Abstract
Phase variably expressed (randomly switching) methyltransferases associated with type III restriction-modification (R-M) systems have been identified in a variety of pathogenic bacteria. We have previously shown that a phase variable methyltransferase (Mod) associated with a type III R-M system in Haemophilus influenzae strain Rd coordinates the random switching of expression of multiple genes, and constitutes a phase variable regulon—‘phasevarion’. We have now identified the recognition site for the Mod methyltransferase in H. influenzae strain Rd as 5′-CGAAT-3′. This is the same recognition site as the previously described HinfIII system. A survey of 59 H. influenzae strains indicated significant sequence heterogeneity in the central, variable region of the mod gene associated with target site recognition. Intra- and inter-strain transformation experiments using Mod methylated or non-methylated plasmids, and a methylation site assay demonstrated that the sequence heterogeneity seen in the region encoding target site specificity does correlate to distinct target sites. Mutations were identified within the res gene in several strains surveyed indicating that Res is not functional. These data suggest that evolution of this type III R-M system into an epigenetic mechanism for controlling gene expression has, in some strains, resulted in loss of the DNA restriction function.
INTRODUCTION
Restriction-modification (R-M) systems are ubiquitous in bacteria and are involved in protection of the bacterial cell from incoming foreign DNA. R-M systems are comprised of two enzymes: a methyltransferase and a restriction endonuclease. The methyltransferase catalyses the methylation of a specific DNA recognition sequence distinguishing ‘self’ DNA and protecting it from cleavage (1,2). The restriction endonuclease catalyses the double-stranded cleavage of unmethylated ‘non-self’ DNA. R-M systems are classified into three major groups: types I, II and III (3). In type II systems, the most common type of R-M system (3), the methyltransferase activity and restriction activity are performed by two independently acting enzymes (4). In the more complex type I systems, methyltransferase activity and restriction activity are performed by the same holoenzyme, which consists of three types of subunits: S, M and R encoded by hsdS, hsdM and hsdR, respectively. The S and M subunits are required for methyltransferase activity, and all three subunits are necessary for restriction activity (5). Type III systems consist of only two subunits, the methyltransferase (modification, Mod) subunit which can independently function as a methyltransferase (6,7) and the restriction (Res) subunit which must form a complex with Mod to recognize and cleave DNA (8).
Phase variation is the reversible, high frequency switching of gene expression. In Haemophilus influenzae and several other mucosal pathogens, phase variation is commonly mediated by mutations in simple tandem DNA repeats in the open reading frame or promoter region of phase variable genes (9,10). Phase variation is usually associated with genes encoding surface expressed virulence determinants, where switching of expression allows the generation of a diverse population of phenotypically distinct cells—some of which will be better adapted to survival. There are several examples of type III R-M systems in a variety of pathogenic bacteria that have been proven to undergo phase variation [H. influenzae (11) and Helicobacter pylori (12)] or from sequence analysis would be predicted to undergo phase variation [Pasteurella haemolytica (13), Neisseria meningitidis, Neisseria gonorrhoeae (14) and Moraxella catarrhalis (15)]. Several proposals have been made as to the functional significance of phase variable type III R-M systems (16) but these have not been tested experimentally. Classically, R-M systems are thought to provide resistance to invading bacteriophages or foreign DNA acquired via natural transformation (17–20). In this context phase variation may permit temporary removal of this restriction barrier, allowing the acquisition of foreign, potentially beneficial, DNA molecules (19). It has also been suggested that phase variation of methyltransferases may lead to autolytic self-DNA degradation by the cognate restriction enzyme and that such systems may be suicidal (19,21). This would lead to release of DNA into the environment for uptake by other cells, potentially to the benefit of the population (19,21). An alternative function for phase variable methyltransferases may be gene regulation, mediated by differential methylation of the genome (15). It has previously been established that DNA methylation can affect gene expression in several systems (e.g. Dam methylation) (22), however, there were no examples where the methyltransferase affecting gene expression is itself phase variably expressed. Recent work from our laboratory has shown that in H. influenzae strain Rd, a phase variable methyltransferase (Mod) of a type III R-M system coordinates the random switching of expression of multiple genes, and constitutes a phase variable regulon—‘phasevarion’ (23). H. influenzae is an important human pathogen causing invasive diseases, such as meningitis and respiratory tract infections. Functional phasevarions have also recently been confirmed in the human pathogens, N. meningitidis and N. gonorrhoeae, where a mod gene also coordinates expression of a regulon of many genes, several of which have a potential role in pathogenesis (Srikhanta et al., submitted for publication). Since a role in gene regulation is now established in several systems, the question arises of whether during evolution the role of these type III R-M systems has become exclusively gene regulation, or whether their role as DNA restriction systems has been retained.
MATERIALS AND METHODS
Bacterial isolates
Fifty-nine H. influenzae isolates were used in this study, as described in Table 1 of Supplementary Data. Strains included a set of 24 Finnish otitis media isolates that have been used in several previous studies (24–27). The other NTHi isolates have also been described previously (28). Encapsulated strains included type strains from the American Type Culture Collection (ATCC) as well as clinical isolates from the A. Smith laboratory and strains from the E. R. Moxon laboratory. For strains that had not previously been typed, multilocus sequence typing (MLST) was carried out as previously described (26). Strain details and sequence types were submitted to the public H. influenzae MLST website (haemophilus.mlst.net). An UPGMA (unweighted pair group method using arithmetic mean) dendrogram was constructed using the START2 collection of MLST-related software available at http://pubmlst.org/software/analysis/start2/(29).
Table 1.
Assay of type III-specific restriction
| Recipient cells | % Transformation efficiency (±SD) | |||
|---|---|---|---|---|
| Plasmid source | ||||
| Rd mod ON | Rd mod::kan | 162 wt (mod ON) | 162 mod::kan | |
| Rd mod ON | 78 (±14)* | 43 (±9)* | 1.6 (± 1.2) | ND |
| 162 wt (mod ON) | 0.1 (±0.2) | ND | 83 (± 16)** | 43 (±6)** |
Transformation efficiency of plasmid DNA isolated from mod ON and mod::kan cells, from H. influenzae strain Rd or 162, transformed into H. influenzae strain Rd or 162 recipient cells. A statistically significant difference (P < 0.05) was shown between the transformation efficiency using plasmid isolated from mod ON and mod::kan cells for both strains using the two-tailed Student's t-test; *P = 0.028; **P = 0.013; SD, standard deviation. ND not determined.
Bacterial growth conditions
Haemophilus influenzae was grown at 37°C in brain heart infusion (BHI) broth supplemented with hemin (10 μg/ml) and NAD (2 μg/ml). BHI plates were prepared with 1% (v/v) agar and supplemented with 10% (v/v) Levinthal base (30) and when appropriate kanamycin (10 μg/ml) or tetracycline (5 μg/ml).
DNA preparation, manipulation and analysis
All enzymes were sourced from New England Biolabs. PCR was performed using primers purchased from Sigma Proligo (Table 2 of Supplementary Data). Primers him6A and him11 were used to amplify the variable region of the mod gene, primers him1 and him3 or him4 and him5 were used to amplify the mod repeat tract, and primers HI1059for and HI1052rev were used to amplify across the mod/res region. The region of the res gene containing known frameshift mutations was amplified using primers HI1054for and HI1054rev or HI1054for2 and HI1054rev2. Sequencing reactions were prepared using PCR products as template and Big-Dye sequencing kit (Perkin Elmer). Samples were analysed using a 3130xl Capillary Electrophoresis Genetic Analyser (Applied Biosystems International). Data were analysed using MacVector (version 9.0) and DNA Sequencher. To analyse DNA fragment sizes, PCR products, amplified using a primer set in which the forward primer was labelled with 6-carboxyfluorescein (6-FAM), were analysed using the GeneScan system (Applied Biosystems International). Southern hybridization analysis was carried out as described by Sambrook et al. (31) using a DIG-labelled (Roche) PCR product as a probe. To confirm the presence/absence of the mod gene, primers him7 and him2 (Table 2, Supplementary Data) were used to PCR amplify the 3′ conserved region of mod from strain Rd DNA and this PCR product was used as a probe.
Construction of mod and res mutant strains of H. influenzae
Sheared genomic DNA from the Rdmod::kan mutant strain previously described (23) was used to transform non-typeable H. influenzae isolates R2866 and 162 by the MIV method (32). Mod::kan transformants were selected on BHI plates containing kanamycin and confirmed by PCR and Southern analysis. The res gene was PCR amplified using primers ResF and ResR. The PCR product was cloned into pGEM-Teasy vector (Promega), digested with HindIII and blunted using Klenow polymerase (New England Biolabs). The Tn903 kanamycin resistance cassette from the pUC4K vector (Pharmacia) was excised using HincII and inserted into the blunt HindIII site. The resulting plasmid, pGEMres::kan, was linearized by digestion with NcoI and used to transform non-typeable H. influenzae isolates 162 by the MIV method. Res::kan transformants were selected on BHI plates containing kanamycin and confirmed by PCR analysis.
Transformation of H. influenzae
Bacteria harvested from five plates of confluent growth on BHI agar supplemented with Levinthal base were resuspended in 20 ml of ice-cold sterile water containing 15% v/v glycerol and 272 mM sucrose (pH 7.4). Bacteria were centrifuged for 2 min at 13 000 r.p.m. and resuspended in 1 ml sterile water (containing sucrose and glycerol). This step was repeated 4–5 times, keeping the cells on ice between spins. The OD of the cell suspension was measured and normalized to an OD 600 of 10. One microgram of DNA was added to the cells that were then incubated on ice for 2 min. Cells were electroporated (Bio-Rad micropulser electroporator, 2.5 kV, 0.2 cm cuvettes) and BHI broth was added immediately. After 90 min incubation at 37°C with shaking, cells were plated on BHI agar containing tetracycline. After overnight growth, single colonies were selected, grown in broth and plasmid prepared using the Qiagen Plasmid Midi Kit (Qiagen, Doncaster, Vic, Au). For the quantitative transformation experiments, numbers of transformants were recorded. Twelve colonies were picked for sequencing of the mod repeat tract and a sample of the cell population was subject to fragment size analysis using the GeneScan system, as previously described (33). The transformation efficiency was calculated as number of colonies/μg DNA. The ratio of mod ON to mod OFF cells in the mod ON recipient cell population was checked pre- and post-transformation by fragment size analysis and confirmed to be unchanged.
ApoI cleavage assay
Plasmid pHStet was extracted from H. influenzae strain Rd mod ON and mod::kan cells, or R2866 mod ON and mod::kan cells using the Qiagen Plasmid Midi Kit (Qiagen, Doncaster, Vic, Au). One microgram of each plasmid was digested overnight with ApoI according to manufacturer's instructions and the resulting fragments were separated on a 2% high resolution agarose gel (Nusieve 3:1 Agarose,Cambrex BioScience, Rockland, ME, USA) with TBE at 70 V for 2 h and visualized under UV illumination. Similarly, digests were carried out using TaqI.
SouthWestern analysis
Plasmid pHStet was extracted from H. influenzae strain R2866 mod ON and mod::kan cells using the Qiagen Plasmid Midi Kit (Qiagen, Doncaster, Vic, Au). Mod ON cells were verified by sequencing the repeat tract. Five micrograms of each plasmid was digested overnight with DpnI according to manufacturer's instructions and the resulting fragments were separated on a 1.5% high resolution agarose gel (Nusieve 3:1 Agarose,Cambrex BioScience, Rockland, ME, USA) with TBE at 70 V for 2 h. The separated DNA fragments were transferred to nitrocellulose membrane (GeneScreen, Perkin Elmer, Rowville, Vic, Au) using overnight capillary transfer with 10× SSC. The DNA was cross-linked to the membrane by exposure to UV light, and then the membrane was washed three times in TBST (100 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% Tween 20) for 5 min with gentle agitation. The membrane was blocked for 1 h in 3% BSA in TBST with gentle agitation after which it was incubated for 1 h in 10 ml of a 1:1000 dilution of a rabbit anti-N6-methyl-adenine monoclonal antibody (Megabase Research Products, Lincoln, NE, USA) in the above blocking solution. The membrane was washed three times for 5 min in TBST prior to being incubated in a 1:20 000 dilution of a goat anti-rabbit IgG alkaline phosphatase conjugate antibody (Sigma-Adrich, Castle Hill, NSW, Au) in blocking solution for another hour. After another three washes, the membrane was immersed in 10 ml of Sigma FAST BCIP/NBT (Sigma-Aldrich) substrate.
RESULTS
H. influenzae strain Rd mod recognition site is 5′-CGAAT-3′
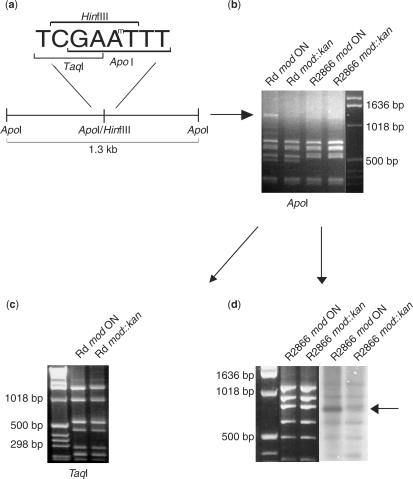
Early work on H. influenzae strain Rf DNA restriction systems characterized the recognition sequence for a type III R-M system (HinfIII) as 5′-CGAAT-3′ (34). The H. influenzae strain Rd genome sequence contains a single R-M system with homology to type III systems, encoded by the mod and res genes (35). The mod gene is subject to phase variable expression due to a 5′-AGTC-3′ tetranucleotide repeat tract within the open reading frame (11,23). To investigate whether the recognition sequence for Mod in strain Rd is the same as that for HinfIII, plasmid pHStet (36) was grown in strain Rd mod ON (40 repeats) and strain Rdmod::kan cells. The resulting methylated or non-methylated plasmids were then digested with an enzyme whose recognition sequence overlaps the proposed methylation sequence. Digestion by ApoI (5′-RAATTY-3′) is known to be inhibited by methylation of either of the adenines of the HinfIII sequence (Figure 1a). Only one of the multiple ApoI sites in pHStet overlaps with a HinfIII site and would potentially be inhibited by methylation. A 1.3-kb ApoI fragment containing this overlapping HinfIII/ApoI site was observed after digestion of the strain Rd mod ON derived (methylated) plasmid with ApoI. This band was absent from the strain Rdmod::kan-derived (non-methylated) plasmid digest (Figure 1b), indicating inhibition of ApoI digestion by methylation of the DNA by Mod. We conclude that Mod from strain Rd has the same site specificity as HinfIII (5′-CGAAT-3′). Here, we also demonstrate that it is the second adenine in the HinfIII sequence that is methylated. This is demonstrated by digestion of pHStet with TaqI. The TaqI recognition site (5′-TCGA-3′) overlaps with the first adenine of the HinfIII sequence. The TaqI cleavage pattern of pHStet, isolated from Rd mod ON and mod::kan cells, is identical, indicating that this base is not methylated by Mod (Figure 1c). This confirms the findings of a previous study, in which DNA methylated by the HinfIII enzyme, isolated from H. influenzae strain Rf, using S-[methyl-3H] adenosyl methionine, was cleaved by TaqI (34).
Figure 1.
Assessment of Mod methylation in H. influenzae strains Rd and R2866. (a) Schematic diagram showing the overlapping recognition sites for restriction endonucleases HinfIII, ApoI and TaqI. Plasmid pHStet contains one overlapping ApoI/HinfIII, located on a 1.3-kb fragment. (b) ApoI digest of plasmid pHStet isolated from Rd mod ON and mod::kan cells, or R2866 mod ON and mod::kan cells. Differential cleavage pattern of the 1.3-kb fragment evident with plasmid isolated from Rd mod ON and mod::kan cells. (c) TaqI digest of plasmid pHStet isolated from Rd mod ON and mod::kan cells. The recognition site for restriction endonuclease TaqI overlaps with the first A of the HinfIII recognition sequence [see schematic in part (a)]. No difference in the cleavage pattern evident between plasmid isolated from Rd mod ON and mod::kan cells. (d) DpnI digest of plasmid pHStet isolated from R2866 mod ON and R2866 mod::kan cells (first panel). Separated DNA fragments were transferred to nitrocellulose membrane and probed with anti-N6-methyladenosine antisera (second panel). Differential binding of the antisera to plasmid isolated from R2866 mod ON and R2866 mod::kan cells is indicated by an arrow. Increased background in the R2866 mod ON sample compared to the R2866 mod::kan sample is due to differential binding of the antisera to chromosomal DNA.
Evidence that the Mod recognition site is distinct in other strains of H. influenzae
An alignment of four mod gene sequences, from the four strains of H. influenzae for which the genomes have been sequenced and made available, indicated division of the mod gene sequence into three domains, typical of mod genes of type III R-M systems (37). The N- and C-terminal regions of the protein show between 90 and 96% similarity in sequence amongst the genome-sequenced strains, and are separated by a central domain that is completely dissimilar between strains, showing only 29–31% amino acid sequence similarity. This is consistent with previous findings that Mod proteins are highly conserved in the N- and C-terminal thirds of the protein with relatively low conservation in the central third (17,37). This central portion of the protein has been proposed to play a role in target sequence recognition and binding, and in binding of the methyl donor (38). The conserved regions are proposed to play a role in protein–protein interactions between Mod and Res.
To test whether the variant mod alleles present amongst genome sequenced strains of H. influenzae encode proteins with the same or distinct DNA recognition sequences, the ApoI restriction inhibition assay described above was carried out on plasmids derived from non-typeable H. influenzae (NTHi) strain R2866 (strain R2866). This strain was chosen since it contains a mod repeat tract length permissive for expression of the gene and it has a distinct DNA site specificity domain to strain Rd. In this strain background, the ApoI restriction pattern of plasmid pHStet was identical for plasmid derived from strain R2866 mod ON cells and mod::kan cells (Figure 1b), indicating that the methylation site specified by strain R2866 Mod, and hence the restriction site, is distinct from that of strain Rd Mod (i.e. not HinfIII; 5′-CGAAT-3′). However to confirm this, it was necessary to demonstrate that Mod is actually active in this strain. Plasmid pHStet isolated from R2866 mod ON and mod::kan cells was digested; separated DNA fragments were transferred to nitrocellulose membrane and probed with anti-N6-methyladenosine antisera. The antisera bound to all of the separated DNA fragments, indicating that endogenous methylases were methylating many sites within the plasmid. With one of the DNA fragments, differential binding of the antisera to plasmid isolated from R2866 mod ON and mod::kan cells was observed, indicating that many Mod methylation sites occur within this fragment allowing for an observable difference in antisera binding between the isogenic strains. This difference in antisera binding between wild-type and mod mutant confirms that Mod is an active methyltransferase in this strain (Figure 1d).
Heterogeneity in mod gene sequence defines 15 groups in H. influenzae strains
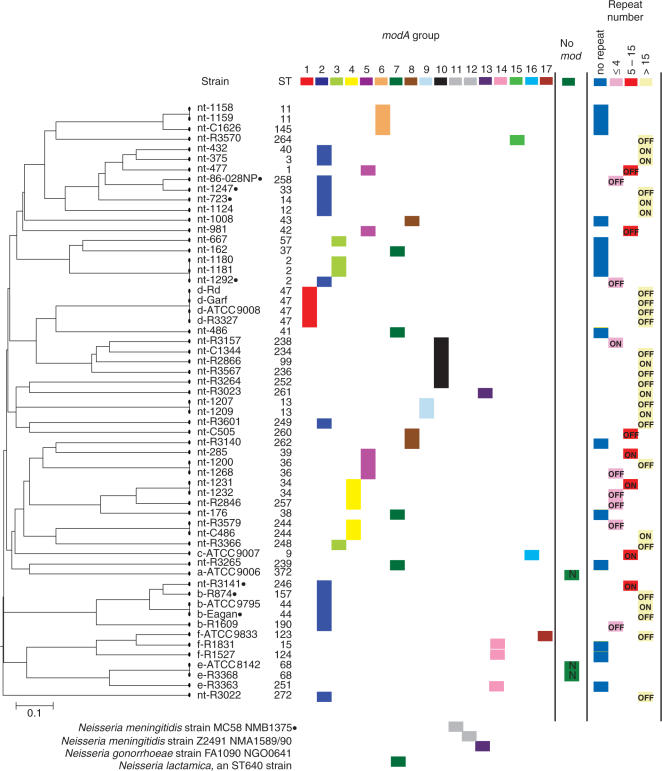
The sequence heterogeneity observed amongst the mod genes of the genome sequenced strains of H. influenzae (see above) and our experimental data supporting the correlation between mod sequence type and recognition site specificity, highlighted the need for a phylogenetic analysis of the mod gene from a genetically diverse set of H. influenzae strains. Encapsulated strains of H. influenzae are associated with invasive diseases such as meningitis and pneumonia, while acapsular or non-typeable H. influenzae (NTHi) strains are associated with otitis media and respiratory tract infections (39). We examined the mod gene in 59 strains, including 43 NTHi and 16 encapsulated strains. The genetic relatedness of these strains had been determined by multilocus sequence typing (MLST). PCR amplification and DNA sequencing of the central variable region of the mod gene and the mod repeat tract indicated significant heterogeneity.
We identified 15 mod sequence groups in the H. influenzae survey, such that within a group the variable region sequence showed >95% amino acid similarity. Between groups, the percentage sequence similarity of the variable region of the protein ranged from 29 to 38%. During the course of this study, Bayliss et al. (40) reported mod sequences of 22 NTHi isolates, constituting a subset of the NTHi collection described above, and proposed mod groupings. To avoid confusion, we have assigned the same numbers for common mod groups. The relationship of the 15 H. influenzae sequence groups to a MLST dendrogram is shown in Figure 2. A correlation is evident between mod sequence type and capsular serotype. Each strain from a particular capsular serotype shares an identical mod sequence type to all other members of that serotype, with the exception of one type f strain, ATCC 9833. All capsular type d strains possess a unique mod sequence type (Group 1). Capsular type e and f strains that contain the mod gene (two type e strains lacked mod and res), share a common mod sequence type that is unique from all other capsular or non-typeable strains (Group 14). Capsular type b strains share a mod sequence type with a number of NTHi isolates (Group 2). The serotype a strain examined does not possess a mod gene and the mod sequence from the one serotype c strain is unlike all other sequences identified (Group 16). The NTHi isolates show significant diversity in their mod gene sequence, with the NTHi isolates being distributed across 12 mod sequence groups. This finding is generally consistent with previous reports of the overall genetic diversity of NTHi strains, relative to encapsulated strains (26,27).
Figure 2.
Phylogenetic relatedness of 59 H. influenzae strains, based on MLST data, and the relationship to mod sequence group, mod repeat tract length and capsular serotype. The capsular serotype of each strain is indicated as a prefix to each strain name (nt, non-typeable). The ON/OFF expression status of the mod gene due to the number of repeats within the mod repeat tract is indicated within each box in the repeat number column. MLST type (ST) is indicated for each strain. Mod groups specific to pathogenic Neisseria are indicated at the bottom of the figure. Black dots next to the strain name indicate that the res gene in that strain has a frame shift mutation and is inactive.
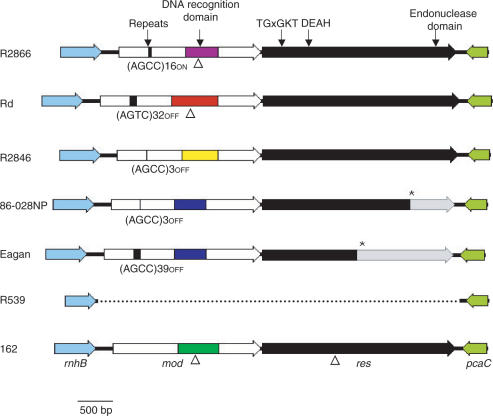
In three capsular H. influenzae strains (ATCC 9006, type a; ATCC 8142, type e; R3368, type e) no mod and res gene could be identified. PCR amplification with primers that bind to the genes flanking mod and res gave a product size consistent with the absence of mod and res. DNA sequencing of this PCR product indicated that only the flanking genes and a small amount of intergenic sequence was present (Figure 3). No remnant of the mod or res gene was found, indicating that either a clean deletion event had occurred or that these strains had never acquired this R-M system. Southern analysis using a probe against the 3′ conserved region of mod indicated that these genes were not present elsewhere within the genome (data not shown).
Figure 3.
Schematic diagram of the region of the genome containing the mod and res genes of six H. influenzae strains. Block arrows represent genes. The white regions of the block arrow representing the mod gene indicate the conserved N- and C-terminal regions. The coloured boxed region indicates the variable central region encoding DNA sequence recognition. The black box represents the position of the mod repeat tract indicated by an arrow, the sequence of the repeated motif, number of repeats and expression state of the gene are indicated below the black box. The block arrow representing the res gene indicates that the gene sequence is full-length in each case, with the transition from black to grey representing the position of a frame shift mutation. An asterisk indicates the position of a stop codon brought in frame due to the frame shift mutation in res. Strain Rd has no frame shift mutation in res. The genome sequence of strain KW20 Rd does have a frame shift mutation in res. It is not known whether this difference represents a sequencing error in the genome sequence or a genuine strain difference. The positions of the conserved motifs that are important in Res function are indicated above Res. Triangles indicate the site of insertion of a kanamycin resistance cassette. The solid black line indicates the intergenic region and the dotted line indicates a deletion.
A correlation is evident between mod repeat tract sequence and capsular serotype. The repeat unit 5′-AGTC-3′ was found in all strains of capsular serotype d, while the repeat unit 5′-AGCC-3′ was found in all other strains containing repeats, including NTHi isolates and strains of capsular serotypes b, c and f. A correlation is also evident between mod sequence group and the length of the mod repeat tract. In three of the 15 sequence groups, there is no repeat tract within the mod gene. Two of these groups are made up of NTHi isolates only (Groups 6 and 7), the other group contains capsular type e and f strains (Group 14). In these strains the DNA sequence 5′-TCAGATAGTCAG-3′ is present in place of the repeat tract. The mod gene is not predicted to be phase variable in these strains. Of the other twelve mod sequence groups, some groups contain predominantly low numbers of repeats or no repeats (Groups 3, 4, 5 and 8) and some contain predominantly high numbers of repeats (Groups 1, 2, 9 and 10). It has previously been reported in strain Rd that the length of the repeat tract correlates to the rate of mod phase variation (11). The phase variability of mod expression and the rate at which this occurs may reflect functional differences amongst different mod alleles. No correlation was detected in this study between mod sequence group or mod repeat tract length and disease phenotype of the corresponding strains (Figure 2).
Noted also are pathogenic Neisseria genes that define mod groups 11, 12 and 13. Previous work by Kroll et al. (41) reported the mod res genes of the type III R-M system as clear examples of horizontally transferred genes between the pathogens H. influenzae and N. meningitidis. In mod group 13 we show an example of NTHi isolate R3023, which has the same DNA recognition domain sequence as N. gonorrhoeae strain FA1090. The four NTHi isolates in mod group 7 have the same DNA recognition domain sequence as the Neisseria lactamica genome strain (Sanger Institute, UK). These genes clearly have a common origin. Due to the high sequence conservation of mod in H. influenzae and Neisseria species, the common DNA repeats mediating phase variation and the common origin via horizontal gene transfer, we propose the following nomenclature. The genes referred to in Figure 2 are hence called modA, followed by a number corresponding to the mod grouping based on the DNA recognition site allele, recognizing the key phenotype of DNA recognition specificity (e.g. the nt-R3023 gene is modA13). This avoids confusion in nomenclature with organisms like pathogenic Neisseria, which contain multiple mod genes that are distinct over the whole length of their sequence.
Inactivating mutations within the res gene occur in multiple strains
Unlike the significant diversity observed in the mod sequence amongst strains of H. influenzae, the res sequence is more highly conserved amongst the genome sequence strains of NTHi and strain Rd. The protein encoded by the res ORF varies in length amongst these strains from 722 amino acids (aa) in strain 86-028NP to 930 aa in strain Rd. The 930 aa Res protein from strain Rd and the 929 aa Res protein from strains R2866 and R2846 are similar in size to the homologous Res protein from other organisms. Alignment of the nucleotide sequence of the res genes from these four strains indicates homology to the full-length gene sequence in all cases although in strain 86-028NP, a single base pair deletion results in a frame-shift mutation truncating 207 aa from the C-terminus of the protein. The Res subunit of type III R-M systems contains several sequence motifs characteristic of DNA and RNA superfamily II helicases (42). These conserved motifs include the ATP-binding motif (TGxGKT) (42,43) and motif II (DEAH or DEPH) (42). In addition, a weakly conserved sequence ‘PD … (D/E)XK’ is found at the C-terminus of the Res protein which represents the endonuclease domain or active site, a signature found in several types of nucleases (44). This region of the protein is involved in metal binding, a requirement for DNA cleavage by several restriction enzymes (45). The truncation at the C-terminus of the strain 86-028NP protein results in loss of the endonuclease domain, and the protein is predicted to be inactive (Figure 3).
To investigate whether the same 86-028NP single base pair frame shift mutation is found in the res gene of other strains, a 1.3-kb region of the res gene encompassing the 86-028NP mutation was sequenced in the 56 strains containing mod and res. The single base pair deletion in the res gene of 86-028NP was also found in strains nt-723 and nt-1247 (mod group 2). Unexpectedly, a different, 32-bp deletion was found in the res gene of four strains in the mod group 2. This mutation introduced a frame-shift resulting in a peptide of only 495 aa,—a more extensive carboxy terminal truncation than the 86-028NP mutation (see Figure 3). All serogroup b strains tested contain this mutation, as does the genome sequence serogroup b strain 10810 (Sanger Institute, UK). Conversely, in the res gene of strains containing a non-phase variable mod gene, no obvious frame-shift mutations were observed.
Transformation experiments using plasmids specifically methylated or non-methylated by Mod to test type III restriction activity
To further investigate restriction activity of the type III R-M systems, we used plasmids specifically methylated or non-methylated by Mod (i.e. isolated from mod ON or mod::kan cells, respectively) to transform H. influenzae strains Rd and 162. Prior to transformation, the methylation state of plasmid isolated from strain Rd was checked using the ApoI inhibition assay described above and by sequencing of the mod repeat tract, this was not necessary for strain 162 since mod is constitutively expressed in this strain. A statistically significant difference in transformation efficiency was shown between Mod methylated plasmids and non-methylated plasmids transformed into the homologous strain (mod ON cells of strain Rd or 162) (Table 1). The Mod methylated plasmids transformed only two times more efficiently into these strains than non-methylated plasmids. This difference in transformation efficiency is attributable entirely to the activity of the type III R-M system and is independent of other R-M systems, which remain unchanged in the isogenic wild-type/mod::kan strain pairs.
When Mod methylated plasmids were transformed into the heterologous strain (i.e. plasmid isolated from Rd mod ON cells transformed into 162, and vice versa), a significant difference in transformation efficiency was observed between plasmids methylated in the same strain and plasmids methylated in a heterologous strain (Table 1). This significant reduction in transformation efficiency is due to differences in other, non-type III methylation systems between these two strains.
The contribution of the Mod-Res system to the overall restriction barrier is indicated by the transformation efficiency of non-methylated plasmids. Although the transformation efficiency is significantly decreased compared to Mod-methylated plasmids, 43% of plasmids are still able to transform the cells, indicating that these type III R-M systems play only a minor role in the overall defence of the cell against invasion by foreign DNA.
DISCUSSION
We have previously shown that H. influenzae strain Rd Mod is a phase variably expressed, epigenetic regulator of multiple genes—a phasevarion (23). Here we report that the recognition sequence for Mod is 5′-CGAAT-3′, the same as previously reported for HinfIII (46). This is a key finding enabling our studies resolving how ON/OFF switching of methylation influences the promoters of genes controlled by the phasevarion of strain Rd. Identification of the strain Rd Mod target site also enabled design of strategies used in this study to address functional and evolutionary questions presented by phase variable type III R-M systems of H. influenzae. Comparisons of the mod and res genes from the genome strains, and our large scale survey of capsulate and NT H. influenzae revealed a wide range of variation in the region of the mod gene thought to dictate sequence specificity (37,40). This variation was also observed in the recent NTHi sequence analysis conducted by Bayliss et al. (40). Here we used an ApoI inhibition assay to demonstrate that the modA10 gene of strain R2866 (mod group 10) does not modify the same sequence as modA1 of strain Rd (mod group 1), confirming these enzymes methylate distinct target sequences. These data provide experimental evidence that sequence variation observed in the putative DNA specificity domain, used to generate mod groupings in Figure 2, reflect Mod proteins with distinct target sequences. Consistent with these findings, our recent work in pathogenic Neisseria has confirmed that phase variation of the ModA11, 12 and 13 methyltransferases (see Figure 2) control expression of distinct genes (Srikhanta, submitted for publication).
These findings raise the question of whether the 15 mod groupings in H. influenzae represent 15 different type III R-M systems or 15 different methyltransferases controlling phasevarions, and whether these functions are mutually exclusive. Sequence analysis revealed three groupings (Groups 6, 7 and 14) in which none of the mod genes contain tetranucleotide repeat sequences. These genes are therefore not phase variably expressed, and by definition these are not phasevarions. These groups include strain 162, in which we have demonstrated a functional type III R-M system. We propose all these groups, and any other strain with a non-phase variable modA, are likely to be functional, dedicated type III R-M systems. In contrast, representatives of modA2 group all have high numbers of tetranucleotide repeats, consistent with phase variable expression. Analysis of the res gene from strains within these groups revealed frame-shift mutations in res in many cases that are inconsistent with expression of a functional Res protein. We propose that these strains, and any other strains with tetranucleotide repeats in mod, and obvious inactivating mutations in res, function exclusively as phasevarions. Hence, of the 41 strains in the survey that contain a phase variable mod gene, seven have an obvious mutation in res and appear to be dedicated phasevarions. This represents 17% of strains containing a phase variable mod gene. The remaining 15 strains that do not contain a phase variable mod gene are likely to be dedicated, functional, type III R-M systems. There were no strains in this group that had a corresponding inactivating mutation in the res gene. In the strains above the distinct functions of regulation and restriction appear to be mutually exclusive.
In a related study, another type of inactivating frame-shift mutation has been identified in the res genes associated with modA11 allele of N. meningitidis (Figure 2) that results in premature truncation of the encoded protein and loss of the endonuclease domain, this mutation is present in 70% of strains with the modA11 allele (Srikhanta et al., submitted for publication). In N. gonorrhoeae, a 250-aa in-frame deletion has been observed in Res associated with the modA13 allele, potentially inactivating restriction function (Srikhanta et al., submitted for publication). These findings illustrate that inactivation of restriction function is common amongst the modA containing phase variable type III R-M systems.
Transformation experiments with strain Rd and strain 162 measuring the effectiveness of type III-specific restriction revealed only a two-fold increase in protection when the system was active. This appears to be a marginal level of functionality and raises the question of the selective advantage of restriction function at this level. More detailed analysis of restriction function is required to determine how many of the remaining strains contain a functional type III restriction system. Sequencing a region corresponding to one-third of the res gene to look for 86-028NP point mutations, we found a different 32-bp deletion mutation that inactivated res. The possibility remains that the obvious, inactivating frame-shift mutations found in res genes associated predominantly with modA2 group, the largest mod group (representing 25% of strains surveyed), may represent the ‘tip of the iceberg’ with many other possible silencing mechanisms either unsurveyed or currently undetectable by simple sequence analysis of these poorly defined enzymes.
The observation of non-functional type III R-M systems in the modA2 strains containing phase variable mod genes suggests two possibilities, first, that the restriction function is redundant and has been lost, and second, that phase variable expression of mod may be inconsistent with a viable organism if an active Mod-Res holoenzyme can form. In non-phase variable type III R-M systems, sites are modified during replication, and any unmodified sites in newly replicated DNA are either in the same orientation or paired with methylated sites, thereby preventing suicidal restriction of cellular DNA (8). In a strain containing an active type III R-M system with a phase variable mod gene, if the mod gene switches OFF for several rounds of replication, then phase varies back to ON, the condition may be lethal or detrimental, as none of the Mod sites in the genome would be methylated and protected from cleavage. Our original work in strain Rd (23), and recent work in pathogenic Neisseria (Srikhanta et al., submitted for publication), have established a role for phase variable mod genes in control of gene expression. This study has revealed that the H. influenzae strains associated with human disease may contain a series of distinct phasevarions. Knowledge of the changes in gene expression that occur as these phasevarions switch the organisms between two distinct cell types will have a major impact on our ability to develop an understanding of the general principles of host pathogen interactions for H. influenzae, and to assess H. influenzae antigens as vaccine candidates. Our current understanding is that the role of the mod-res locus in H. influenzae biology is in transition. In some strains of the H. influenzae population it retains its function as a type III R-M system, while in others it plays a key role as a dedicated, randomly switching, epigenetic mechanism for controlling gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank Kevin Nelson and Nicole Bradbury for their contribution to the MLST typing of strains and DNA sequencing of the mod gene. We also wish to thank Joanne Tan and Sarah-Jane Matthews for technical assistance. This work was supported by National Health and Medical Research Council (Australia) Program Grant 284214, and by AI 46512 from the National Institute of Allergy and Infectious Disease (USA). Funding to pay the Open Access publication charges for this article was provided by National Health and Medical Research Council (Australia) Program Grant 284214.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boyer HW. DNA restriction and modification mechanisms in bacteria. Annu. Rev. Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- 2.Iida S, Meyer J, Bachi B, Stalhammar-Carlemalm M, Schrickel S, Bickle TA, Arber W. DNA restriction–modification genes of phage P1 and plasmid p15B. Structure and in vitro transcription. J. Mol. Biol. 1983;165:1–18. doi: 10.1016/s0022-2836(83)80239-3. [DOI] [PubMed] [Google Scholar]
- 3.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 5.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadi SM, Bachi B, Iida S, Bickle TA. DNA restriction – modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J. Mol. Biol. 1983;165:19–34. doi: 10.1016/s0022-2836(83)80240-x. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad I, Rao DN. Interaction of EcoP15I DNA methyltransferase with oligonucleotides containing the asymmetric sequence 5′-CAGCAG-3′. J. Mol. Biol. 1994;242:378–388. doi: 10.1006/jmbi.1994.1588. [DOI] [PubMed] [Google Scholar]
- 8.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiser JN, Williams A, Moxon ER. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 1990;58:3455–3457. doi: 10.1128/iai.58.10.3455-3457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ham SM, van Alphen L, Mooi FR, van Putten JP. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73:1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- 11.De Bolle X, Bayliss CD, Field D, van de Ven T, Saunders NJ, Hood DW, Moxon ER. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 2000;35:211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 12.de Vries N, Duinsbergen D, Kuipers EJ, Pot RG, Wiesenekker P, Penn CW, van Vliet AH, Vandenbroucke-Grauls CM, Kusters JG. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 2002;184:6615–6623. doi: 10.1128/JB.184.23.6615-6623.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan KA, Lo RY. Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res. 1999;27:1505–1511. doi: 10.1093/nar/27.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R, Moxon ER. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 15.Seib KL, Peak IR, Jennings MP. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 2002;32:159–165. doi: 10.1111/j.1574-695X.2002.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 16.Fox KL, Srikhanta YN, Jennings MP. Mol. Microbiol. 2007. Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. in press. [DOI] [PubMed] [Google Scholar]
- 17.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dybvig K, Yu H. Regulation of a restriction and modification system via DNA inversion in Mycoplasma pulmonis. Mol. Microbiol. 1994;12:547–560. doi: 10.1111/j.1365-2958.1994.tb01041.x. [DOI] [PubMed] [Google Scholar]
- 19.Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl Acad. Sci. USA. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson IR, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J. Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 22.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl Acad. Sci. USA. 2005;102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hood DW, Makepeace K, Deadman ME, Rest RF, Thibault P, Martin A, Richards JC, Moxon ER. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 1999;33:679–692. doi: 10.1046/j.1365-2958.1999.01509.x. [DOI] [PubMed] [Google Scholar]
- 25.Bolduc GR, Bouchet V, Jiang RZ, Geisselsoder J, Truong-Bolduc QC, Rice PA, Pelton SI, Goldstein R. Variability of outer membrane protein P1 and its evaluation as a vaccine candidate against experimental otitis media due to nontypeable Haemophilus influenzae: an unambiguous, multifaceted approach. Infect. Immun. 2000;68:4505–4517. doi: 10.1128/iai.68.8.4505-4517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 2003;41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cody AJ, Field D, Feil EJ, Stringer S, Deadman ME, Tsolaki AG, Gratz B, Bouchet V, Goldstein R, et al. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 2003;3:57–66. doi: 10.1016/s1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, Unrath WC, Campos J, Crook DW, et al. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 2005;73:5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 30.Alexander HE. The Haemophilus group. In: Dubos RJ, Hirsch JG, editors. Bacterial and Mycotic Infections of Man. London, England: Pittman Medical Publishing Co. Ltd; 1965. [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Herriott RM, Meyer EM, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erwin AL, Allen S, Ho DK, Bonthuis PJ, Jarisch J, Nelson KL, Tsao DL, Unrath WC, Watson ME, Jr, et al. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect. Immun. 2006;74:6226–6235. doi: 10.1128/IAI.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piekarowicz A, Bickle TA, Shepherd JC, Ineichen K. The DNA sequence recognised by the HinfIII restriction endonuclease. J. Mol. Biol. 1981;146:167–172. doi: 10.1016/0022-2836(81)90372-7. [DOI] [PubMed] [Google Scholar]
- 35.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 36.Lancashire JF, Terry TD, Blackall PJ, Jennings MP. Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 2005;49:1927–1931. doi: 10.1128/AAC.49.5.1927-1931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humbelin M, Suri B, Rao DN, Hornby DP, Eberle H, Pripfl T, Kenel S, Bickle TA. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 1988;200:23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- 38.Rao DN, Page MG, Bickle TA. Cloning, over-expression and the catalytic properties of the EcoP15 modification methylase from Escherichia coli. J. Mol. Biol. 1989;209:599–606. doi: 10.1016/0022-2836(89)90597-4. [DOI] [PubMed] [Google Scholar]
- 39.Turk DC. The pathogenicity of Haemophilus influenzae. J. Med. Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 40.Bayliss CD, Callaghan MJ, Moxon ER. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 2006;34:4046–4059. doi: 10.1093/nar/gkl568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroll JS, Wilks KE, Farrant JL, Langford PR. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl Acad. Sci. USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbalenya AE, Koonin EV. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991;291:277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- 43.Saha S, Ahmad I, Reddy YV, Krishnamurthy V, Rao DN. Functional analysis of conserved motifs in type III restriction-modification enzymes. Biol. Chem. 1998;379:511–517. doi: 10.1515/bchm.1998.379.4-5.511. [DOI] [PubMed] [Google Scholar]
- 44.Sistla S, Rao DN. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 45.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kauc L, Piekarowicz A. Purification and properties of a new restriction endonuclease from Haemophilus influenzae Rf. Eur. J. Biochem. 1978;92:417–426. doi: 10.1111/j.1432-1033.1978.tb12762.x. [DOI] [PubMed] [Google Scholar]