mayrand and Marty Jacobson for important suggestions. This investigation was supported by National Institutes of Health Grant GM-21595-21 to T.P. and by National Institutes of Health Institutional Postdoctoral Training Grant HD-07312 to P.A.F.
Footnotes
Abbreviations: hnRNP, heterogeneous nuclear ribonucleoprotein particle protein; CKII, casein kinase II.
References
- 1.Pederson T. J Cell Biol. 1983;97:1321–1326. doi: 10.1083/jcb.97.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 3.Economidis I V, Pederson T. Proc Natl Acad Sci USA. 1983;80:1599–1602. doi: 10.1073/pnas.80.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayrand S H, Dwen P, Pederson T. Proc Natl Acad Sci USA. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayrand S H, Fung P A, Pederson T. Mol Cell Biol. 1996;16:1241–1246. doi: 10.1128/mcb.16.3.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson M S, Nakagawa T Y, LeVan K, Dreyfuss G. Mol Cell Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y D, Dreyfuss G. J Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayrand S H, Pederson T. Nucleic Acids Res. 1990;18:3307–3318. doi: 10.1093/nar/18.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Temsamani J, Pederson T. J Biol Chem. 1996;271:24922–24926. doi: 10.1074/jbc.271.40.24922. [DOI] [PubMed] [Google Scholar]
- 11.Holcomb E R, Friedman D L. J Biol Chem. 1984;259:31–40. [PubMed] [Google Scholar]
- 12.Friedman D L, Kleiman N J, Campbell F E., Jr Biochim Biophys Acta. 1985;847:165–176. doi: 10.1016/0167-4889(85)90017-5. [DOI] [PubMed] [Google Scholar]
- 13.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 14.Davis P D, Hill C H, Keech E, Lawton G, Nixon J S, Sedgwick A D, Wadsworth J, Westmacott D, Wilkinson S E. FEBS Lett. 1989;259:61–63. doi: 10.1016/0014-5793(89)81494-2. [DOI] [PubMed] [Google Scholar]
- 15.Gadbois D M, Hamaguchi J R, Swank R A, Bradbury E M. Biochem Biophys Res Commun. 1992;184:80–85. doi: 10.1016/0006-291x(92)91160-r. [DOI] [PubMed] [Google Scholar]
- 16.Ohta Y, Ohba T, Miyamoto E. Proc Natl Acad Sci USA. 1990;87:5341–5345. doi: 10.1073/pnas.87.14.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel C E. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 18.Kleinschmidt A M, Pederson T. Mol Cell Biol. 1987;7:3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piñol-Roma S, Dreyfuss G. Mol Cell Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosser R, Faura M, Sernatosa J, Renau-Piqueras J, Pruschy M, Bachs O. Mol Cell Biol. 1995;15:661–670. doi: 10.1128/mcb.15.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 22.Jackson S, Gottlieb T, Hartley K. Adv Second Messenger Phosphorylation Res. 1993;28:279–286. [PubMed] [Google Scholar]
- 23.Robertson H D, Manche L, Matthews M B. J Virol. 1996;70:5611–5617. doi: 10.1128/jvi.70.8.5611-5617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Matthews M B. RNA. 1996;2:937–951. [PMC free article] [PubMed] [Google Scholar]
- 25.Lipmann F. Adv Enzymol. 1941;1:99–162. [Google Scholar]
- 26.Lorsch J R, Szostak J W. Nature (London) 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- 27.Lohse P A, Szostak J W. Nature (London) 1996;381:442–444. doi: 10.1038/381442a0. [DOI] [PubMed] [Google Scholar]
- 28.Hermann C H, Rice A P. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westheimer F H. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 30.Huang M, Rech J E, Northington S J, Flicker P F, Mayeda A, Krainer A R, LeStourgeon W M. Mol Cell Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAfee J G, Soltaninassab S R, Lindsay M E, LeStourgeon W M. Biochemistry. 1996;35:1212–1222. doi: 10.1021/bi951974k. [DOI] [PubMed] [Google Scholar]