Abstract
Objective
To specify and identify disease and patient care processes represented by temporal patterns in clinical events and observations, and retrieve patient populations containing those patterns from clinical data repositories, in support of clinical research, outcomes studies, and quality assurance.
Design
A data processing method called PROTEMPA (Process-oriented Temporal Analysis) was developed for defining and detecting clinically relevant temporal and mathematical patterns in retrospective data. PROTEMPA provides for portability across data sources, “pluggable” data processing environments, and the creation of libraries of pattern definitions and data processing algorithms.
Measurements
A proof-of-concept implementation of PROTEMPA in Java was evaluated against standard SQL queries for its ability to identify patients from a large clinical data repository who show the features of HELLP syndrome, and categorize those patients by disease severity and progression based on time sequence characteristics in their clinical laboratory test results. Results were verified by manual case review.
Results
The proof-of-concept implementation was more accurate than SQL in identifying patients with HELLP and correctly assigned severity and disease progression categories, which was not possible using SQL only.
Conclusions
PROTEMPA supports the identification and categorization of patients with complex disease based on the characteristics of and relationships between time sequences in multiple data types. Identifying patient populations who share these types of patterns may be useful when patient features of interest do not have standard codes, are poorly-expressed in coding schemes, may be inaccurately or incompletely coded, or are not represented explicitly as data values.
Introduction
Health care institutions store large volumes of patient data that represent phenotypic responses associated with disease presentation and progression, and response to therapy. 1,2 These responses may be reflected by time sequences of laboratory test results, medical observations, clinical orders, coded diagnoses and other time-stamped data elements that are mathematically and temporally related. 3–7 Common clinical data retrieval systems that are implemented in standard relational databases do not provide tools for characterizing such data sequences or retrieving groups of patients whose data sequences have similar features. 1,8–12 As a result, patient characteristics that are not explicitly coded and are represented as mathematical patterns or temporal relationships between data elements are difficult to include in clinical, quality assurance or outcomes research studies.
The need for an improved ability to represent and query temporal relationships in databases has been recognized for many years. 13 Unfortunately, attempts to add standard temporal extensions to Structured Query Language (SQL) 14 have been unsuccessful and the current SQL standard supports only limited temporal features (for a brief review, see Adlassnig et al. 2006). 13 Separate development efforts specifically targeting clinical data have resulted in several systems that provide sophisticated temporal query layers on top of relational databases. Such systems, for example AMAS, 9 DXtractor 15 and Chronus II, 16 generally implement a query language with temporal capabilities. While these languages are expressive for temporal and sequential relationships between specified data elements, they typically do not support recognizing and querying for time series that express shared characteristics such as frequency relationships or trends. SQL-based temporal query languages also embed database schema information within queries, limiting their portability unless an intermediate relational representation is used to abstract local databases. 17 Though designed for a different purpose, the Arden Syntax 18,19 can specify rules (medical logic modules, MLM) that identify patients based on temporal relationships between individual data values. Detecting some types of patterns within data sequences is technically possible, though awkward, using Arden’s limited set of mathematical functions. Arden MLMs also suffer limited portability resulting from embedded database-specific information. 20
A second group of temporal data analysis systems has been developed specifically to recognize the characteristics of time sequences, using a method called temporal abstraction. 21 Temporal abstraction systems (for a recent survey, see Stacey and McGregor) 22 infer states or processes implicit in subsequences of time-stamped data and represent them with abstractions that specify an interval of time over which a state or process exists. Abstractions may be inferred from raw data sequences based on mathematical relationships in the data (low-level abstractions), or from combinations of previously-inferred abstractions based on temporal relationships between their intervals (high-level abstractions). Existing systems generally offer a built-in set of low-level abstraction primitives (e.g., state and trend) that are used by knowledge experts as templates for defining low-level abstractions. To date, temporal abstraction has been employed primarily for the identification of data patterns of interest in individual patients for data summarization in patient monitoring and decision support, 7,23–29 and information visualization. 30–32
Temporal abstraction systems have the potential to identify patient populations that show patterns in time sequences of interest for research and quality assurance studies, but this possibility has not been demonstrated. Chronus II (mentioned above) contains a temporal abstraction module and can combine temporal query with temporal abstraction, 16,33 but the performance of this approach for identifying groups of patients with similar temporal profiles has not been described. The IDAN architecture 34 supports specifying high-level abstractions in temporal queries, but its use has been demonstrated only for monitoring tasks in individual patients. Existing temporal abstraction implementations also have limitations for application to research queries. The built-in primitives from which low-level abstractions are constructed appear to reflect the intended use of those systems (e.g., patient monitoring) and may not be designed optimally for other settings. In addition, primitives and low-level abstractions may not be accessible to users for modification and experimentation. Clinical research queries are difficult to predict and may be problem-specific, they may involve processing longer time sequences using more complex algorithms than monitoring tasks, and they may involve data exploration in which abstraction definitions must be iteratively optimized.
We believe that temporal abstraction can be an important complement to standard database query methods in clinical research settings. This complementary relationship 35 has been recognized in decision support, but has not been clearly articulated or demonstrated with respect to research queries. As a proof of this concept, we present a method, called PROTEMPA (Process-oriented Temporal Analysis), for specifying temporal and mathematical relationships between data elements in standard time-stamped databases, and retrieving populations of patients who show those relationships. PROTEMPA builds on the features of the temporal abstraction systems described above but is applied here to retrospective analysis. It does not depend on built-in temporal abstraction primitives, but instead provides a framework for defining, storing, and modifying a library of primitives and abstractions that may be general-purpose or task-specific. To determine whether our approach facilitates identification of patient populations containing temporal intervals of interest, we implemented PROTEMPA in software and evaluated its ability to 1) identify a subset of patients from a large clinical data repository who show the features of HELLP (Hemolysis, Elevated Liver enzymes, and Low Platelets) syndrome and 2) stratify those patients by disease severity and progression, based on the time sequence characteristics in their clinical laboratory test results.
Design Objectives
Our goal is to create tools that identify sequences of clinical events and observations based on mathematical and temporal relationships between their data elements, and retrieve patient populations containing those sequences from clinical data repositories. These tools should support a variety of tasks, including clinical research, and should not constrain future research questions. They should therefore provide substantial flexibility in specifying the relationships that are used to define data sequences, and these definitions should be portable across underlying databases of varying structure.
We believe these goals are best met by extending existing temporal abstraction strategies to support retrospective query and user management of abstraction definitions. To simplify the creation and maintenance of knowledge about data sequences and to allow reuse of abstraction definitions across multiple settings, the system should provide an internal model for time series data, and a framework for building and modifying a library of temporal abstraction primitives and abstraction definitions. Abstraction primitives containing algorithms that recognize time series characteristics should be straightforward to construct and use in defining low-level temporal abstractions. Because disease and clinical care processes may be reflected by multiple related data sequences, each with characteristic features (e.g., time courses of change in several laboratory tests following a clinical procedure), the framework must also allow specification of temporal relationships between groups of abstractions to define higher-level abstractions. To be broadly useful, the design must support interoperability with existing data stores and integration into standard networked computing environments.
Design
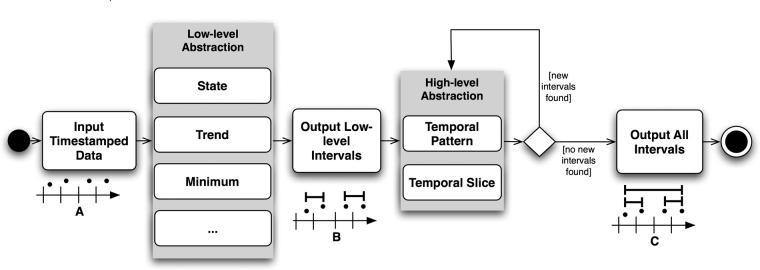
PROTEMPA is a software library with a modular architecture that is callable through defined Application Programming Interfaces (APIs). The following description provides a generic overview of PROTEMPA’s structure and the subsequent section describes our implementation in Java. PROTEMPA has four modules, shown in ▶, that provide 1) a framework for defining temporal abstraction primitives and processing time-stamped data with those primitives (the Algorithm Source), 2) a framework for specifying algorithm parameters and interval relationships that define abstractions of interest (the Knowledge Source), 3) a connection to an existing data store (the Data Source), and 4) a data processing environment for managing the abstraction-finding routines (the Abstraction Finder). The first three modules have back ends that implement environment- or application-specific features.
Figure 1.
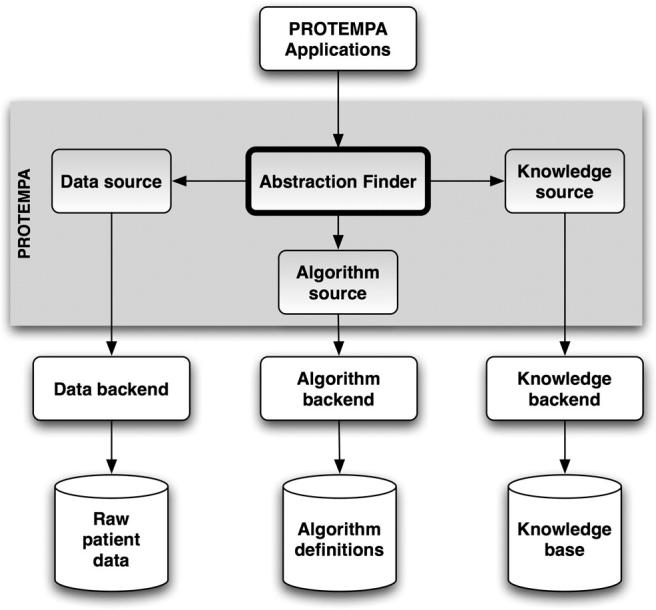
PROTEMPA architecture. PROTEMPA is a modular software library that implements an extension of the temporal abstraction method 40 for retrospective clinical data retrieval. The Abstraction Finder controls data processing and is supported by the Data Source, Knowledge Source, and Algorithm Source modules. Backends provide interfaces to specific data or knowledge stores. Arrows represent dependencies between modules. A PROTEMPA application is a program that calls the PROTEMPA library through a defined API.
A program using PROTEMPA initiates a temporal abstraction search by passing a list of abstraction names and a date range to an API provided by the Abstraction Finder. The Abstraction Finder retrieves definitions and data requirements for each abstraction from the Knowledge Source. It then extracts the data needed to compute those abstractions from the Data Source, performs temporal abstraction (calling the Algorithm Source as required), and returns a chronological list of named time interval objects.
PROTEMPA’s Abstraction Finder decomposes temporal abstraction into three mechanisms: a low-level mechanism that applies temporal abstraction primitives defined in the Algorithm Source to time-stamped data, and two high-level mechanisms (discussed below) that apply interval relationships defined in the Knowledge Source to temporal intervals previously identified during processing. The low-level mechanism scans a sequence of time-stamped data using a sliding window to select successive data subsequences, 36,37 as shown in ▶. A default length for the sliding window is specified as part of the primitive definition and may be modified in an abstraction definition (see below). The subsequence beginning with each data element is passed to the Algorithm Source for processing, and found abstractions that are overlapping or adjacent are joined (see interval combination procedure, below). The identity of the temporal abstraction primitive, arguments for the primitive’s algorithm and constraints on permissible sequence lengths and temporal spans for each abstraction are specified in abstraction definitions in the Knowledge Source. When a data subsequence matches the mathematical constraints specified by an algorithm, argument and constraint set, the low-level mechanism returns an interval element that includes the name of the abstraction and start and end times corresponding to the timestamps of the earliest and latest data values in the matching subsequence.
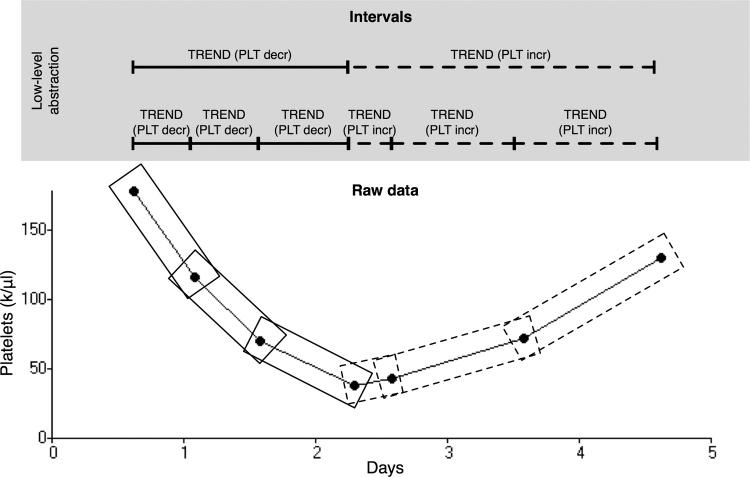
Figure 2.
Illustration of the low-level abstraction mechanism scanning a time series of platelet counts. In this example a trend primitive (TREND) is used with criteria for determining if adjacent platelet values are increasing (PLT incr) or decreasing (PLT decr). The sliding window mechanism (see Design) selects successive sequences with lengths specified in the primitive or abstraction definition. After the intervals are identified, the interval combination procedure (see Design) combines adjacent TREND (PLT incr) intervals and adjacent TREND (PLT decr) intervals.
Processing for the high-level abstraction mechanisms is contained within the Abstraction Finder module. The first mechanism, temporal pattern, scans for groups of intervals with sequential, overlap, or co-occurrence temporal relationships. Relationships are specified in the Knowledge Source as minimum and maximum temporal distances between the endpoints of pairs of participating intervals, 38,39 and constraints may also be specified on the minimum and maximum duration of each interval in the group. When a group of intervals is found that satisfies the defined relationships and constraints, 38 a named interval is created that typically encompasses the temporal extent of the group but may alternatively be temporally offset relative to the intervals in the group. A returned interval might best correspond to the full temporal extent of the group, to one of the contributing intervals, or to a newly-defined time span that represents the anticipated extent of a clinical response or a period of risk. The ability to offset the returned interval using existing intervals as references supports this flexibility.
The second high-level mechanism, temporal slice, processes all intervals of a given type as a chronological list and returns new intervals that are copies of an ordinal range, or “slice,” of the intervals in the list (e.g., the first two intervals of therapy with a particular drug among multiple courses of therapy). This mechanism can return the first, last or other intervals of a particular abstraction based on arguments specified in the Knowledge Source. The high-level mechanisms are similar to the temporal pattern matching 40 and cardinality constraint 41 mechanisms described previously.
The Abstraction Finder also implements an interval combination procedure that joins pairs of intervals of the same type if their nearest endpoints are within a defined maximum time limit, similar to previously described horizontal temporal inference and temporal interpolation mechanisms. 40 When the limit is satisfied, the pair of intervals is replaced by a new interval of the same type that spans the two intervals’ temporal extent. These optional combination limits are specified for the low-level and temporal pattern mechanisms as part of the definition of an abstraction in the Knowledge Source.
The Abstraction Finder module implements a data processing sequence that incorporates the three abstraction mechanisms (▶). When processing is initiated, the low-level mechanism scans time-stamped data and identifies intervals corresponding to low-level abstractions. The temporal pattern and slice mechanisms then operate on these intervals, repeatedly adding new intervals until no more are found. When new intervals are created, they become available for further processing. Intervals are cached so that subsequent calls to the Abstraction Finder’s API do not cause the same intervals to be re-computed. Specific examples of the low- and high-level abstractions are discussed in the Evaluation section, below.
Figure 3.
Activity diagram of PROTEMPA’s processing sequence. The low-level abstraction mechanism scans time-stamped data sequences (A) for time intervals that correspond to low-level abstractions defined in the Knowledge Source (▶). Found intervals (B) are subsequently scanned by the high-level mechanisms, which add new intervals corresponding to defined temporal pattern and temporal slice abstractions (C), repeatedly processing all intervals until no more are found.
The Abstraction Finder is supported by the Algorithm Source, Knowledge Source and Data Source modules (▶). The Algorithm Source provides a method for storing, and a run-time environment for executing, temporal abstraction primitives that support the low-level temporal abstraction mechanism described above. Primitives consist of an algorithm in an arbitrary programming language, a default sequence length for the sliding window mechanism and an optional set of parameters that define arguments the algorithm can receive at run time. Algorithms without parameters identify a specific mathematical data pattern and are essentially self-contained; parameters allow for algorithms that detect a general pattern (e.g., a trend) and are passed arguments (starting or ending cutoff values, slope, etc.) at run time that define particular instances of the pattern for different settings. Low-level abstractions that use primitives with arguments include the name of the primitive and specific arguments for it in their definition (see below). The Algorithm Source backend manages the transfer of data to and from the primitive storage and algorithm run-time environments.
The Knowledge Source provides for storage and retrieval of abstraction definitions, each of which specifies an abstraction mechanism, a set of mechanism-specific parameters as shown in ▶A, and the data or interval types it uses. Abstractions that use the low-level mechanism specify the name of a temporal abstraction primitive, a set of constraints for the abstraction that include duration and sequence length limits (which may override the default sequence length in the primitive), and, if appropriate, arguments for the primitive’s parameters. Abstractions that use the temporal pattern mechanism specify a set of temporal relationships between a group of previously defined abstractions and, if appropriate, constraints on the durations of the abstractions in the group. Abstractions that use the temporal slice mechanism specify a previously defined abstraction and an ordinal range. Abstractions that use the low-level or temporal pattern mechanisms also optionally specify interval combination limits for use by the interval combination procedure. A Knowledge Source backend connects to a knowledge base for storage of these definitions.
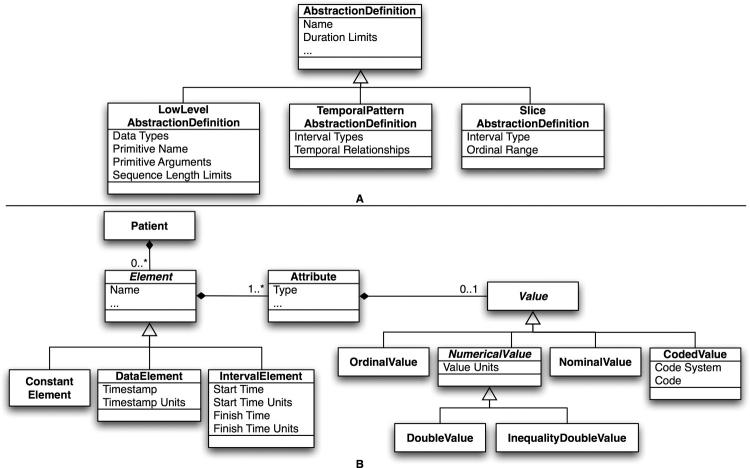
Figure 4.
Class diagrams of PROTEMPA’s temporal abstraction definitions (A) and data model (B). Three types of classes for abstractions (A) are provided to represent low level, temporal pattern and temporal slice abstractions. Data elements and abstracted intervals (Elements, B) belong to Patients and have one or more Attributes, each of which may have a numerical or textual Value, including floating point (DoubleValue) and inequality numerical (InequalityDoubleValue) values with associated units, categorical values (NominalValue), ordinal values (OrdinalValue) and values represented as codes from standard vocabularies (CodedValue). Time-stamped data (DataElement) are associated with a time point with time units; intervals (IntervalElement) are associated with start and finish times with time units. Atemporal data such as gender and race are represented as ConstantElements.
The Knowledge Source additionally defines a general-purpose model, shown in ▶B, for representing time-stamped data and intervals within PROTEMPA’s modules and abstraction definitions. Time-stamped data types have one or more attributes, each of which may have a numerical or textual value. In addition to, for example, a laboratory test result, these attributes provide data “slots” for information about the event and designators such as standard terminologies. Interval types may have one such attribute for representing a value associated with the interval, and start and finish times with time units. PROTEMPA supports standard time units from second to year, automatically converts between time units, and automatically resolves time granularity in comparison operations to the coarsest unit. 42
The Data Source provides a connection to a physical database containing time-stamped clinical data, a mapping from the schema and terminologies of the database to the data model described above, and any necessary data processing methods (e.g., unit conversion routines). The database connection, mapping and processing methods are implemented in the Data Source backend. The other PROTEMPA modules call the Data Source’s API to request specific patient data over a defined date range.
Implementation
We implemented PROTEMPA using the Java Software Development Kit version 1.4.1 (java.sun.com) on Apple Power Mac G5 hardware running Mac OS X 10.3 (Apple Computer, Inc.). PROTEMPA’s modules (▶) are Java classes that run in a single process, and the PROTEMPA API can be called from any Java program. Abstraction finding incrementally matches intervals and raw data to abstraction definitions using the Rete algorithm 43 as implemented by the Drools rules engine. 44 PROTEMPA has been successfully deployed on Mac OS X and on standard PC hardware running Windows XP (Microsoft Corp.).
The backends (▶) are also Java classes; their class names are specified in a configuration file and loaded dynamically into the Java virtual machine. Abstraction definitions are stored in a knowledge base implemented in the Protégé ontology environment, 45 which provides for object-oriented storage, representation of complex interrelationships between objects, and evolution of schemas over time. The Knowledge Source backend connects to Protégé using its built-in Java API. The Data Source backend maps PROTEMPA’s data type designators to local terminologies and implements a connection to a MySQL relational database (www.mysql.org) via JDBC. The Algorithm Source backend provides the R statistical computing system 46 as the execution environment for temporal abstraction primitive algorithms, using software from the RoSuDa project 47 that supports execution of R source code from Java programs. The algorithms are written as functions in the R language and stored as text in another Protégé knowledge base that is accessed through the Algorithm Source backend.
Evaluation
We evaluated PROTEMPA’s performance by using it to identify patients with laboratory signs of HELLP syndrome 48 in a clinical data repository and categorize those patients according to specific features of their platelet count profiles. HELLP is a dangerous complication of pregnancy that appears during the latter part of the third trimester or after childbirth. 48 There is no standard diagnosis code (ICD-9) for HELLP. Diagnosis and management are based on monitoring clinical symptoms and three clinical laboratory tests: platelet count (PLT), serum lactate dehydrogenase (LDH), and serum aspartate aminotransferase (AST). HELLP syndrome has been defined as pre-eclampsia with PLT < 100,000/μL, LDH > 600 U/L, and AST > 70 U/L, 49,50 and rising PLT indicates recovery. 50 The PLT nadir can be used to classify a HELLP patient by disease severity: class 1 HELLP, PLT < 50,000/μL; class 2 HELLP, PLT >= 50,000/μL and < 100,000/μL. 51 There is clinical interest in our facility in the characteristics and therapeutic circumstances of HELLP patients who show a partial recovery of PLT followed by a second suppression. Formerly, identifying these patients required extracting patients by pregnancy diagnosis code and inspecting their laboratory results by hand.
To determine whether PROTEMPA could be used to identify and categorize patients with laboratory signs of HELLP, we retrieved a set of cases from the University of Virginia’s Clinical Data Repository (CDR) 12 that occurred between 2000 and 2005, had ICD-9 codes indicating pregnancy and had at least one LDH > 300 U/L. All available laboratory test results and diagnosis codes for each case were exported from the CDR and imported into our test implementation’s data store. Laboratory data were originally obtained as part of routine clinical care using standard analysis methods. Institutional Review Board approval was obtained.
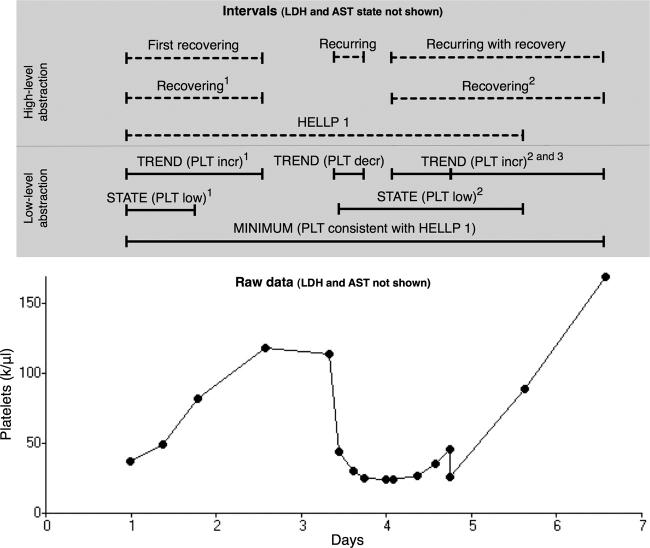
We defined five temporal abstractions (▶A) that distinguish between the two disease severity categories (HELLP 1 and HELLP 2, as defined above) and four PLT response categories: 1) those that recovered after PLT suppression (First recovering), 2) those that partially recovered, recurred and then recovered (Recurring with recovery), 3) those that partially recovered, recurred and then did not recover (Recurring), and 4) those that showed no evidence of recovery (all others). These five abstractions are defined using general primitives for detecting states, trends, and the minimum value in a data sequence, with arguments specific for PLT, LDH, and AST. The HELLP 1 and HELLP 2 abstractions use the temporal pattern mechanism, and specify intervals of elevated AST, elevated LDH, and low PLT occurring within 7 days of each other with a minimum PLT value of less than 50,000/μL or between 50,000/μL and 100,000/μL, respectively. The First recovering abstraction uses the temporal slice and temporal pattern mechanisms. It specifies the first trend interval in platelet values, beginning after the start of a class 1 or 2 HELLP interval, that has an increase of more than 9,000/μL per day and a duration of at least 12 hours. The Recurring abstraction uses the temporal pattern mechanism and specifies an interval of First recovering followed by a platelet trend with a decrease of more than 9,000/μL per day to an endpoint of less than 100,000/μL. The Recurring with recovery abstraction uses the temporal pattern mechanism and specifies an interval of Recurring followed by a platelet trend with an increase of more than 9,000/μL per day to an endpoint of at least 100,000/μL. A platelet profile with these abstractions is shown in ▶, and the definition of the Recurring abstraction is illustrated in ▶.
Figure 5.
Example time series of platelet (PLT) counts in class 1 HELLP syndrome with the intervals that PROTEMPA identified. Intervals found by the low-level abstraction mechanism (solid lines) were inferred from mathematical patterns in the raw time-stamped data shown at the bottom of the figure, and are labeled as in ▶. Intervals found by the high-level mechanism (dashed lines) were identified as in ▶ and are labeled with the corresponding abstraction’s name. Superscripts refer to the ordinal position of the intervals accessible through the temporal slice mechanism (see Design). AST and LDH test results and associated intervals are omitted for clarity. This profile was classified Recurring with recovery (▶).
Figure 6.
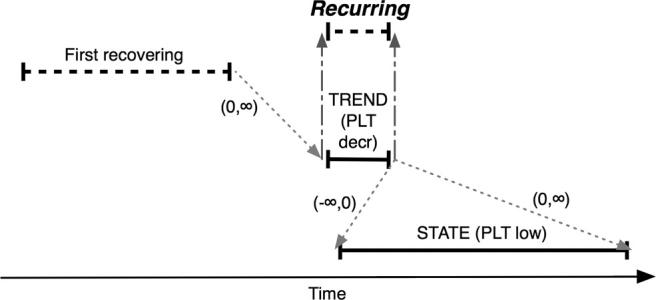
Composition of the Recurring abstraction (see Evaluation and ▶), defined as an interval of decreasing platelet values (PLT decr) occurring after the first interval of recovering platelets (First recovering) and overlapping an interval of platelet values less than 100,000/μL (PLT low). Intervals are illustrated as in ▶. Recurring intervals have the same endpoints as the contributing PLT decr interval (gray dashed arrows). Gray dotted arrows denote temporal relationships defined between endpoints of intervals and are labeled with minimum and maximum time constraints. For example, (0, ∞) indicates that the second time point in the relationship must occur on or after the first point (∞ = largest possible time distance). This type of abstraction is recognized by the temporal pattern high-level abstraction mechanism (see Design).
A simple Java application invoked the PROTEMPA library with database connection information, Protégé knowledge bases defining the abstractions and algorithms, and a list of abstractions to find; and it output found intervals for each case to a text file. The abstraction definitions were adjusted after a preliminary processing run to optimize patient categorization and accommodate typical variations in laboratory values. After PROTEMPA identified a set of cases and their abstractions, its output file was passed to a post-processing script, which categorized the cases according to the types and sequence of intervals found (▶).
Table 1.
Table 1 HELLP Syndrome Cases Categorized by Laboratory Result Profile
| Severity | Category | Number (percent of total) |
|---|---|---|
| HELLP 2 ∗ | Recurring† | 5 (6.2%) |
| Not recurring‡ | 33 (40.7%) | |
| Total | 38 (46.9%) | |
| HELLP 1 ∗ | Recurring with recovery§ | 7 (8.7%) |
| Recurring without recovery¶ | 4 (4.9%) | |
| Not recurring with recovery‡ | 31 (38.3%) | |
| Not recurring without recovery# | 1 (1.2%) | |
| Total | 43 (53.1%) | |
| All HELLP | Total | 81 (100%) |
∗ Patients were categorized as HELLP 1 or HELLP 2 based on co-occurring intervals of elevated LDH and AST, and suppressed platelet count (see Evaluation). All HELLP 2 patients recovered based on normalization of platelet counts.
† Final platelet interval is Recurring or Recurring with recovery.
‡ Final platelet interval is First recovering.
§ Final platelet interval is Recurring with recovery.
¶ Final platelet interval is Recurring.
# All patients not classified as above.
The data set included 761 eligible cases (pregnancy and LDH above 300 U/L). A standard SQL query identified 87 cases as potential HELLP diagnoses based on the presence of at least one low PLT, high AST, and high LDH consistent with the HELLP definition. In comparison, PROTEMPA identified 81 cases as likely HELLP. The PROTEMPA cases were all included in the SQL-identified data set; in the six additional SQL-identified cases, laboratory results meeting the required thresholds were more than seven days apart. Of the 761 eligible cases, 190 included an ICD-9 code for severe pre-eclampsia. In this case subset, SQL and PROTEMPA both detected the same 72 cases as having HELLP, and their correctness was confirmed by manual inspection of all 190 cases. Of the nine cases identified by both PROTEMPA and SQL that were not part of the pre-eclampsia subset, five were consistent with HELLP based on manual review and four were complex patients whose laboratory patterns could have resulted from multi-system disease without HELLP.
The post-processing step correctly classified the 81 cases found by PROTEMPA into severity and platelet response pattern categories (see ▶), as compared with manual inspection of each case’s PLT data sequence. Overall, PROTEMPA identified the “recurring” pattern (partial platelet recovery followed by a subsequent platelet suppression) in about 20% of the patients (6.2% of HELLP 2 and a total of 13.6% of HELLP 1 patients, ▶). Stratifying patients into platelet response categories based on the profile of the PLT data sequence was not possible using SQL.
Discussion
We have developed a data processing strategy and software library (PROTEMPA) that meet most of our primary design goals, including flexible specification of low- and high-level temporal abstractions for clinical data sequences, and retrieval of groups of patients from a retrospective clinical data repository based on these abstractions. We tested PROTEMPA in a proof-of-concept implementation that was more accurate than SQL in identifying patients with HELLP syndrome, a disease with no ICD-9 code, and accurately assigned severity and disease progression categories based on the temporal characteristics of laboratory test profiles (platelet count, AST, and LDH; see ▶ and ▶). This clinical setting favors SQL because substantial elevation in AST and LDH, and platelet count suppression, are relatively unlikely to occur within the same hospital admission in pregnancy apart from HELLP. PROTEMPA could well perform even better against SQL in other scenarios. Furthermore, the assignment of disease progression categories based on the temporal pattern of laboratory results was not possible using SQL only, and highlights the unique capabilities of temporal abstraction systems. Previous systems 9,15,52 have applied temporal abstraction to the identification of temporal characteristics in data sequences of individual patients, using pre-defined temporal abstraction primitives. Our study extends this work by demonstrating a general-purpose framework for specifying and optimizing temporal abstractions to successfully query a retrospective data repository for patient populations that would be difficult to identify by other methods.
PROTEMPA’s temporal abstraction primitives and abstraction definitions are managed in knowledge bases separate from the abstraction-processing environment, to allow their convenient creation and modification. Primitives may employ algorithms that are fully specified (“hard coded”), or that receive arguments. The latter supports the use of “template” algorithms that can be configured to identify general types of data sequence patterns (e.g., state, trend, peak, trough, frequency) with desired specific characteristics. Primitives may be written to target relatively short intervals, similar to primitives in other systems, 40 but may also process longer data sequences. Theoretically, the algorithms may be of arbitrary complexity and may be implemented in any programming language, as long as an appropriate runtime environment is provided by the Algorithm Source backend (▶). We used the R statistical processing environment in our proof-of-concept, but other interpreted or compiled programming environments suitable for statistical, scientific or modeling applications (e.g., Mathematica, Matlab, Python and Ruby) could be implemented similarly via the Algorithm Source backend using existing connectors. 53–57
New temporal abstraction primitives are defined by storing an algorithm and a listing of its parameters in a knowledge base record in the Algorithm Source backend. Low- and high-level abstractions (▶A) are defined by creating entries in the Knowledge Source knowledge base with duration parameters and the identity of a primitive with appropriate arguments (low-level) or a set of interval types and their relationships (high-level). Currently, these definitions are created directly within PROTEMPA’s knowledge bases. Future development will include end-user interfaces for defining primitives and abstractions (see further discussion below). With such interfaces, statisticians or others familiar with the locally-implemented run-time environment could develop or modify primitives, and researchers and other domain experts could use these primitives and existing abstraction definitions to specify new abstractions. Thus PROTEMPA’s design supports the notion of a library of primitive and abstraction definitions, implemented in our proof-of-concept as Protégé knowledge bases, which is modifiable and extensible by users. Because abstractions refer to PROTEMPA’s internal data model (▶) rather than site-specific database terminology, schema and connection details, abstraction libraries should be portable between PROTEMPA deployments that implement similar knowledge bases. PROTEMPA’s support for an extensible and portable library of primitives and abstractions appears to be unique among reported temporal abstraction systems.
Abstraction of the details of the local environment by the Data Source backend also supports portability of the PROTEMPA framework across different data sources. Currently, deploying PROTEMPA in a local environment requires creating a site-specific Data Source backend that 1) incorporates local terminology into its terminology translation table, 2) adds any necessary data-conversion methods (e.g., unit conversion), and 3) implements appropriate SQL calls to the local clinical data store to retrieve data for processing. Installations will also provide a front end that calls the Abstraction Finder API with the desired abstraction retrieval requests, and implements any necessary post-processing steps. Once the site-specific install is complete, temporal primitive and abstraction definitions are entered or imported into the Algorithm and Knowledge Source backends. Future development of PROTEMPA is intended to provide a default (but replaceable) front end that offers query creation and results review capabilities. Because data sources for clinical research currently use a variety of terminologies and data models, manually mapping local environments to PROTEMPA’s internal model through a backend adapter is necessary for portability. Standardization of terminologies and data models in the future may allow pre-built backends to provide more of the necessary mappings a priori, and decrease the requirement for site-specific work. Using a back end developed as above, PROTEMPA was previously deployed at the University of Pittsburgh Medical Center for detecting quality assurance problems in clinical laboratory testing. 58 Other systems use analogous techniques for portability: IDAN 34 provides a generic data model and a pluggable database access module, and Chronus II 16 may be deployed against an intermediate abstract database representation for portability as described above. DXtractor 15 and the Arden Syntax 20 are less portable because some details of the underlying local database schema and terminology are incorporated into their queries.
PROTEMPA is a code library that may provide developers with knowledge representation expertise a useful foundation for building systems to specify and detect temporal sequences. It is designed to be general-purpose, and therefore could be incorporated into several different types of temporal query processing systems. In clinical practice settings, where decision support, patient monitoring and quality assurance needs may be relatively well-defined, PROTEMPA could identify abstractions that are pre-defined by knowledge experts and are used to trigger system responses, or stored in a database for query using standard techniques. Alternatively, PROTEMPA can support retrospective queries in a clinical research setting. In this application, queries are less predictable and may be developed in the setting of data exploration. In addition, useful abstractions may be relatively specific to a particular application (for example, the Recurring with recovery abstraction we define in the Evaluation section) and therefore difficult for knowledge experts to pre-construct. Thus a research query system should support construction, execution and evaluation of queries directly by research users. Temporal abstraction systems with these characteristics have not been described previously.
The proof-of-concept system that we demonstrate here is a first step toward a clinical research query system based on temporal abstraction. In its current implementation, constructing and executing queries requires significant technical knowledge and thus PROTEMPA does not completely fulfill our design goals. To do so, PROTEMPA would need to be incorporated into a system that provided 1) a query interface that allowed research users to define temporal abstractions and combine them to construct complex queries, 2) a reporting interface with temporal display and summarization features that allowed users to evaluate data sets returned from queries as part of an iterative process of data exploration and query refinement, and 3) a management interface that allowed users to classify, store, recall and share abstraction definitions. Each of these system components would have novel elements and each would be a significant challenge to construct.
Though temporal abstraction can be a powerful method for specifying and identifying the characteristics of time sequences in clinical data, it also has several limitations. PROTEMPA uses an approach similar to Knowledge-Based Temporal Abstraction 40 and therefore key details of temporal primitive algorithms, abstraction definitions and abstraction relationships such as cutoff values, rates of change in trends, and both qualitative and quantitative interval relationships are based on literature values and expert experience. These relationships and cutoff values are generally estimates and have not been verified or optimized for use with temporal abstraction. In addition, while a temporal relationship between a set of clinical observations may be characteristic of a particular condition, a similar relationship might occur for other reasons (as, for example, in the four patients we identified with multi-system disease, see Evaluation). The prevalence of these “false positives” is difficult to predict and will generally depend on the patient population and research question under study. Thus the sensitivity and specificity of temporal abstractions for distinguishing between different groups of patients, or appropriately distinguishing patients with borderline values, are not well established. PROTEMPA may correctly identify temporal abstractions, but misidentify cases because of suboptimal abstraction definitions or actual similarities between different types of patients. A related problem would occur if abstraction definitions were shared without modification between sites that use different measurement techniques for clinical observations, resulting in inadvertent misapplication of abstractions. PROTEMPA also does not distinguish between cases in which an abstraction is not identified because the data is present but fails to satisfy the abstraction definition, or because the data is not present. Thus it may miss cases in which key data about the patient was not collected. Future development may be able to address some of these limitations by providing tools for clinical researchers that support, for example, query optimization and validation, and summarization and exploration of the hierarchy of abstractions contributing to a final query result.
PROTEMPA’s performance characteristics have not yet been fully evaluated. Its data processing mechanism (▶) has exponential worst-case complexity, but other temporal abstraction systems show significantly better average-case complexity 21,40 and we expect this to be true for PROTEMPA. Performance issues may be mitigated in part because clinical research questions usually require searching for a limited set of abstractions, as was the case in our proof-of-concept. Intelligently sequencing the abstraction search might also optimize performance. For example, dynamic interpretation contexts might be used to trigger searching for some abstractions only after certain other abstractions or data have been found. 59 Finally, for large data sets or high search volumes, PROTEMPA’s algorithms are parallelizable by patient 23 and could be implemented in a straightforward manner on clustered hardware.
Conclusions
PROTEMPA is a flexible temporal abstraction software library designed for querying implicit temporal relationships in clinical data. Its design provides a number of features useful for retrospective research, including definable algorithms for abstracting raw data with “pluggable” data processing environments, iterative higher-level abstraction, flexible knowledge-based temporal abstraction definition with abstraction configuration via arguments, abstraction storage and reuse, standard connectivity mechanisms for relational databases and an internal data model that is independent of local database details. In a preliminary study focused on retrieval of patient populations in retrospective data, it supported the development of abstractions that accurately identified and categorized patients with a complex disease based on temporal relationships between multiple laboratory results. Temporal abstraction has not been commonly applied to clinical data retrieval, but it may provide significant advantages when used to augment standard data retrieval methods. The ability to automate identification of patient populations based on the temporal characteristics of clinical data may substantially decrease the effort required to retrieve and classify patients for a wide range of clinical studies, outcomes research, and quality assurance evaluations. This capability may be particularly useful when patient features of interest are those, such as clinical severity, disease progression, and response to therapy, that do not have standard codes, are poorly expressed in commonly used coding schemes, may be inaccurately or incompletely coded, or are not represented explicitly as data values.
Footnotes
This work was supported in part by National Library of Medicine grant R01 LM008192 and National Library of Medicine Training Grant T15 LM007059.
The authors thank Vanathi Gopalakrishnan for valuable advice.
References
- 1.Safran C, Chute CG. Exploration and exploitation of clinical databases Int J Biomed Comput 1995;39(1):151-156. [DOI] [PubMed] [Google Scholar]
- 2.Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper J Am Med Inform Assoc 2007;14(1):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahar Y. Dimensions of time in illness: an objective view Ann Intern Med 2000;132(1):45-53. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki S, Ho TB, Nguyen DT. Abstraction of Long-Term Changed Tests in Mining Hepatitis Data 2003. pp. 366-3727th Int Conf on Knowledge-Based Intelligent Information and Engineering Systems (KES 2003);.
- 5.Weydert JA, Nobbs ND, Feld R, Kemp JD. A simple, focused, computerized query to detect overutilization of laboratory tests Arch Pathol Lab Med 2005;129(9):1141-1143. [DOI] [PubMed] [Google Scholar]
- 6.Ridgeway JJ, Weyrich DL, Benedetti TJ. Fetal heart rate changes associated with uterine rupture Obstet Gynecol 2004;103(3):506-512. [DOI] [PubMed] [Google Scholar]
- 7.Carrault G, Cordier MO, Quiniou R, Wang F. Temporal abstraction and inductive logic programming for arrhythmia recognition from electrocardiograms Artif Intell Med 2003;28(3):231-263. [DOI] [PubMed] [Google Scholar]
- 8.Murray MD, Smith FE, Fox J, Teal EY, Kesterson JG, Stiffler TA, et al. Structure, functions, and activities of a research support informatics section J Am Med Inform Assoc 2003;10(4):389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorda W, Gall W, Duftschmid G. Clinical data retrieval: 25 years of temporal query management at the University of Vienna Medical School Methods Inf Med 2002;41(2):89-97. [PubMed] [Google Scholar]
- 10.Bui AA, Weinger GS, Barretta SJ, Dionisio JD, Kangarloo H. An XML Gateway to Patient Data for Medical Research Applications Ann NY Acad Sci 2002;980:236-246. [DOI] [PubMed] [Google Scholar]
- 11.van Mulligen EM, Timmers T, van Bemmel JH. User evaluation of an integrated medical workstation for clinical data analysis Methods Inf Med 1993;32(5):365-372. [PubMed] [Google Scholar]
- 12.Schubart JR, Einbinder JS. Evaluation of a data warehouse in an academic health sciences center Int J Med Inf 2000;60(3):319-333. [DOI] [PubMed] [Google Scholar]
- 13.Adlassnig KP, Combi C, Das AK, Keravnou ET, Pozzi G. Temporal representation and reasoning in medicine: research directions and challenges Artif Intell Med 2006;38(2):101-113. [DOI] [PubMed] [Google Scholar]
- 14.Elmasri R, Navathe SB. Fundamentals of database systems3rd ed.. New York: Addison-Wesley; 2000.
- 15.Nigrin DJ, Kohane IS. Temporal expressiveness in querying a time-stamp—based clinical database J Am Med Inform Assoc 2000;7(2):152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor MJ, Tu SW, Musen MA. The Chronus II Temporal Database Mediator Proc AMIA Symp 2002:567-571. [PMC free article] [PubMed]
- 17.Das AK, Musen MA. SYNCHRONUS: A Reusable Software Module for Temporal Integration Proc AMIA Symp 2002:195-199. [PMC free article] [PubMed]
- 18.Karadimas HC, Chailloleau C, Hemery F, Simonnet J, Lepage E. Arden/J: an architecture for MLM execution on the Java platform J Am Med Inform Assoc 2002;9(4):359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman EH, Hripcsak G, Starren J, Jenders RA, Clayton P. Using intermediate states to improve the ability of the Arden Syntax to implement care plans and reuse knowledge Proc Annu Symp Comput Appl Med Care 1995:238-242. [PMC free article] [PubMed]
- 20.Jenders RA, Sujansky W, Broverman CA, Chadwick M. Towards improved knowledge sharing: assessment of the HL7 Reference Information Model to support medical logic module queries Proc AMIA Annu Fall Symp 1997:308-312. [PMC free article] [PubMed]
- 21.Augusto JC. Temporal reasoning for decision support in medicine Artif Intell Med 2005;33(1):1-24. [DOI] [PubMed] [Google Scholar]
- 22.Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: A survey Artif Intell Med 2007;39(1):1-24. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor MJ, Grosso WE, Tu SW, Musen MA. RASTA: a distributed temporal abstraction system to facilitate knowledge-driven monitoring of clinical databases Medinfo 2001:508-512. [PubMed]
- 24.Haimowitz IJ, Kohane IS. Managing temporal worlds for medical trend diagnosis Artif Intell Med 1996;8(3):299-321Jul. [DOI] [PubMed] [Google Scholar]
- 25.Larizza C, Moglia A, Stefanelli M. M-HTP: a system for monitoring heart transplant patients Artif Intell Med 1992;4:111-126. [Google Scholar]
- 26.Shahar Y, Musen MA. Knowledge-based temporal abstraction in clinical domains Artif Intell Med 1996;8(3):267-298. [DOI] [PubMed] [Google Scholar]
- 27.Bellazzi R, Larizza C, Magni P, Montani S, Stefanelli M. Intelligent analysis of clinical time series: an application in the diabetes mellitus domain Artif Intell Med 2000;20(1):37-57. [DOI] [PubMed] [Google Scholar]
- 28.Kahn MG, Fagan LM, Sheiner LB. Combining physiologic models and symbolic methods to interpret time-varying patient data Methods Inf Med 1991;30(3):167-178. [PubMed] [Google Scholar]
- 29.Hayes-Roth B, Washington R, Ash D, Hewett R, Collinot A, Vina A, et al. Guardian: a prototype intelligent agent for intensive-care monitoring Artif Intell Med 1992;4:165-185. [Google Scholar]
- 30.Martins SB, Shahar Y, Galperin M, Kaizer H, Goren-Bar D, McNaughton D, et al. Evaluation of KNAVE-II: A Tool for Intelligent Query and Exploration of Patient Data Medinfo 2004:648-652. [PubMed]
- 31.Shahar Y, Cheng C. Model-based visualization of temporal abstractions Comput Intell 2000;16(2):279-306. [Google Scholar]
- 32.Fackler J, Kohane I. Monitor-driven data visualization: SmartDisplay Proc Annu Symp Comput Appl Med Care 1994:939-943. [PMC free article] [PubMed]
- 33.O’Connor MJ, Shankar RD, Das AK. An ontology-driven mediator for querying time-oriented biomedical dataSalt Lake City, UT: 9th IEEE Intl Symp Comp Based Med Sys; 2006. pp. 264-269.
- 34.Shahar Y, Goren-Bar D, Boaz D, Tahan G. Distributed, intelligent, interactive visualization and exploration of time-oriented clinical data and their abstractions Artif Intell Med 2006;38(2):115-135. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen JH, Shahar Y, Tu SW, Das AK, Musen MA. A temporal database mediator for protocol-based decision support Proc AMIA Annu Fall Symp 1997:298-302. [PMC free article] [PubMed]
- 36.Gall W, Duftschmid G, Dorda W. Moving time window aggregates over patient histories Int J Med Inform 2001;63(3):133-145. [DOI] [PubMed] [Google Scholar]
- 37.Perng C-S, Parker DS. Temporal Coupling Verification in Time Series Databases J Intell Inf Syst 2000;15:29-49. [Google Scholar]
- 38.Dechter R, Meiri I, Pearl J. Temporal constraint networks Artif Intell 1991;49:61-95. [Google Scholar]
- 39.Allen JF. Maintaining Knowledge about Temporal Intervals Commun ACM 1983;26(11):832-843. [Google Scholar]
- 40.Shahar Y. A framework for knowledge-based temporal abstraction Artif Intell 1997;90:79-133. [DOI] [PubMed] [Google Scholar]
- 41.Chakravarty S, Shahar Y. Acquisition and analysis of repeating patterns in time-oriented clinical data Methods Inf Med 2001;40(5):410-420. [PubMed] [Google Scholar]
- 42.Combi C, Pinciroli F, Pozzi G. Managing different time granularities of clinical information by an interval-based temporal data model Methods Inf Med 1995;34(5):458-474. [PubMed] [Google Scholar]
- 43.Forgy CL. Rete: A Fast Algorithm for the Many Pattern/Many Object Pattern Match Problem Artif Intell 1982;19:17-37. [Google Scholar]
- 44.JBoss. Drools—Home. Available at: http://legacy.drools.codehaus.org. Accessed Apr 4, 2007.
- 45.Stanford Medical Informatics. The Protege Ontology Editor and Knowledge Acquisition System. Available at: http://protege.stanford.edu/. Accessed Apr 4, 2007.
- 46.R Development Core Team R: A Language and Environment for Statistical Computing 2006. Vienna, Austria.
- 47.Universitat Augsburg Dept. of Computer Oriented Statistics and Data Analysis. Interactive statistical software—RoSuDa. Available at: http://stats.math.uni-augsburg.de/software/. Accessed Apr 4, 2007.
- 48.Sibai BM. The HELLP syndrome (hemolysis, elevated liver enzymes, and low platelets): much ado about nothing? Am J Obstet Gynecol 1990;162(2):311-316. [DOI] [PubMed] [Google Scholar]
- 49.Sibai BM, Barton JR. Dexamethasone to improve maternal outcome in women with hemolysis, elevated liver enzymes, and low platelets syndrome Am J Obstet Gynecol 2005;193(5):1587-1590. [DOI] [PubMed] [Google Scholar]
- 50.Fonseca JE, Mendez F, Catano C, Arias F. Dexamethasone treatment does not improve the outcome of women with HELLP syndrome: a double-blind, placebo-controlled, randomized clinical trial Am J Obstet Gynecol 2005;193(5):1591-1598. [DOI] [PubMed] [Google Scholar]
- 51.Martin JN, Rinehart BK, May WL, Magann EF, Terrone DA, Blake PG. The spectrum of severe preeclampsia: comparative analysis by HELLP (hemolysis, elevated liver enzyme levels, and low platelet count) syndrome classification Am J Obstet Gynecol 1999;180(6):1373-1384. [DOI] [PubMed] [Google Scholar]
- 52.Snodgrass R, Bohlen MH, Jensen CS, Steiner A. Transitioning temporal support in TSQL2 to SQL3In: Etzion O, Jajodia S, Sripada S, editors. Temporal Databases: Research and Practice. Berlin, NY: Springer; 1998. pp. 150-194.
- 53.Wolfram Research. Java Toolkit: J/Link: Integrating Mathematica and Java. Available at: http://www.wolfram.com/solutions/mathlink/jlink/. Accessed Apr 4, 2007.
- 54.Muller S. JMatLink Site. Available at: http://jmatlink.sourceforge.net/. Accessed Apr 4, 2007.
- 55.Jython Development Team. The Jython Project. Available at: http://www.jython.org. Accessed Apr 4, 2007.
- 56.Johnson M. Jepp—Java Embedded Python. Available at: http://jepp.sourceforge.net/. Accessed Apr 4, 2007.
- 57.JRuby Development Team. JRuby—Home. Available at: http://jruby.codehaus.org. Accessed Mar 5, 2007.
- 58.Post AR, Ho J, Blank G, Harrison Jr JH. Web-based Implementation of a Modular General Purpose Temporal Abstraction Framework for Pattern Identification in Clinical Laboratory Data Arch Pathol Lab Med 2004;128(10):1102. [Google Scholar]
- 59.Shahar Y. Dynamic temporal interpretation contexts for temporal abstraction Ann Math Artif Intell 1998;22(1–2):159-192. [Google Scholar]