Abstract
Objective
Inefficiencies in clinical trial data collection cause delays, increase costs, and may reduce clinician participation in medical research. In this proof-of-concept study, we examine the feasibility of using point-of-care data capture for both the medical record and clinical research in the setting of a working clinical trial. We hypothesized that by doing so, we could increase reuse of patient data, eliminate redundant data entry, and minimize disruption to clinic workflow.
Design
We developed and used a point-of-care electronic data capture system to record data during patient visits. The standards-based system was used for clinical research and to generate the clinic note for the medical record. The system worked in parallel with data collection procedures already in place for an ongoing multicenter clinical trial. Our system was iteratively designed after analyzing case report forms and clinic notes, and observing clinic workflow patterns and business procedures. Existing data standards from CDISC and HL7 were used for database insertion and clinical document exchange.
Results
Our system was successfully integrated into the clinic environment and used in two live test cases without disrupting existing workflow. Analyses performed during system design yielded detailed information on practical issues affecting implementation of systems that automatically extract, store, and reuse healthcare data.
Conclusion
Although subject to the limitations of a small feasibility study, our study demonstrates that electronic patient data can be reused for prospective multicenter clinical research and patient care, and demonstrates a need for further development of therapeutic area standards that can facilitate researcher use of healthcare data.
Introduction
Technological advances in the last 20 years have the potential to strengthen links between patient healthcare and clinical research. Many authors have endorsed secondary uses of healthcare data, including for research purposes; 1–18 however, this potential remains largely untapped. Data that could benefit patients, physicians, investigators, regulators, and the biopharmaceutical industry remain sequestered in disparate databases, stored as narrative text, or confined to paper records. 19 The current system also impedes research activities by creating significant amounts of inefficiency and delay, the expense of which threaten to make clinical research prohibitively expensive and slow. 8 Further complicating this problem is a lack of therapeutic area data content standards, perpetuating a lack of semantic specificity at the content and clinical definition levels needed to support interoperability. 2,3,20–23 Given the existing technological capacity to support interoperability and the availability of a core set of data standards for clinical research, it is essential to explore those critical factors still needed in order to foster the convergence of healthcare and clinical research informatics. 4
The Single Source project, an initiative sponsored by the Clinical Data Standards Interchange Consortium (CDISC), 24 seeks to reduce burdens associated with clinical data capture at investigational sites. A standards-based, technology-enabled process using electronic source (eSource) data collection and interchange (eSDI) 25 has the potential to reduce transcription errors, increase sponsor and site personnel efficiency, facilitate information flow, and improve timeliness of data.
In the Single Source proof-of-concept study, we sought to better understand the challenges of using data captured in the healthcare setting in conjunction with appropriate standards to directly support clinical trials. Our goals were to 1) examine the feasibility of using Single Source data capture to increase reuse of clinical information; 2) eliminate redundant data collection, medical record abstraction, and multiple data transcription and entry processes; 3) minimize disruption to clinical workflow; and 4) assess the capability of open standards to enable reuse of clinical information for large-scale prospective multicenter clinical research.
We hypothesized that by introducing data standards and technology that enabled single source data capture within the existing clinical workflow, we would eliminate or reduce redundant data collection and entry without disrupting existing clinical practice.
Background
Existing paper-based processes for collecting clinical trial data are inefficient and error-prone, employing workflows that may involve multiple iterations of data transcription, entry, and validation (▶). Increasing use of current electronic data capture (EDC) methods may improve certain aspects of the eClinical trial process, but each EDC system requires medical record abstraction and has different processes and requirements. Worse, an investigational site conducting five clinical trials may have to use five different EDC systems, one hosted by each sponsor (or their representative).
Table 1.
Table 1 Common Clinical Trial Data Processing Procedures
| Data entry step | Purpose |
|---|---|
| Initial data capture | Record data from patient encounter |
| Transcription | Patient encounter data transcribed from notes, worksheets, and dictation and entered into primary medical record |
| Medical record abstracted | Data transcribed to CRF |
| CRF sent to data center | - |
| First data entry | Data from CRF entered into database |
| Second data entry | Data from CRF re-entered into database in separate step as quality assurance measure 8 |
| Data validation and “cleaning” | Data entered in center database programatically checked for error/inaccuracy. Discrepancies are sent to clinical sites in the form of queries requiring response. |
| Query response | Sites respond to queries by checking source documents and (if necessary) updating or amending CRF; response is submitted to Data Center. Database is updated. |
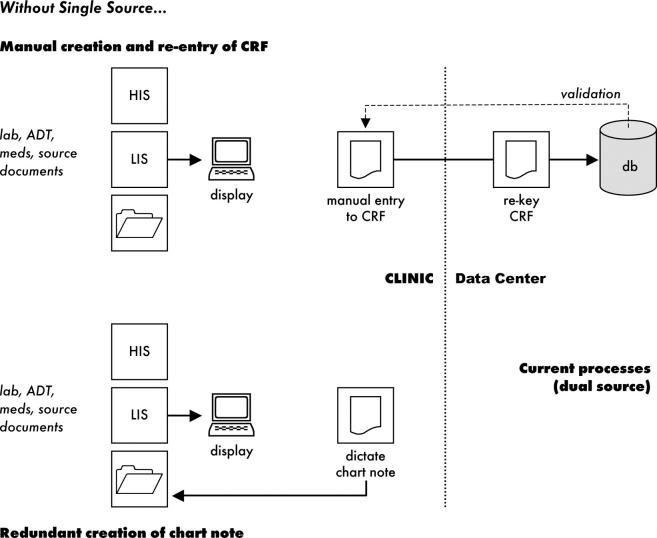
In a parallel process used in many clinics (including the subject of our research), data collected for clinical research patients are also dictated and recorded for transcription to the patient’s medical record. To create the final clinic note documenting a patient encounter, the transcribed draft encounter note is edited by the clinician, using paper notes produced during the encounter as well as electronic notes (including lab reports) available via an institutional browser interface. Once the clinic note is finalized, it is posted to the medical center’s clinical document management system and added to the clinical record. Thus, data that may already be available electronically are keyed twice in the healthcare setting, transcribed to a paper case report form (CRF), and then keyed twice in the research setting (▶).
Figure 1.
Without Single Source
Large-scale direct use of healthcare data for research, although advocated by many, 1–18 has thus far eluded researchers. 9 Successful implementations described in the literature 6,26 cite workflow incompatibility, additional research data requirements, and regulatory differences as challenges. 7 Successful integration of research into the patient care setting depends on a full understanding of inherent challenges, and on finding solutions adapted to the realities of the clinical, technological, and regulatory environment.
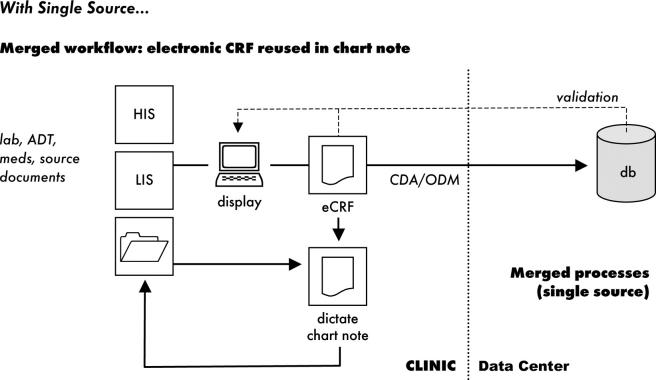
Use of data standards is essential for any successful implementation of a data collection system designed to reuse patient data. Many systems allow reuse of patient care data, 6,7,10,12,13,18,27–36 but with rare exceptions 30,36 most examples consist of institution-specific approaches that do not use standards and thus lack the broad interoperability required for multicenter trials. Multicenter trials require data from different sites to be submitted to a central data center, with whom the site’s relationship may exist for only a single trial; for reuse of patient care data to be feasible, data collection methods must be easy to implement and use, and must minimize disruption at the clinical site. Standards have the potential to make this possible by allowing investigational sites to use existing systems without the burden of data transformation (▶).
Figure 2.
With Single Source
Methods
Study Design
To investigate these issues, this project was designed as a proof-of-concept study. Following the example of other informatics implementation research, we employed a single case-study design. 37 In doing so, we hoped to assess the underlying causes, possibly process-related and social in nature, of the phenomenon under study. 38 We therefore used a qualitative, participant observer approach to identifying the challenges of using a single source of data for research and patient care. Our study was developed in 3 stages, yielding 3 corresponding areas of findings: 1) document analysis (including study of the data-containing documents themselves as well as the workflow associated with their creation, management, and dissemination; 2) technical implementation, including tools and use of existing data standards; and 3) data collection on two test cases. Because we sought to gain detailed information on the challenges of data reuse in prospective multicenter trials, the process and logistics of designing and implementing the proof-of-concept were of primary interest, while the application of the system to a large number of cases was considered more appropriate for a later study. We examined five cases as part of the document analysis, and used two live test cases. Throughout the design, we were conscious of ensuring compliance with relevant regulations for regulated clinical research.
The STARBRITE Study
The Single Source proof-of-concept study was conceived as a parallel study to an ongoing clinical trial, the Strategies for Tailoring Advanced Heart Failure Regimens in the Outpatient Setting: Brain Natriuretic Peptide versus the Clinical Congestion Score (STARBRITE) clinical trial. 39 By incorporating our proof-of-concept into an existing multicenter clinical trial, we were able to explore challenges arising from a working trial design and data-collection environment, rather than using simulations or “dummy” data sets. By design, our data collection and management system was redundant to those already in place for the STARBRITE trial.
Selection criteria were: 1) a small, investigator-initiated study that was not being conducted for the purpose of regulatory approval for marketing authorization (although the design will support regulated clinical studies); 2) a relatively small number of sites (one of which was in-house at Duke University Medical Center); 3) a substantial patient enrollment and follow-up period that would accommodate the 1.5-year project plan; and 4) an investigator willing to actively collaborate on the project. Although STARBRITE was a multicenter clinical trial, the proof-of-concept was implemented at only one site to facilitate immersion in the workflow and challenges of the setting. The proof-of-concept study was performed over a period starting in March of 2003 and ending in December of 2004.
The design and aims of the STARBRITE study have been previously described. 39 Briefly, patients with acute decompensated heart failure at participating centers were screened for participation in the study, which randomly assigned subjects to one of two fluid management strategies tailored to specific symptoms and to levels of brain natriuretic peptide (BNP). Subjects were then followed to examine the effect of fluid management strategy on the endpoint of 90-day death and rehospitalization. Patient follow-up consisted of a series of monitored outpatient visits and telephone calls. The STARBRITE study used several data collection forms designed for these different stages of data collection; however, the proof-of-concept addressed only the data gathering requirements of the clinic form used for follow-up visits.
Standards Used
The CDISC Operational Data Model (ODM) and the Health Level Seven (HL7) Clinical Document Architecture (CDA) are standards for the exchange of clinical information. Both use Extensible Markup Language (XML) syntax from the World Wide Web Consortium. 40
The CDA standard was chosen because it is designed for the exchange of clinical documents and offers ease of implementation over a wide range of technical infrastructures, from primitive document imaging systems to sophisticated electronic health records (EHRs). The document exchange concept used by the CDA rests on the premise that there is inherent unity in the set of information to which a clinician affixes a signature, and that this, consequently, creates a natural unit of data exchange. All clinically relevant, legal information in the CDA can be displayed directly in a Web browser using a single style sheet. The degree to which this information is coded for machine processing is variable according to the sophistication of the document-generation application. 41,42 These features and workflows fit the use case for the proof-of-concept.
The ODM is a database insertion schema that uses XML to mimic the data fields of a clinical research database; there is by design a great deal of latitude for accommodating different content. The ODM standard was chosen because it is designed for the reporting of results between data collection sites, data centers, other research organizations, and the sponsor and the FDA, with the goal of easing data management burdens at both ends of the transaction. Application of ODM to data collection also allows sites to deploy data-gathering applications across disparate studies among research organizations capable of supporting automated management of collected data, and the ODM readily supports regulations relevant to electronic record retention, including audit trails and archival requirements for electronic data.
CDA and ODM differ in purpose, application, and scope. For example, CDA requires human-readable reproduction of legally-authenticated content, without fidelity regarding page layout. ODM does not prescribe layout of authenticated content, but can provide appropriate layout by linking to a defined style sheet. Sequence is significant within CDA, but not in ODM. Other differences stem from the life cycles and constituencies of the two different specifications. ODM references the study identifier and the current version of the study metadata. These data may exist in a patient chart, but are not currently expected or explicitly identified within CDA. CDA, on the other hand, in addition to a document title contains a clinical document type code that classifies the document for retrieval (Progress Note, History & Physical, etc.); such metadata are not relevant within a clinical trial database. Such differences need not be bridged; rather, it should be recognized that CDA and ODM form two overlapping sets of data, neither of which subsumes the other.
The ODM is bound to the actual implementation database schema used to collect data for statistical analysis. Thus, the same concept can be represented in multiple ways in different clinical trials, depending on the decisions of the team that designed the database and upon the trial itself. However, CDA uses the HL7 Reference Information Model (RIM) to represent semantics independently from the implementation applications consuming, generating, and storing data. The two standards approach the use of controlled vocabulary from different perspectives. CDA uses RIM structures to supply context to externally-defined codes, whereas ODM defines its own structures and references clinical database or SAS field names with optional translation to external code sets. Consider representation of the following data in the CDA and ODM sample fragment (▶). Note that the fundamental construct of a data type for the physical quantity of “weight” differs in CDA and ODM. In ODM, weight is a locally-defined concept integrated with the type of unit; in CDA, weight is a type of physical quantity with units defined by an external vocabulary. 43 The goals of our study did not require a full mapping between generic ODM and generic CDA; such a mapping, given the different approaches to semantics within the two specifications, would have been impossible. Harmonization efforts subsequent to this research have been undertaken between ODM and HL7 RIM. 44
Figure 3.
CDA and ODM sample fragment
Use of XML syntax is essential to the design of this study but is not itself sufficient for semantic interoperability beyond the constraints of a single trial. Given our relatively limited goals, we decided to use the unique strings established for the clinical data management database as unique keys that would allow reuse of data between an electronic Case Report Form (eCRF) for the clinical study and the clinic note. Thus, in the final information design, the “transform” between CDA and ODM is less a transform than an “extract and populate,” acting on active data fields and not on predefined structures.
Scope of the Proof-of-Concept Study
Study objectives constrained the degree of integration with legacy tools. Potential integration points were 1) administrative data, 2) laboratory data, 3) the clinical trials data management system (CDMS), and 4) the Duke Clinic clinical document repository. The first two points of integration could have been achieved with routine (although labor-intensive) interfaces to patient data maintained in hospital scheduling and laboratory information systems; however, these were considered to be outside the scope of our project. The third and fourth points of integration formed the critical core of the study. Integration with CDMSs had been previously achieved via CDISC ODM. Thus, our endpoint was production of the ODM. Because the CDISC Submission Data Model had not been defined at project inception, we used the semantics defined in the CDMS and carried their keys as identifiers into both the eCRF and the clinic note.
The Duke clinical document repository stores simple text notes. For this project, these text notes were stored as XML to support extensive reuse. Notes from the most recent prior visit were used to pre-populate data fields in the current note, allowing clinicians to enter only those data that had changed from one visit to the next, thus reducing transcription and data entry burdens.
Study Conduct
We describe study conduct in 3 phases: 1) document and workflow analysis, 2) technical implementation, and 3) data collection. The document and workflow analysis was conducted first to define the requirements for the proof-of-concept system. After data content was defined, we then chose standards and integrated them into the workflow. Lastly, a proof-of-concept was built and used to collect data on two live cases.
Document and Workflow Analysis
Document analysis is the canonical method for specifying requirements when transforming paper-based records into machine-processable electronic records. 45,46 Document analysis for information design performs a function parallel to that of use case analysis for application design, and is typically conducted on information artifacts and workflow processes to create a set of requirements for the design of structured electronic text, a category that includes both the eCRF and the standards-based, structured electronic clinical note. For our study, five samples of data-populated CRFs and associated clinic notes were obtained, and the clinic was visited to observe use of the forms, and document analysis sessions were held both before and after on-site visits. During these sessions, we examined samples, itemized CRF data fields, and created a table indicating fields that were used in both CRFs and clinic notes, and those that were unique to one or the other. During this stage of the study design, all documents were redacted according to HIPAA anonymization requirements.
Aside from examining the documents themselves, the most critical component of document analysis lies in understanding the business processes surrounding those documents. We therefore observed documentation processes for the CRF and clinic note at the Duke Clinic. We initially assumed that the clinic note would be considered the source document and that the CRF was the derivative. We also expected to design a Single Source process that extracted data from the note and used that data to pre-populate the CRF.
Technical Implementation
Although technology was not the focus of our research, a standards-based system intended to facilitate reuse of patient care data was built to support the workflow described in the document analysis. We first created an eCRF that resembled the paper CRF used in the STARBRITE study. Unlike many EDC tools currently used in clinical trials, the Single Source process does not rely on a proprietary data format. Instead, it gathers data in an industry-standard CDA document. This process formed the “single source” used to pre-populate a clinic note, which was then completed in the usual fashion. The same source data were transformed to a CDISC ODM for transfer to the clinical trial database at the Duke Clinical Research Institute (DCRI). The design of the clinic note application was predicated on reuse of CRF data based on our findings from the workflow analysis. For overall clinic efficiency, we aimed to pre-populate data available from prior clinic notes, as well as data previously collected on CRFs.
It should be noted that different clinicians may contribute to a single encounter and document patient data at varying times. Within the life cycle of a single document, nurses collected the patient’s vital signs, investigators interviewed the patient, and a single electronic document was opened, modified, saved, and closed multiple times before being finalized. Thus, the system was designed to support different clinicians as they collaborated on the same case, preventing them from overwriting each other’s edits and tracking changes to the form by different team members.
The implementation matched identified clinic workflow with four primary tasks: 1) CRF data collection; 2) transformation to ODM and clinic note; 3) achieving the ODM necessary for CDMS integration; and 4) integration of the completed clinic note back into the clinic document repository. Although the CRF data entry form information design could theoretically have been ODM, in practice, the logical choice was CDA: ODM data was more easily extracted from CDA than the converse because of the manner in which CDA handles narrative text and RIM semantics, and the initial single-source data collection instrument more closely resembled the clinic note than the database structure reflected in the ODM. The source CDA (CDA CRF) was transformed to the ODM for integration into the CDMS and was then transformed into a clinic note (CDA CN) for integration with data from the previous clinic note. The pre-populated clinic note was completed by the physician and stored in the document repository.
It was evident during system design that the nature of the workflow and the kinds of data collected would influence the tools used. We sought to capture information that ranged from highly structured and constrained data collected for the clinical trial to the more unstructured and textual data captured during clinic visits. For this reason, we allowed the system’s tools to be adapted to varying needs and data collection practices at different stages of the workflow (▶). Microsoft InfoPath, a structured data capture application, replaced the paper CRF used to collect data for the clinical trial. Implementation mimicked the paper design, thus minimizing training requirements. Further, the application provided business and data rules enforcement; e.g., checking that required entries were present and that data were within prescribed ranges. As an XML application based on XML Schema Definition (XSD), the InfoPath design supported simple transformation to the industry-standard CDA output (▶).
Table 2.
Table 2 Tools Used
| Task | Tool |
|---|---|
| Client data collection (CRF) | Microsoft Office InfoPath |
| Document management | Microsoft Windows SharePoint Services, included in Microsoft Windows Server 2003 |
| Transformation and business process management | Microsoft BizTalk Server 2004 |
| Clinic notes editor | Microsoft Word |
Figure 4.
Microsoft Office InfoPath Clinical Research Form
The SharePoint Services tool supported check-in/check-out for editing and managed multiple document versions for tracking and audit. The BizTalk Server 2004 supported integration between different applications and the transformation of data using industry standards such as Web Services, XML, XSD and XSLT. MS Word included XML support, so that clinicians could use the familiar word processing interface to modify and complete clinic notes, while the underlying structure and data definition supported CDA output (▶).
Figure 5.
Business process integrating the document management system with the CTMS and the clinic notes repository
Results
This proof-of-concept case study produced a wealth of detailed information about reuse of patient care data. The proof-of-concept was successful, in that the process for reuse of patient data was tested in two live patient encounters and worked seamlessly within the clinic workflow. In addition, capturing data via the CDA allowed automated transformation and transfer of data within the CDISC ODM. The proof-of-concept also succeeded in eliminating redundant data capture and entry for the two test cases.
However, the most valuable contribution of this study lies in the information gained during design and implementation. From our preliminary analysis, we determined that there was significant overlap between clinic note and CRF (approximately 75% of data fields from five sample cases), but that neither constituted a superset of the other. Some data critical for the STARBRITE study were not included in the clinic note (e.g., “most recent BNP,” and observations such as “greater than or equal to 2 pounds weight gain from dry weight”). In general, data that appeared in the CRF but were absent from the clinic note included: 1) study-specific metadata, such as visit number in the sequence of protocol-required visits; 2) data specific to the clinical trial was not typically gathered in the course of routine clinic visits (e.g., BNP value); or 3) data typically incorporated in patient charts via different report (e.g., test results that might be included as a laboratory report but not necessarily incorporated into a clinical note). A detailed summary of our findings are contained in ▶. These findings from the five clinic notes sampled confirmed our general expectations; i.e., there was less consistency in the structure and contents of narrative notes than in structured data entry on a paper CRF, and that any attempt to automate the reuse of data must take this into consideration.
Table 3.
Table 3 Document Analysis: Results Summary
| Document Section | Finding |
|---|---|
| Problem list | Present in all |
| Current medications | Present in all |
| Physical exam | Present in all |
| Laboratory findings | Present in all |
| Assessment and plans | Present in all |
| Patient profile | Present in all but 2 (both were 10-day visits) |
| Cardiovascular risk factors | Present in all but 2 (both were 10-day visits) |
| Procedures | Present in all but 2 (both were 10-day visits) |
| Interval history/history of present illness | Categories do not coexist, but all notes have one or the other |
| Referring physician | Present in all but 1 |
| Family history/Past medical history | Present in 1 (a 10-day note) |
| Allergies | Present in all but 1 |
| Social history | No pattern |
| Echocardiogram | Present in 1 |
Our analysis also showed that even in cases where the same data were present in both clinic note and CRF, presentation and sometimes even values differed (▶). For example, medications might be recorded using either generic or brand names. Different units of dosing might also be used (the CRF used total daily dosage, whereas the clinic note expressed dosage in quantity per dose × doses per day). Exposition of data also differed widely: for instance, vital signs were recorded in a table on the CRF but were captured as part of a narrative in the clinic note.
Figure 6.
Data representation in Case Report Form and Clinic Note
A second major finding from our document analysis was that the degree of data reuse between clinic note and preceding chart note was greater than between clinic note and CRF. During the design phase, clinicians reported that they found it useful to have the form pre-populated with patient data from the previous visit. Thus, from an implementation perspective, we focused on integration with the full flow of clinical information, between successive notes, as well as between note and CRF. Pre-populating screens with data from the previous visit proved to be a helpful feature to clinic staff during test cases and was preferred over screens that did not pre-populate.
It is important to balance the natural tension between the potential dangers of pre-population and the benefits of making data capture as efficient as possible for clinicians. As Hirschstick notes, there are a number of potential problems associated with pre-populated clinic data. 47 Our approach to this issue required the clinician to verify all pre-populated data. This requirement was enforced by the system workflow, in which each data field had to be visited by the clinician and verified or changed. Our workflow analysis showed that, contrary to our initial assumptions, the CRF is completed before the clinic note is finalized, sometimes days in advance. Not only was the CRF more granular (i.e., more detailed and stringent in information-gathering requirements), but it constituted a fixed data set. The clinic note, on the other hand, had a core set of expected data but could contain other material (e.g., findings from diagnostic procedures ordered on the day of the clinic visit, but which were not available until several days later). Thus, the eCRF serves as the source document and the clinic note is a derivative of the eCRF, previous clinic notes, and other data sources.
Personal idiosyncrasy also played a role in the workflow: the principal investigator reported that she preferred to have all results in hand before composing the note, and indicated that she often preferred to wait until a few days after the patient encounter in order to consider the case before finalizing her diagnosis, assessment, and plan. Thus, the final note (the permanent record of the encounter) was often completed a few days after the clinic visit.
The logistics of accomplishing the design and implementation of the study also yielded interesting findings. We found that involvement of the clinic study coordinator was essential at each stage of the project. Document analysis required the study coordinator to redact CRFs and corresponding clinic notes, and to provide input at document and workflow analysis meetings. In addition, the study coordinator was instrumental in providing in-service training for clinical staff, coordinating technology installation, and scheduling test cases. During these stages, approximately 20% of the study coordinator’s time was required for conducting the proof-of-concept. Although this was expected given the decision to conduct the proof-of-concept in parallel with the ongoing STARBRITE trial, it underscores the importance of clinician engagement with the design and implementation of such systems. Our success in integrating new technology into the clinic workflow is directly attributable to the high degree of clinician involvement in the design effort, and to a design team that realized the importance of clinician involvement and had the experience to leverage it for the design.
Discussion
Our findings regarding the proportions of data found in both CRF and clinic note are specific to the STARBRITE study. Because of wide variation in design (including CRF design), study aims, areas of therapeutic interest among different protocols, and the degree to which protocols collect data outside of the clinical standard of care, such findings may not be generalizable and will vary according to each implementation environment.
The finding that the CRF was completed before the clinic note has implications for near-term reuse of patient care data. At present, there are two different basic approaches to clinical data capture. Some institutions have chosen structured data capture and record encounter data at the time of the patient visit. In this scenario, such structured data is used to generate the text encounter report. For institutions that capture structured data, extracting data from the EHR or other systems is a viable approach. However, institutions that do not capture structured data from patient encounters instead record narrative text either electronically or via dictation. Reuse of healthcare data at such institutions will require a different approach: a structured form of data capture or natural language processing will be necessary. The fact that multiple workflows exist (and will for some time) complicates implementation of a general approach for multicenter clinical trials.
In the course of this study, we experimented with various approaches to ODM, but eventually selected a design that reflected the single-document concept of CDA and that identified semantics using STARBRITE field identifiers already used in the actual study database. If the ODM for STARBRITE had used the data definitions in the CDISC submission data model, a semantic mapping to CDA and the RIM would have been possible, although it would still have been specific to STARBRITE. Use of a submission dataset could have provided a common semantic; this would constitute a good test for future projects. The lack of a higher-level definition of semantic structures to create a reusable map that would retain its usefulness beyond a single trial protocol led directly to efforts to create a Clinical Research domain model under the RIM. Partly as a result of this project, the CDISC ODM has now been mapped to the HL7 RIM. 48
The semantics, however, warrant consideration at a higher level of abstraction. A key finding is that in the structured narrative note, the metadata lacked the semantic structures and controlled terminology required for direct machine processing. Thus, a manual mapping of data elements was necessary. Unfortunately, this situation exists for healthcare data in narrative text and structured form, as standards for the clinical content and the expression of that content in an electronic format reusable across computer applications do not yet exist for most therapeutic areas in healthcare. As long as this situation persists, there will be no large-scale interoperability, and reuse of healthcare data in general will exist as isolated implementations.
During the course of planning and implementing this study, several unanticipated challenges were encountered. First, there was an unforeseen impact on resources at the clinic. Our proof-of-concept study was conducted in parallel to an ongoing clinical trial. We had initially anticipated that clinic personnel would be available and willing to volunteer time and efforts for this project. However, in the busy setting of the clinic, this proved not to be the case. It was necessary for this project to fund a research assistant at the site to carry out study-related activities to off-set the study coordinator time expended on the proof-of-concept. This proved crucial to successfully using the parallel data collection process central to this study. As mentioned above, a second interesting finding arose from the fact that, contrary to our initial expectations, the study CRF was completed several days in advance of the clinic note, which meant that our anticipated methods of extracting data from the health record would not be practicable. We therefore redesigned our system to match actual clinic workflow, as described in the Methods section. A final problem arose when our system was developed as a “thick-client,” requiring software to be installed on the computer at the site. To maintain our validated clinic environment, the entire proof-of-concept application was installed on a separate server and laptop for data collection, an arrangement that would be suboptimal for a large-scale multicenter trial.
Limitations
Because this study was designed as a proof-of-concept with limited implementation, we are not able to report performance of the system throughout an entire clinical trial or for hundreds of patient encounters, or at multiple sites. Thus, results may not be generalizable outside of this particular therapeutic setting. This proof-of-concept focused on only one of several types of data collection instrument used in the study, and correspondingly examined only one workflow pattern. Our study had limited technical objectives and was not intended to replace existing systems and methods. The technology used was built outside the clinical and research environments and then tested on-site in parallel with existing technology. Live feeds from existing laboratory and hospital information systems or EHRs were not attempted.
In addition, reusing healthcare data required a manual nonsemantic mapping between the CRF, Clinic Note, ODM, and CDA for the data collected by the study. This is neither desirable nor scalable for use in prospective multicenter clinical research; however, until standards for content and computable semantics exist for health care data, it will remain a necessity. The environment of an academic research organization (such as the DCRI) that is part of the same medical center as the clinic where the study is performed does not necessarily reflect environments where most care is delivered or most research is accomplished. Although we did not test the accuracy of pre-populated data as such testing lay outside the scope of our study, future pilot studies should address the potential challenges of using pre-populated data in the clinical research environment. Also, conditions may have changed in the two years since the last test case was examined.
Future Directions
Three major ongoing projects within CDISC and the HL7 Regulated Clinical Research and Information Management (RCRIM) Technical Committee will raise the level of automation and semantic interoperability for future single-source projects: 1) the development of an abstract information model, specifically a domain analysis model for clinical research; 2) the definition of a standard for electronic protocol representation; and 3) the development of an integration profile by CDISC, working through Integrating the Healthcare Enterprise (IHE). In addition, within HL7, individual therapeutic areas and corresponding stakeholder groups are pursuing work to specify and define the content and relationships for individual therapeutic area domains.
The STARBRITE Single Source proof-of-concept study helped prompt the development of the domain analysis model (now called the Biomedical Research Integrated Domain Group [BRIDG] model 44 ) and will benefit from its completion and implementation. The elements comprising the standard for protocol representation are all being modeled into the BRIDG, in addition to all CDISC and RCRIM standards, to ensure harmonization among these standards, between the relevant HL7 and the CDISC clinical research standards and, in general, across clinical research and healthcare.
Multiple follow-up pilots aimed at building upon the successes of this CDISC Single Source project are underway. An Integrating the Healthcare Enterprise (IHE) profile was demonstrated within various commercial EHR systems for five use cases (entering data once for secondary or reuse downstream): clinical research using an EDC system, clinical research with lab and imaging data, pharmacovigilance, clinical study registry, biosurveillance (HIMSS 2007-New Directions Interoperability Demonstration); 20 organizations participated. In the future, Single Source projects will drive the design of both CRF and clinic note from an electronic study protocol. (▶).
Table 4.
Table 4 Future Directions
| Development and implementation of content standards including clinical definition of terms and ontologies for medical specialty areas |
| Implementation of Single Source in additional sites (including vendors, CROs and sponsors) |
| Investigation of single source methodology for reuse of EHR data |
| Development of CDISC submission data set and semantic markup for more general reuse |
| Development and implementation of terminology code lists to increase semantic interoperability |
| As electronic protocol specification develops, derive CRF and note input templates from protocol |
| Explore use of large repositories of industry-standard clinic notes to encompass inclusion/exclusion testing |
| Add hypertext link (URL) from collected research data to the full clinic note (eSource) |
| Investigate use of natural language processing to extract data from narrative notes to support clinical trials |
| Develop guides for testing Single Source in a platform-independent integration profile |
Conclusions
Despite limitations inherent to a small proof-of-concept design, our study provides evidence that, given appropriate industry-standard data design, readily available desktop tools can promote reuse of data gathered either during clinical trials or in the course of patient care. In order to ensure success, Single Source implementation must be flexible with regard to data collection (so that individual trials can be accommodated), comprehensive in its approach to data reuse, and must be integrated with minimal disruption into existing clinical workflows at the investigational site. Therapeutic area content and messaging standards are crucial to the integration of research and patient care. Considering the elimination of redundant data collection, data entry, and medical record abstraction realized with this proof-of-concept, it is reasonable to predict that improvements in efficiency in data gathering for research may speed implementation of EHRs and generally contribute to increased quality of health care and clinical research.
Footnotes
This work was sponsored by the Duke Clinical Research Institute (DCRI), Microsoft Corporation, the Clinical Data Interchange Standards Consortium (CDISC); and three pharmaceutical companies, including Novartis and Merck & Co. Technology partners included Microsoft Corporation (primary), Digital Infuzion, and Topsail Technologies. The production of the manuscript was partially supported by the Duke Clinical and Translational Science Award, UL1-RR024128. The authors wish to acknowledge Jonathan McCall of the Duke Clinical Research Institute, who edited this manuscript through revision and review stages.
References
- 1.Justice AC, Erdos J, Brandt C, Conigliaro J, Tierney W, Bryant K. The Veterans Affairs Healthcare System Med Care 2006;44(S8). [DOI] [PubMed]
- 2.Jordan K, Porcheret M, Kadam UT, Croft P. Use of general practice consultation databases in rheumatology research Rheumatol 2006;45:126-128. [DOI] [PubMed] [Google Scholar]
- 3.Lusignan S, van Weel C. The use of routinely collected computer data for research in primary care: opportunities and challenges Fam Pract 2005;23:253-263. [DOI] [PubMed] [Google Scholar]
- 4.Joffe M, Chapple J, Beard RW. Making routine data adequate to support clinical auditData collection should be integrated with patient care. BMJ 1995;310:655d-666d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellings P. A rich source of clinical research data: medical records and telephone logs J Pediatr Health Care 2004;18:154-155. [DOI] [PubMed] [Google Scholar]
- 6.Murray MD, Smith FE, Fox J, Teal EY, Kesterson JG, Stiffler TA, et al. Structure, functions, and activities of a research support informatics section J Am Med Inform Assoc 2003;10:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm MB, Rogers JC, Burgio LD, McDowell BJ. Observational data collection using computer and manual methods: Which informs best? Top Health Inf Manag 1999;19:15-25. [PubMed] [Google Scholar]
- 8.Musen MA, Carlson RW, Fagan LM, Deresinski SC, Shortliffe EH. T-HELPER: Automated support for community-based clinical research Proc Annu Symp Comput Appl Med Care 1992:719-723. [PMC free article] [PubMed]
- 9.Powell J, Buchan I. Electronic health records should support clinical research J Med Internet Res 2005;7:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrino AC, Luther MA, Phillips DB, Levin FL. A multimedia perioperative record keeper for clinical research J Clin Monit 1996;12:251-259. [DOI] [PubMed] [Google Scholar]
- 11.Pfister M, Akyildiz S, Gunhan O, Maassen M, Rodriguez JJ, Zenner H, Apaydin F. A patient database application for Hereditary Deafness Epidemiology and Clinical Research (H.E.A.R.): an effort for standardization in multiple languages Eur Arch Otorhinolaryngol 2003;260:81-85. [DOI] [PubMed] [Google Scholar]
- 12.Tierney WM, Miller ME, Hui SL, McDonald CJ. Practice randomization and clinical researchThe Indiana Experience. Med Care 1991;29:JS57-JS64. [PubMed] [Google Scholar]
- 13.Ward NS, Snyder JE, Ross S, Haze D, Levy MM. Comparison of a commercially available clinical information system with other methods of measuring critical care outcomes J Crit Care 2004;19:10-15. [DOI] [PubMed] [Google Scholar]
- 14.Nordyke RA, Kulikowski CA. An informatics-based chronic disease practice: case study of a 35-year computer-based longitudinal record system J Am Med Inform Assoc 1998;5:88-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christo GG, Marianus BVP, Krishnan L, Iyer RS, Venkatesh A. Computerised neonatal case records: A four year experience Indian Pediatr 1992;29:173-180. [PubMed] [Google Scholar]
- 16.Holmin C, Krakau CE, Strom B. Computerized patient record system for the glaucoma ward Acta Ophthalmol 1991;69:444-449. [DOI] [PubMed] [Google Scholar]
- 17.Drolsum L, Davanger M, Haaskjold E. Cataract surgery computer-based registration and analysis of data Acta Ophthalmol 1993;71:477-481. [DOI] [PubMed] [Google Scholar]
- 18.Wong IC, Murray ML. The potential of UK clinical databases in enhancing paediatric medication research Br J Clin Pharmacol 2005;59:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman TB, Brown A, Easterling MJ. Obstacles and approaches to clinical database research: experience at the University of California, San Francisco Proc Annu Symp Comput Appl Med Care 1994:568-572. [PMC free article] [PubMed]
- 20.President’s Information Technology Advisory Committee Revolutionizing Health Care Through Information Technology: Report to the President 2004. June. Available at: http://www.nitrd.gov/pitac/reports/20040721_hit_report.pdf. Accessed on 01/19/2007.
- 21.Cimino JJ. Review paper: coding systems in health care Methods Inf Med 1996;35:273-284. [PubMed] [Google Scholar]
- 22.Los RK, Roukema J, van Ginneken AM, de Wilde M, van der Lei J. Are structured data structured identically? Methods Inf Med 2005;44:631-638. [PubMed] [Google Scholar]
- 23.McKee M, Dixon J, Chenet L. Making routine data adequate to support clinical audit BMJ 1994;309:1246-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Single Source project. Clinical Data Interchange Standards Consortium Web site. Available at: http://www.cdisc.org/single%5Fsource/. Accessed on 01/19/2007.
- 25.Electronic Source Data Interchange Group Leveraging the CDISC standards to facilitate the use of electronic source data within clinical trials 2006. Version1.0, November 20.
- 26.Los RK, Van Ginneken AM. Experiences with extracting structured patient data for use in clinical research Stud Health Technol Inform 2002;93:119-126. [PubMed] [Google Scholar]
- 27.Collen MF. Clinical research databases—a historical review J Med Syst 1990;14:323-344. [DOI] [PubMed] [Google Scholar]
- 28.Kahn MG. Clinical databases and critical care research Crit Care Clin 1994;10:37-51. [PubMed] [Google Scholar]
- 29.Nordyke RA, Kulikowski CA. An informatics-based chronic disease practice JAMIA 1998;5:88-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor GS, Muhlestein JB, Wagner GS, Bair TL, Li P, Anderson JL. Implementation of a computerized cardiovascular information system in a private hospital setting Am Heart J 1998;136:792-803. [DOI] [PubMed] [Google Scholar]
- 31.Ricciardi TN, Lieberman MI, Kahn MG, Masarie Jr FE. Clinical terminology support for a national ambulatory practice outcomes research network Proc AMIA Symp 2005:629-633. [PMC free article] [PubMed]
- 32.Embi PJ, Jain A, Clark J, Harris CM. Development of an electronic health record-based clinical trial alert system to enhance recruitment at the point of care Proc AMIA Symp 2005:231-235. [PMC free article] [PubMed]
- 33.Chambers IR, Barnes J, Piper I, Citerio G, Enblad P, Howells T, et al. BrainIT Group BrainIT: a trans-national head injury monitoring research network Acta Neurochir Suppl 2006;96:7-10. [DOI] [PubMed] [Google Scholar]
- 34.Mosis G, Vlug AE, Mosseveld M, Dieleman JP, Stricker BC, van der Lei J, Sturkenboom MC. A technical infrastructure to conduct randomized database studies facilitated by a general practice research database JAMIA 2005;12:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy SN, Rabbani UH, Barnett GO. Using software agents to maintain autonomous patient registries for clinical research Proc AMIA Annu Fall Symp 1997:71-75. [PMC free article] [PubMed]
- 36.Flack JR. Seven years experience with a computerized diabetes clinic database Medinfo 1995(8 Pt 1):332. [PubMed]
- 37.Pipino LL, Lee YW, Wang RY. Data quality assessment Comm ACM 2002;45:211-218. [Google Scholar]
- 38.Koppel R, Metlay J, Cohen A, Abaluck B, Localio A, Kimmel S, Strom B. Role of computerized physician order entry systems in facilitating medication errors JAMA 2005;293:1197-1203. [DOI] [PubMed] [Google Scholar]
- 39.Shah MR, Claise KA, Bowers MT, Bhapkar M, Little J, Nohria A, et al. Testing new targets of therapy in advanced heart failure: the design and rationale of the Strategies for Tailoring Advanced Heart Failure Regimens in the Outpatient Setting: BRain NatrIuretic Peptide Versus the Clinical CongesTion ScorE (STARBRITE) trial Am Heart J 2005;150:893-898. [DOI] [PubMed] [Google Scholar]
- 40.World Wide Web Consortium (W3C) Extensible Markup Language (XML), 1.0. Recommendation 1998. February 10. Available at: http://www.w3c.org/TR/REC-xml/. Accessed on 04/24/2007.
- 41.In: Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shabo A, editors. HL7 Clinical Document Architecture, Release 2.0. ANSI-approved HL7 Standard. Ann Arbor, MI: Health Level Seven, Inc; 2005. May 2005.
- 42.Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Amnon S. The HL7 Clinical Document Architecture, Release 2 J Am Med Inform Assoc 2006;13:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadow G, Clement J. Unified Code for Units of Measure (UCUM). The Regenstrief Institute For Health Care, Indianapolis, IN. Available at: http://aurora.regenstrief.org/UCUM/. Accessed on 01/19/2007.
- 44.Biomedical Research Integrated Domain Group (BRIDG) Web site. The BRIDG Model—What is it? Available at: http://www.bridgproject.org/. Accessed on 01/19/2007.
- 45.Maler E, El Andaloussi J. Developing SGML DTDs: From text to model to markupNew Jersey: Prentice Hall; 1996.
- 46.Glushko RJ, McGrath T. Document Engineering: Analyzing and Designing Documents for Business Informatics and Web ServicesCambridge, Massachusetts: MIT Press; 2005.
- 47.Hirschtick RE. A piece of my mind JAMA 2006;20:2335-2360. [DOI] [PubMed] [Google Scholar]
- 48.Health Level Seven, Inc HL7 Reference Information ModelAnn Arbor, MI: Health Level Seven, Inc; 1994. Available at: http://www.hl7.org/library/data-model/RIM/modelpage_non.htm. Accessed on 01/19/2007.