Abstract
Young meristematic plant cells contain a large number of small vacuoles, while the largest part of the vacuome in mature cells is composed by a large central vacuole, occupying 80% to 90% of the cell volume. Thus far, only a limited number of vacuolar membrane proteins have been identified and characterized. The proteomic approach is a powerful tool to identify new vacuolar membrane proteins. To analyze vacuoles from growing tissues we isolated vacuoles from cauliflower (Brassica oleracea) buds, which are constituted by a large amount of small cells but also contain cells in expansion as well as fully expanded cells. Here we show that using purified cauliflower vacuoles and different extraction procedures such as saline, NaOH, acetone, and chloroform/methanol and analyzing the data against the Arabidopsis (Arabidopsis thaliana) database 102 cauliflower integral proteins and 214 peripheral proteins could be identified. The vacuolar pyrophosphatase was the most prominent protein. From the 102 identified proteins 45 proteins were already described. Nine of these, corresponding to 46% of peptides detected, are known vacuolar proteins. We identified 57 proteins (55.9%) containing at least one membrane spanning domain with unknown subcellular localization. A comparison of the newly identified proteins with expression profiles from in silico data revealed that most of them are highly expressed in young, developing tissues. To verify whether the newly identified proteins were indeed localized in the vacuole we constructed and expressed green fluorescence protein fusion proteins for five putative vacuolar membrane proteins exhibiting three to 11 transmembrane domains. Four of them, a putative organic cation transporter, a nodulin N21 family protein, a membrane protein of unknown function, and a senescence related membrane protein were localized in the vacuolar membrane, while a white-brown ATP-binding cassette transporter homolog was shown to reside in the plasma membrane. These results demonstrate that proteomic analysis of highly purified vacuoles from specific tissues allows the identification of new vacuolar proteins and provides an additional view of tonoplastic proteins.
Young meristematic plant cells contain a large number of small vacuoles known to fuse during cell expansion and finally form a single large central vacuole, which occupies 80% to 90% of the cell volume (Marty, 1999). Cell expansion is, therefore, closely linked with increase of vacuolar volume. To reach the full cell size without expending too much energy, large amounts of inorganic ions are transported into the vacuole during vacuolar growth, concomitantly followed by a substantial water influx (Maeshima, 2001; Martinoia et al., 2007). In adult cells the vacuole plays a central role in cytosolic homeostasis. The concentration of metabolites in the cytosol and other metabolic active compartments has to be tightly regulated to facilitate the optimal functioning of a large number of metabolic pathways. Uptake, storage, and release of solutes into and out of the vacuole are thus of pivotal importance for a plant cell. Our knowledge of vacuolar transport processes is mainly derived from studies on vacuoles and tonoplast vesicles isolated from fully expanded cells (Maeshima, 2001; Martinoia, et al., 2007).
Two vacuolar proton pumps, the vacuolar H+-ATPase (V-ATPase) and the vacuolar pyrophosphatase (V-PPase) generate an electrochemical gradient, which can be used to drive solute uptake into the vacuole (Rea and Sanders, 1987; Maeshima, 2001). The membrane potential across the tonoplast enables the accumulation of anions. In the case of organic acids they may be additionally trapped within the vacuole by changing their protonation state within the acidic lumen. Cations, such as sodium or calcium, and uncharged solutes such as Suc can be accumulated by use of antiporters, which exchange protons with the corresponding solute. In addition, glutathionated, glucuronated, or glucosylated compounds may be transported into the vacuole by ATP-binding cassette-type transporters (Rea et al., 1998; Theodoulou, 2000; Martinoia et al., 2002).
Several vacuolar membrane proteins have been identified during the last years. In terms of abundance in the membrane the aquaporins are by far the most prevalent tonoplastic proteins (Maeshima, 2001). The sodium/proton antiporter has probably attracted most interest since overexpression of this transporter leads to salt-tolerant plants (Apse et al., 1999; Zhang and Blumwald, 2001). The members of the CAX family exchange calcium against protons and at least some of these transporters are also able to exchange cadmium against protons and thus are of potential interest for phytoremediation (Hirschi et al., 2000; Hirschi, 2001; Cheng et al., 2003).
NRAMP is a transporter implicated in iron export from the vacuole, since deletion mutants in this transporter exhibit a slow growth phenotype under low iron conditions, and double deletion mutants AtNRAMP3xAtNRAMP4 cannot mobilize iron from the seed vacuoles (Thomine et al., 2000, 2003; Lanquar et al., 2005). Recently two vacuolar key transporters have been identified at the molecular level: a K+/Ca2+ channel (Peiter et al., 2005) corresponding to the well-described slow vacuolar channel and a nitrate antiporter responsible for the accumulation of nitrate, belonging to the family of the ClC channels (De Angeli et al., 2006). Furthermore, a malate transporter has been identified (Emmerlich et al., 2003). This transporter does not correspond to the described vacuolar dicarboxylate channel (Hurth et al., 2005) but plays an important role in vacuolar storage of malate. Glutathionated, glucuronated, and possibly malonylated compounds are substrates of ATP-binding cassette transporters. In view of the multitude of compounds identified in the vacuole it must be postulated that a large number of vacuolar transporters still await identification and characterization. One attempt to gain more insights into the vacuolar proteome is to isolate highly enriched vacuolar preparations and subject them to a proteomic approach. Such an approach has been undertaken for Arabidopsis (Arabidopsis thaliana) and barley (Hordeum vulgare; Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004; Endler et al., 2006; Jaquinod et al., 2007).
Each group of researchers used different purification and separation methods. Interestingly, the overlap between identified proteins is relatively low. Between 34 and 650 different proteins, including soluble and membrane proteins were detected. From these 23 to 195 were reported to be membrane proteins. This discrepancy can be attributed to, for example, different amounts of proteins, and individual techniques used in the respective approaches. All six groups of researchers identified at least three subunits of the V-ATPase. Besides tonoplastic proteins, nonvacuolar proteins from other organelles like mitochondria, plasma membrane, and chloroplasts were additionally detected. Therefore, for all newly identified putative vacuolar proteins final proof for vacuolar localization has to be shown. However, this aspect has only been addressed by Endler et al. (2006) and Jaquinod et al. (2007). An alternative strategy to identify membrane proteins from organelles has been presented by Dunkley et al. (2006). Using density gradients in combination with isotope tagging they could differentiate membrane proteins from different organelles.
As mentioned above, vacuoles undergo different developmental stages. To analyze vacuoles from growing tissues we isolated corresponding organelles from cauliflower (Brassica oleracea) buds, which are mainly constituted of small and only partially expanded cells. Here we show that using different treatments for the purification of the tonoplast and analyzing these against the Arabidopsis database, 102 membrane proteins and 214 soluble cauliflower proteins could be identified. From the 102 identified membrane proteins 32.3% of the peptides could be assigned as vacuolar membrane proteins, comprising nine vacuolar membrane proteins. We identified 57 proteins containing at least one membrane spanning domain with an unknown subcellular localization. To verify whether some of the newly identified proteins are indeed tonoplastic we constructed and expressed five GFP fusions, four of which are localized in the tonoplast.
RESULTS
Purification of Tonoplast Proteins
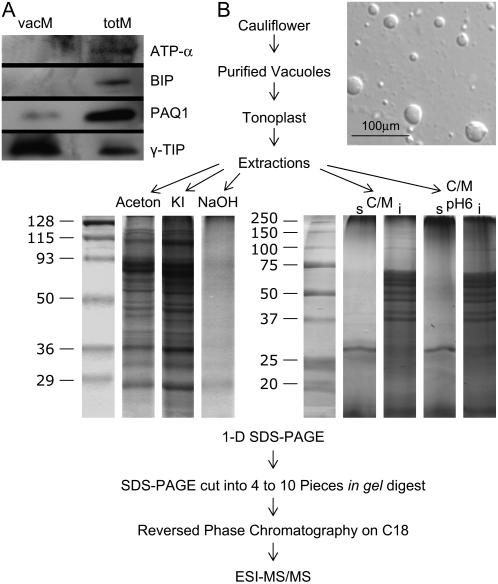
Using the procedure published by Marty-Mazars et al. (1995), an average of 1 mg tonoplast proteins was purified from 750 g of cauliflower heads. The purity of the Nycodenz-purified fraction was assessed by using marker enzymes and the ATPase assay in the presence of specific inhibitors (Table I) as well as by western blots using specific antibodies (Fig. 1A). Based on the enzymatic analyses we determined that the enriched tonoplast fractions were contaminated up to 1.3% with plasma membranes, chloroplasts, mitochondria, Golgi, and endoplasmic reticulum (ER; see Table I). Estimating the contamination of the vacuolar fraction using the western-blot technique shows that this fraction is only slightly contaminated (see Fig. 1A). The vacuolar marker γ-TONOPLAST INTRINSIC PROTEIN (TIP) is strongly enriched in the vacuolar preparation compared to the microsomal fraction. In contrast, a strong signal for the PLASMA MEMBRANE AQUAPORIN1 (PAQ1) could be detected in the microsomal fraction. The corresponding signal was only very week in the vacuolar fraction and based on the ratio of the bands between the microsomal fraction and the purified vacuoles a contamination of less than 5% can be estimated. The signals of the ER and plastidic markers BIP and ATPase (subunit α) were weaker than that of the PAQ1 and could not be detected in the vacuolar preparation. Therefore, it can be concluded that the vacuolar preparation is only marginally contaminated with other membranes. This highly purified vacuolar membrane preparation was used to perform the vacuolar proteomic analysis.
Table I.
Enzymatic characterization of the tonoplast fraction
Specific activities of marker enzymes in membrane fractions isolated from cauliflower inflorescence. Marker enzyme activities for mitochondria (CCO), plastids and mitochondria (F-ATPase), Golgi apparatus (IDPase), ER (CCR), and plasma membrane ATPase (P-ATPase) are presented together with the marker enzyme activity for tonoplast (V-ATPase) in microsomal and tonoplast-enriched fractions.
| Marker Enzyme | Specific Enzyme Activity
|
% in Tonoplast Fraction | |
|---|---|---|---|
| Total Microsomes | Tonoplast Fraction | ||
| pkat mg−1 protein | |||
| CCO | 41 × 103 | n.m.a | 0 |
| CCR | 9 × 103 | 7 × 102 | 1.1 |
| Latent IDPase | 29 × 103 | 2 × 103 | 0.9 |
| F-ATPase | 650 | 66 | 1.3 |
| P-ATPase | 1,450 | 77 | 0.7 |
| V-ATPase | 277 | 2,085 (7.5×)b | 100 |
Not measurable.
Relative specific activity (degree of enrichment).
Figure 1.
Evaluation of vacuolar membrane purity by western blot analysis (A) and fractionation strategy for cauliflower tonoplast proteins (B). A, Purified vacuoles were treated with five different extraction methods: KI and alkaline (NaOH) washing, acetone and C/M extraction, and treatment with sodium phosphate pH 6 followed by C/M pH 6 extraction, subjected to one-dimensional SDS-PAGE followed by in gel digestion and LC/MS/MS. B, For analysis of purification 10 μg of total membrane proteins (totM) and tonoplast proteins (vacM) were loaded on each lane. Compartment specific antibodies were used for western-blot analysis: chloroplastic ATPase α-subunit (ATP-α; 55 kD), luminal binding protein (BIP; 73 kD), plasma membrane aquaporin (PAQ1; 30 kD), and γ-TIP (25 kD).
Identification of Membrane Proteins from Different Extraction Methods
Ferro et al. (2003) already demonstrated the importance of using complementary methods to identify as many membrane proteins as possible. However, such an approach has not been undertaken using plant vacuoles. Therefore, we used five different extraction procedures: namely alkaline (NaOH) and saline (KI) washing, acetone and chloroform/methanol (C/M) extraction, and treatment with sodium phosphate (at pH 6) followed by C/M extraction (C/M pH 6; Fig. 1B).
To identify the hydrophobic proteins present in the different extracts, proteins were further fractionated by SDS-PAGE (Fig. 1B). Prior to mass spectrometry (MS) we separated the tryptic peptides by reversed phase chromatography (C18) and analyzed them by nanospray ionization MS/MS with an ion trap. Proteins were identified by SEQUEST searches using the National Center for Biotechnology Information Arabidopsis database (http://www.ncbi.nlm.nih.gov/). Protein identifications were based on the Peptide and Protein-Prophet data analysis software (Keller et al., 2002; Supplemental Table S1; cutoff of 0.9). The Arabidopsis database was chosen because cauliflower and Arabidopsis are closely related Brassicacea with highly conserved protein sequences. The Arabidopsis database is well annotated and has a size that is compatible with statistical tools for data analysis. Using higher search spaces (e.g. the Viridiplantae section of the National Center for Biotechnology Information database) increases the risk of false-positive protein identifications and severely hampers the use of statistical tools such as Peptide and Protein-Prophet.
Using the membrane fractions described above we identified 102 cauliflower membrane proteins with our proteomic approach (Table II). Further information on additional identified proteins (214 soluble proteins) is available in the Supplemental Table S1. Table II lists all identified membrane proteins sorted according to the number of their putative transmembrane spanning domains (TMDs) that were determined using the Aramemnon database (http://aramemnon.botanik.uni-koeln.de/) and the MIPS database (http://mips.gsf.de/). We included information about the molecular masses, the pI, the subfractions in which each protein was identified, i.e. acetone, KI, NaOH, C/M, or C/M pH 6, the number of analyzed gel slices, and the overlap to other vacuolar proteomic studies (Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004; Endler et al., 2006; Jaquinod et al., 2007). The number of unique peptides detected in the different treated fractions is indicated. A total of 35 of 102 membrane proteins (34.3%) are identified by a unique peptide hit.
Table II.
Putative vacuolar membrane proteins sorted according to the number of their putative TMDs
AGI Acc. No., Arabidopsis Genome Initiative accession number; AGIs in italics correspond to other possible proteins matching with the same peptides or part of them, AGIs in bold correspond to novel localized membrane proteins. TMD, Putative transmembrane domains were assumed according to the Aramemnon database (http://aramemnon.botanik.uni-koeln.de/) and the MIPS database (http://mips.gsf.de/). Loc., Subcellular localization predictions according to the Aramemnon database [CL, Chloroplast; ED, endosomes; MT, mitochondria; PM, plasma membrane; (PM), novel plasma membrane protein; PR, peroxisomes; T, tonoplast; (T), novel tonoplast protein; U, unknown]. PP, Protein-Prophet scores above 0.9 were accepted; at this cutoff, the rate of false-positive protein identifications is less than 10%; l, lower PP score but additional localization experiments (in bold); kD, predicted Mr in kD; tonoplast fraction treated with KI, potassium iodide; NaOH, sodium hydroxide; C/M pH 6, treatment with sodium phosphate buffer at pH 6 followed by C/M; numbers in parentheses mean the number of gel slices. The fraction in which each protein was identified is indicated by the unique number of peptides. Ref, Indicates Arabidopsis homolog(s) found by: a, Carter et al. (2004); b, Endler et al. (2006); c, Sazuka et al. (2004); d, Shimaoka et al. (2004); e, Szponarski et al. (2004); and f, Jaquinod et al. (2007).
| AGI Acc. No. | Protein Function | TMD | Loc. | PP | kD | pI | KI (8) | Ac (8) | NaOH (4) | NaOH (10) | C/M (4) | C/M pH6 (4) | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At1g16780 | Vacuolar proton-translocating pyrophosphatase | 16 | T | 1 | 85.3 | 5.4 | 3 | 1 | d | ||||
| At1g78920 | Vacuolar proton-translocating pyrophosphatase (AtAVP2) | 16 | T | 1 | 85.1 | 5.4 | 2 | d | |||||
| At3g62700 | Multidrug resistance-related protein (AtMRP14) | 16 | U | 1 | 172.1 | 8.7 | 2 | 7 | 2 | a, b, d, e, f | |||
| At2g47800 | |||||||||||||
| At3g07160 | Callose synthase (1,3-β-glucan synthase) family | 15 | U | 1 | 222.1 | 8.6 | 4 | ||||||
| At1g15690 | V-PPase (AtAVP-3) | 14 | T | 1 | 80.8 | 4.9 | 9 | 21 | 21 | 2 | 4 | a, b, c, d, e, f | |
| At1g01790 | Potassium efflux antiporter, putative (AtKEA1) | 12 | U | 1 | 65.0 | 6.5 | 3 | ||||||
| At3g28860 | Multidrug resistance protein (AtMDR11) | 12 | U | 1 | 136.8 | 8.4 | 4 | 4 | |||||
| At4g38350 | Patched protein family | 12 | U | 1 | 116.1 | 5.1 | 4 | a, d, e, f | |||||
| At1g16390 | Organic cation transporter related | 11 | (T) | 1 | 57.4 | 5.9 | 1 | ||||||
| At3g49310 | Expressed protein | 11 | U | 0.93 | 50.7 | 6.7 | 1 | f | |||||
| At1g64650 | |||||||||||||
| At4g27720 | Expressed protein | 11 | U | 0.93 | 50.9 | 7.2 | 1 | 1 | |||||
| At1g27770 | Ca2+-ATPase 1 (AtACA1) | 10 | ER | 1 | 103.7 | 5.4 | 2 | ||||||
| At1g29310 | Flower pigmentation protein | 10 | U | 0.97 | 52.2 | 9.3 | 3 | 3 | |||||
| At2g34250 | |||||||||||||
| At1g53210 | Drought-induced protein, putative | 10 | U | 1 | 63.4 | 5.0 | 2 | 3 | 3 | a, e, f | |||
| At2g07560 | P-ATPase (AtAHA6) | 10 | PM | 1 | 105.0 | 6.1 | 5 | ||||||
| At2g18960 | P-ATPase (AtAHA1) | 10 | PM | 0.98 | 104.2 | 6.7 | 2 | 4 | 1 | 6 | a, c, f | ||
| At4g30190 | |||||||||||||
| At2g41560 | Ca2+-ATPase 4 (AtACA4) | 10 | T | 1 | 112.7 | 5.9 | 2 | a, b, f | |||||
| At3g57330 | Ca2+-ATPase 11 (AtACA11) | 10 | T | 1 | 111.9 | 6.3 | 3 | a, b, d, e, f | |||||
| At4g35300 | Monosaccharide transporter, putative | 10 | U | 0.97 | 79.7 | 5.4 | 1 | 1 | a, b, f | ||||
| At1g10950 | Endomembrane protein 70, putative | 9 | ER | 0.98 | 66.9 | 7.7 | 2 | 2 | |||||
| At3g13772 | Endomembrane protein 70, putative | 9 | ER | 0.92 | 73.6 | 8.2 | 1 | f | |||||
| At1g55130 | |||||||||||||
| At2g01970 | Endomembrane protein 70, putative | 9 | ER | 0.95 | 68.0 | 7.1 | 2 | ||||||
| At5g45370 | Nodulin N21 family protein | 9 | (T) | 1 | 38.4 | 8.5 | 1 | ||||||
| At3g45870 | |||||||||||||
| At4g19185 | |||||||||||||
| At5g46110 | Phosphate/triose-P translocator (AtTPT) | 9 | CL | 0.99 | 44.6 | 10.3 | 2 | 2 | |||||
| At1g06470 | Integral membrane protein, putative | 8 | (T) | 1 | 46.4 | 4.7 | 1 | ||||||
| At2g25520 | |||||||||||||
| At4g32390 | |||||||||||||
| At5g11230 | |||||||||||||
| At5g25400 | |||||||||||||
| At5g54800 | Glc-6-P/phosphate translocator (AtGPT1) | 7 | CL | 1 | 42.3 | 10.1 | 2 | ||||||
| At1g01620 | Plasma membrane intrinsic protein 1c (AtPIP1.3) | 6 | PM | 1 | 30.6 | 9.1 | 1 | c | |||||
| At1g17840 | White-brown complex homolog, putative (AtWBC11) | 6 | (PM) | l | 78.4 | 8.8 | 1 | ||||||
| At2g16850 | Plasma membrane intrinsic protein 3b (AtPIP2.8) | 6 | PM | 1 | 29.5 | 9.1 | 1 | 1 | 1 | ||||
| At2g21410 | V-ATPase subunit a2 (AtVHA-a2) | 6 | T | 1 | 93.1 | 5.2 | 7 | 8 | 15 | a, b, c, d, f | |||
| At2g39010 | Plasma membrane intrinsic protein 2e (AtPIP2.6) | 6 | PM | 0.98 | 31.1 | 8.3 | 1 | 1 | 1 | c | |||
| At2g45960 | Plasma membrane intrinsic protein 1b (AtPIP1.2) | 6 | PM | 1 | 30.5 | 9.3 | 1 | 5 | 1 | 1 | a, c | ||
| At3g26520 | Tonoplast intrinsic protein 2 γ (AtTIP1.2) | 6 | T | 1 | 25.8 | 4.7 | 1 | 1 | 1 | 1 | 1 | a, b, f | |
| At3g61430 | Plasma membrane intrinsic protein 1a (AtPIP1.1) | 6 | PM | 1 | 30.7 | 9.3 | 2 | 2 | 1 | 7 | 1 | a, c | |
| At4g35100 | Plasma membrane intrinsic protein 3a (AtPIP2.7) | 6 | PM | 1 | 29.7 | 9.1 | 3 | 4 | 2 | 2 | 2 | 2 | c, d |
| At4g39080 | V-ATPase subunit a3 (AtVHA-a3) | 6 | T | 1 | 92.8 | 5.8 | 4 | 8 | 1 | 11 | 1 | 1 | a, b, c, d, e, f |
| At1g09330 | Expressed protein | 4 | U | 0.97 | 21.2 | 9.2 | 1 | ||||||
| At1g44575 | Chloroplast precursor (AtPSBS) | 4 | CL | 0.99 | 28.0 | 10.0 | 2 | a | |||||
| At1g61250 | Secretory carrier membrane protein family | 4 | U | 1 | 32.6 | 9.0 | 1 | ||||||
| At2g26650 | Potassium channel (AtAKT1) | 4 | PM | 0.92 | 97.0 | 7.3 | 2 | ||||||
| At4g14230 | CBS domain-containing protein related | 4 | U | 0.93 | 43.8 | 5.3 | 1 | ||||||
| At4g14240 | |||||||||||||
| At4g39220 | ER protein-retrieval protein (AtRER1A) | 4 | ER | 0.98 | 22.0 | 9.4 | 2 | ||||||
| At1g19910 | V-ATPase subunit c2 (AtVHA-c2) | 3 | T | 1 | 16.7 | 8.7 | 1 | 2 | 6 | 3 | 4 | a, b, c, d, f | |
| At1g75630 | |||||||||||||
| At2g16510 | |||||||||||||
| At4g34720 | |||||||||||||
| At4g38920 | |||||||||||||
| At1g32210 | Defender against cell death protein | 3 | U | 0.97 | 12.7 | 8.5 | 2 | ||||||
| At1g32400 | Senescence-associated protein family | 3 | U | 0.97 | 31.6 | 5.1 | 2 | a, d | |||||
| At1g72960 | Root hair defective | 3 | U | 0.97 | 83.8 | 5.5 | 1 | ||||||
| At3g08580 | Mitochondrial adenylate translocator (AtAAC1) | 3 | MT | 1 | 41.5 | 10.3 | 3 | 2 | 20 | 1 | a, d, f | ||
| At3g10915 | RING zinc finger protein, putative | 3 | U | 0.98 | 25.1 | 9.5 | 2 | ||||||
| At3g45600 | Senescence-associated protein family | 3 | U | 1 | 31.9 | 8.7 | 1 | 1 | |||||
| At4g21150 | Expressed protein | 3 | U | 1 | 74.7 | 7.6 | 2 | f | |||||
| At4g23630 | Expressed protein | 3 | U | 1 | 30.5 | 8.6 | 6 | 2 | a | ||||
| At1g64090 | |||||||||||||
| At4g11220 | |||||||||||||
| At5g41600 | |||||||||||||
| At4g28390 | Mitochondrial adenylate translocator (AtAAC3) | 3 | MT | 1 | 40.7 | 10.3 | 1 | 3 | 1 | ||||
| At4g28770 | Senescence-associated protein, putative | 3 | (T) | 0.97 | 31.0 | 4.7 | 1 | 1 | a, d, f | ||||
| At5g13490 | Mitochondrial adenylate translocator (AtAAC2) | 3 | MT | 1 | 41.7 | 10.3 | 2 | 2 | 1 | 2 | 3 | f | |
| At5g46800 | Mitochondrial Arg-Orn translocator (AtBOU) | 3 | MT | 0.93 | 31.0 | 10.0 | 1 | ||||||
| At1g06950 | Chloroplast inner envelope protein (AtTic110) | 2 | CL | 1 | 19.8 | 5.2 | 2 | 6 | 2 | ||||
| At1g08480 | Expressed protein | 2 | U | 1 | 15.8 | 6.8 | 2 | a, f | |||||
| At1g15820 | Chlorophyll a/b-binding protein (AtLHCb6) | 2 | CL | 1 | 27.5 | 7.7 | 1 | 2 | 2 | 1 | a | ||
| At1g65820 | Microsomal glutathione S-transferase, putative | 2 | U | 1 | 16.6 | 9.3 | 2 | 3 | 1 | a, d | |||
| At2g07698 | Expressed protein | 2 | U | 1 | 85.9 | 5.2 | 7 | 4 | 11 | a, d, f | |||
| At2g40800 | Expressed protein | 2 | U | 0.98 | 41.7 | 10.4 | 2 | 2 | e | ||||
| At3g56430 | |||||||||||||
| At3g08640 | CCT motif-containing protein (AtCIL) | 2 | U | 0.97 | 35.2 | 9.0 | 1 | ||||||
| At5g01500 | Expressed protein | 2 | U | 1 | 45.1 | 10.4 | 5 | ||||||
| At5g19760 | Mitochondrial dicarboxylate/ tricarboxylate carrier (AtDTC) | 2 | MT | 1 | 31.9 | 9.6 | 2 | 2 | 4 | 6 | a, f | ||
| At5g24650 | Expressed protein | 2 | U | 1 | 27.8 | 10.3 | 5 | 3 | 1 | e, f | |||
| At5g25940 | Nodulin-like protein | 2 | U | 1 | 12.3 | 11.0 | 3 | ||||||
| At5g40820 | FAT domain-containing phosphatidylinositol 3- and 4-kinase family | 2 | U | 0.92 | 302.4 | 7.0 | 2 | ||||||
| At1g04630 | Expressed protein | 1 | U | 1 | 16.1 | 9.6 | 3 | ||||||
| At2g33220 | |||||||||||||
| At1g04750 | R-SNARE synaptobrevin, putative (AtVAMP721) | 1 | ED | 1 | 16.1 | 10.0 | 4 | f | |||||
| At2g33120 | |||||||||||||
| At1g34610 | Ulp1 protease family | 1 | U | 0.96 | 111.4 | 6.0 | 2 | ||||||
| At3g09170 | |||||||||||||
| At1g42960 | Expressed protein | 1 | U | 1 | 17.8 | 8.7 | 1 | ||||||
| At1g53840 | Pectinesterase family | 1 | U | 1 | 64.1 | 8.5 | 1 | 1 | f | ||||
| At1g63830 | Expressed protein | 1 | U | 0.99 | 25.7 | 5.7 | 2 | ||||||
| At5g41390 | |||||||||||||
| At1g70680 | Ca2+-binding EF-hand common family protein | 1 | U | 1 | 21.5 | 9.3 | 2 | ||||||
| At1g70770 | Expressed protein | 1 | U | 0.90 | 66.9 | 9.3 | 1 | f | |||||
| At1g76400 | Ribophorin I, putative | 1 | U | 0.91 | 68.6 | 8.8 | 1 | ||||||
| At2g27730 | Expressed protein | 1 | U | 1 | 11.9 | 10.3 | 1 | 1 | a | ||||
| At2g33380 | RD20 protein | 1 | U | 1 | 26.6 | 5.0 | 1 | a | |||||
| At2g36650 | Expressed protein | 1 | U | 0.90 | 43.0 | 5.0 | 1 | ||||||
| At3g09740 | Qc-SNARE domain protein (AtSYP71) | 1 | U | 1 | 30.0 | 4.8 | 3 | ||||||
| At3g11820 | Qc-SNARE domain protein (AtSYP121) | 1 | U | 0.95 | 38.0 | 9.4 | 1 | ||||||
| At3g17840 | Receptor-like kinase RLK, putative | 1 | U | 0.96 | 70.4 | 6.5 | 1 | ||||||
| At3g19820 | Cell elongation protein, Dwarf1 | 1 | U | 1 | 65.4 | 8.1 | 8 | 4 | 2 | 11 | a, d, f | ||
| At3g27240 | Cytochrome c, putative | 1 | U | 1 | 33.7 | 6.1 | 4 | d, f | |||||
| At3g48850 | Mitochondrial phosphate transporter, putative (AtPHT3-2) | 1 | MT | 1 | 39.0 | 9.7 | 3 | 1 | 8 | 6 | f | ||
| At5g14040 | |||||||||||||
| At3g48890 | Progesterone-binding protein homolog Atmp2, putative (AtMSBP1) | 1 | U | 0.96 | 25.4 | 4.3 | 1 | 2 | |||||
| At5g52240 | |||||||||||||
| At3g49720 | Expressed protein | 1 | U | 1 | 28.5 | 9.7 | 1 | f | |||||
| At3g49750 | Expressed protein | 1 | U | 1 | 30.0 | 8.1 | 1 | ||||||
| At3g54110 | Mitochondrial uncoupling protein 1 (AtUCP1) | 1 | MT | 1 | 32.7 | 10.1 | 2 | 2 | f | ||||
| At3g60750 | Transketolase, putative | 1 | U | 0.94 | 80.0 | 6.3 | 1 | ||||||
| At3g62360 | Expressed protein | 1 | U | 1 | 132.9 | 6.4 | 1 | ||||||
| At3g63410 | Chloroplast inner envelope protein (AtAPG1) | 1 | CL | 1 | 37.9 | 9.5 | 2 | 3 | 1 | f | |||
| At4g21105 | Expressed protein | 1 | U | 0.95 | 7.7 | 10.5 | 2 | f | |||||
| At4g35000 | Peroxisomal ascorbate peroxidase (AtAPX3) | 1 | PR | 0.99 | 31.6 | 7.0 | 2 | e, f | |||||
| At4g37830 | Expressed protein | 1 | U | 1 | 11.2 | 9.5 | 1 | 1 | |||||
| At5g01530 | Light-harvesting chlorophyll a/b-binding protein | 1 | CL | 1 | 31.1 | 6.0 | 2 | a | |||||
| At5g07340 | Calnexin, putative | 1 | ER | 1 | 60.5 | 4.5 | 1 | 1 | f | ||||
| At5g08080 | Qa-SNARE domain protein (AtSYP132) | 1 | U | 0.96 | 34.2 | 6.3 | 2 | ||||||
| At5g16660 | Expressed protein | 1 | U | 1 | 18.2 | 8.7 | 1 | ||||||
| At5g47570 | Expressed protein | 1 | U | 1 | 13.2 | 8.5 | 3 | ||||||
| At5g61790 | Calnexin 1 (CNX1) | 1 | ER | 1 | 60.5 | 4.5 | 1 | 3 | a, e, f | ||||
| At5g66680 | Dolichyl-di-phosphooligosaccharide- protein glycotransferase | 1 | U | 1 | 48.7 | 6.3 | 2 | 2 | a, f |
We detected 28 different membrane proteins of 258 peptides in the subfraction of KI (eight slices), 36 of 290 peptides in the subfraction of acetone (eight slices), 13 of 33 peptides in the subfraction of NaOH (four slices), 72 of 1,107 peptides in the subfraction of NaOH (10 slices), 27 of 109 peptides in the subfraction of C/M (four slices), and 18 of 75 peptides in the subfraction of C/M pH 6 (four slices; Table II; Supplemental Table S1). A total of 55% (56 membrane proteins out of 102) of the membrane proteins were found solely in one of the washing or extraction procedures applied (Table II). Comparing the replicates of membranes washed with NaOH (Table II) we identified 83% more membrane proteins by cutting the gel into 10 slices (75 membrane proteins) than when cutting the gel into only four (13 membrane proteins). Interestingly, in all subfractions the total number of peptides from known tonoplast proteins ranged between 30% and 50%. However, in the C/M subfraction the total number of peptides from known tonoplast proteins was only 12%. These results demonstrate the importance of high resolution protein fractionation and that the application of different extraction methods increases the number of peptides that can be identified.
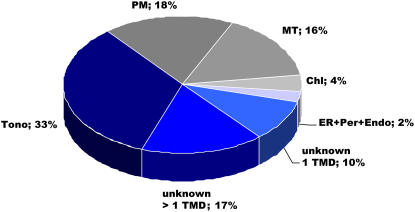
We identified nine intrinsic membrane proteins, which are already known to be localized in the tonoplast. Calculating the total number of peptides we detected 33.5% (626 of 1,870) of all peptides belonging to tonoplast proteins (Fig. 2). If the nonmembrane spanning subunits of the V-ATPase are included, 57.1% (1,655 of 2,899) of the peptides could be attributed to the tonoplast. Based on the number of total peptides detected, the three major tonoplast proteins are the V-PPase (AtAVP-3; 345 peptides), the integral V0-ATPase subunit a3 (89 peptides), and the integral V0-ATPase subunit a2 (86 peptides). In addition, two other V-PPases (the homologs to AtAVP2 and AtAVPL1), the tonoplast intrinsic protein (the homolog to AtTIP1.2), and the integral V0-ATPase subunit c and two Ca2+-ATPases (the homolog to AtACA4 and AtACA11; Geisler et al., 2000; Baxter et al., 2003) were identified. Moreover, 57 membrane proteins (26.4% peptides) are still not localized (Fig. 2) and 36 membrane proteins (40.1% peptides) could be attributed to already known nonvacuolar proteins. Consequently, for more than 56% (57 proteins of 102) of the membrane proteins no function has yet been attributed (Fig. 2).
Figure 2.
Organellar distribution of membrane protein isolates. Classification according to the total number of peptides. Tono, Tonoplast (excluding the V1 domain of the V-ATPase); unknown > 1 TMD, unknown proteins with two or more transmembrane domains; unknown 1 TMD, unknown proteins with one transmembrane domain; PM, plasma membrane; Chl, chloroplast; MT, mitochondria; Endo, endosomes; Per, peroxisomes. TMDs were predicted using ARAMEMNON database accessible at www.aramemnon.botanik.uni-koeln.de.
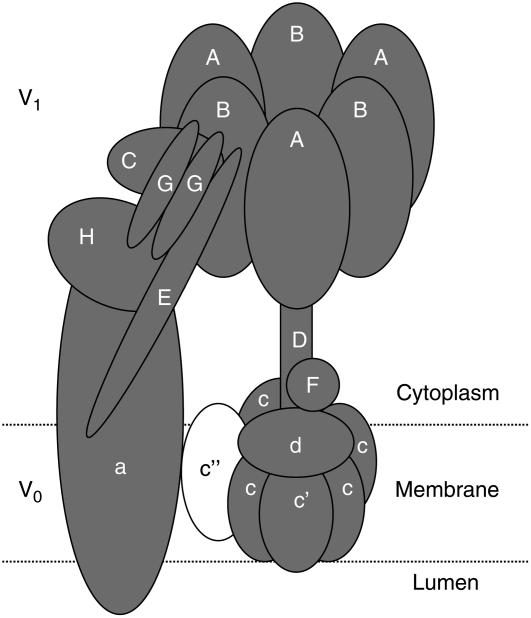
To obtain an overview of the coverage of our proteomic data we had a closer look at the vacuolar ATPase. The V-ATPase is a large membrane bound multisubunit enzyme complex composed of two functional domains: a water soluble V1 domain and a membrane embedded V0 domain (Fig. 3). The Arabidopsis V-ATPase contains 13 distinct functional subunits (Sze et al., 2002). We detected all subunits of the V-ATPase complex except subunits VHA-c" and VHA-e (Fig. 3; Supplemental Table S1). The identification of 11 out of 13 V-ATPase subunits indicates the high coverage of the vacuole proteome reported here.
Figure 3.
Identified proteins from the V-ATPase. Identified proteins are depicted in gray and missing components in white. The V1 catalytic domain consists of subunits A and B in an A3B3 arrangement, C, D, E, F, G, and H. The V0 membrane sector is composed of a and c subunits in an ac6 complex. The detailed position of subunit e is not clear. Except for subunits c″ and e, all known components of the V-ATPase were identified.
Specification of New Tonoplast Proteins
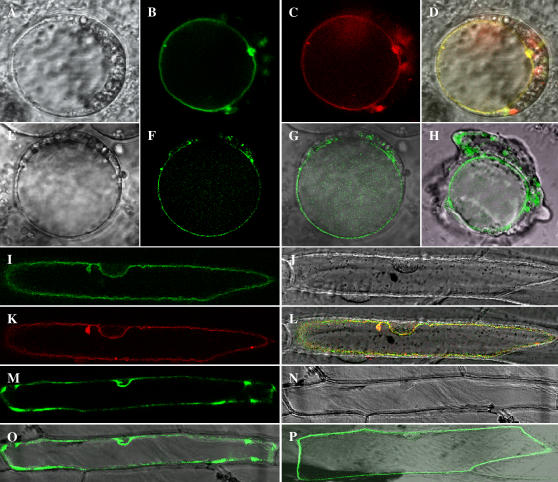
In our search for new transporters and channels in the tonoplast we were interested in novel vacuolar membrane proteins with more than two transmembrane domains. To provide further independent evidence for a vacuolar localization of the newly identified putative tonoplast proteins we transiently transformed both Arabidopsis protoplasts and onion (Allium cepa) epidermal cells with five GFP fusion proteins and analyzed their respective localization (Fig. 4). As a vacuolar control we used the C-terminal DsRed-tagged KCO1 channel from Arabidopsis (At5g55630), known to reside in the vacuolar membrane (Czempinski et al., 2002).
Figure 4.
Subcellular localization of five novel identified proteins. Tonoplast localization of At5g45370 (A–D) and At1g16390 (E–H) in Arabidopsis protoplasts; bright-field image of an Arabidopsis protoplast (A and E), fluorescence of At5g45370∷GFP (B), fluorescence of KCO1∷DsRed (C), colocalization of the KCO1∷DsRed and At5g45370∷GFP fluorescence (D), fluorescence of At1g16390∷GFP (F), overlay of transmission and At1g16390∷GFP fluorescence (G), and lysed protoplast expressing At1g16390∷GFP showing the intact vacuole surrounded by cytosolic material from the broken cell (H). Vacuolar membrane localization of At4g28770 (I–K) and At1g06470 (N and O) in onion epidermal cells; fluorescence of At4g28770∷GFP (I), bright-field image (J and N), fluorescence of KCO1∷DsRed (K), colocalization of KCO1∷DsRed and At4g28770∷GFP (L), fluorescence of At1g06470∷GFP (M), and overlay of transmission and At1g06470∷GFP fluorescence (O). Localization of At1g17840 in the plasma membrane; overlay of transmission and At1g17840∷GFP fluorescence (P).
Synthesis of the recombinant KCO1∷DsRed fusion protein in both Arabidopsis protoplasts or epidermal onion cells resulted in red fluorescence at the tonoplast (Fig. 4, C and K). The selected putative vacuolar membrane proteins (At5g45370, At1g16390, At4g28770, and At1g06470) fused to GFP are targeted to the vacuolar membrane (Fig. 4, B, F, I, and M). In transformed Arabidopsis protoplasts fluorescence is clearly surrounding the central vacuole (Fig. 4, B and F). The tonoplast localization of two proteins (At5g45370 and At4g28770) was additionally demonstrated by colocalizing of the GFP fusion proteins and the KCO1∷DsRed fusion protein (Fig. 4, D and L). In contrast to the four membrane proteins described above, one GFP fusion protein identified in our analysis, namely At1g17840, was localized at the plasma membrane (Fig. 4P), indicating the importance of localization studies in proteomic analysis.
The function of the four newly localized tonoplast proteins is still unknown. The gene At1g16390 encodes a putative organic cation transporter with 11 TMDs. Its product was detected in the KI subfraction as a single hit in two different gel slices. Proteins At1g06470 and At5g45370 were also detected as single hits in the acetone subfraction. The gene At1g06470 encodes an unknown integral membrane protein with eight putative TMDs and At5g45370 codes for a protein belonging to the nodulin N21 family with nine TMDs. All three proteins, At1g16390, At1g06470, and At5g45370 were manually identified. At4g28770 was detected with Protein-Prophet (score 0.97). This vacuolar protein was identified with single hits from the C/M and C/M pH 6 subfractions. In these fractions 10 peptides could be attributed to this protein. The protein is a putative senescence-associated protein with three TMDs.
Our data demonstrates that it is possible to identify new tonoplast proteins from cauliflower by comparison with peptides from the Arabidopsis databank.
DISCUSSION
The main goals of this work were, first to identify new putative vacuolar proteins of a vacuolar preparation containing a large number of small vacuoles from developing cells, second, to use plant material of which large amounts of vacuolar membranes can be quickly and easily isolated, and third, to prove that new candidate proteins are indeed targeted to the vacuolar membrane. Despite the important role of the vacuole for temporary storage, detoxification, and cytosolic homeostasis in plant cells only a limited number of vacuolar proteins have been identified so far.
Since the vacuolar membrane contains less than 1% of the total cellular proteins and as this membrane is less protein dense compared to most other cellular membranes, small contaminations may result in the detection of a large number of nonvacuolar proteins. The heterogeneity observed in previous studies (Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004; Jaquinod et al., 2007) may therefore be due to the different plant material like Arabidopsis mesophyll cells (Carter et al., 2004; Sazuka et al., 2004) or cell suspension culture (Shimaoka et al., 2004; Szponarski et al., 2004), or to different preparation techniques. The fact that a small contamination of the vacuolar preparation can result in the detection of a considerable number of nonvacuolar proteins is also shown in this work (35.3% proteins corresponding to 40.1% peptides; see Fig. 2). In this context it should be mentioned that the inner mitochondrial membrane, as well as the plasma membrane and other internal membranes, contain far more proteins compared to the vacuolar membrane. In density gradients, the latter membranes together with non-rRNA containing ER are the lightest membranes. The fact that more peptides that could be attributed to PIPs rather than TIPs were identified is astonishing. It may be explained that small, nonexpanding vacuoles contain low amounts of aquaporins, since the water permeability in lytic vacuoles is mainly regulated at the transcription level. In contrast, plasma membrane aquaporins are regulated by phosphorylation and appear to be less regulated at the transcription level (Hu et al., 2003; Luu and Maurel, 2005). This hypothesis is sustained by the western blots showing that the PAQ1 signal is much stronger than the TIP signal in the microsomal fraction. This is also in line with experiments analyzing PIPs and TIPs in developing fruits (Hu et al., 2003).
Furthermore, for most other membranes many more membrane proteins have been identified and characterized. In addition, the vacuole is a single continuous compartment transected by tubular-like transvacuolar strands and coupled with the intimate associations with other organelles during stress adaptation (Reisen et al., 2005). Another possible source of contaminants is the basal level of autophagy and the internalization of cytosolic proteins, as well as organelles into the vacuole. Therefore, a comparison of peptides obtained from a vacuolar proteomic will overestimate the number of nonvacuolar proteins.
A further limiting aspect in a vacuolar proteomic approach is the minute amount of membranes that can be isolated. An in-depth proteomic approach where low abundance components of the membrane will also be identified requires washing steps and fractionation. Starting with Arabidopsis mesophyll cells, which probably allow a better purification than suspension cultured cells due to the larger differences in the density of vacuoles and protoplasts, only limited material can be isolated. However, techniques are improving fast and will allow to detect more proteins even with limited amounts of membranes.
The scope of our study was also to identify membrane proteins expressed in the early phase of vacuole development. As a model plant for this study we have chosen cauliflower. Using cauliflower buds, vacuoles from different developmental stages were isolated, resulting in a preparation of small and large vacuoles (see Fig. 1; Dozolme et al., 1995). A further advantage of cauliflower is that the plant material is not a limiting factor and that the mechanical slicing technique allows isolation of large amounts of highly purified vacuoles (Marty and Branton, 1980; Marty-Mazars et al., 1995). However, a database for comparison is required to allow a complete or almost complete coverage of the peptides. Cauliflower is a Brassicacea as is Arabidopsis and thus shares very high sequence identity (95% identity for aquaporins [Barrieu et al., 1998], 87% to 97% identity for other membrane proteins). Therefore, it is not surprising that most of the peptides we identified could be assigned to an Arabidopsis homolog, proving that cauliflower is a suitable model plant. Different washings and extractions of the vacuolar preparation resulted in the identification of different proteins, underlining the necessity to isolate large amounts of membranes to obtain a high coverage.
In total, we identified 102 membrane and 214 soluble proteins, including seven nonmembrane spanning domains of the V-ATPase (see Table II; Supplemental Table S1). This implies that 32.3% of our tonoplast fractions belong to membrane proteins. Of these, three membrane proteins (2.9% of all membrane proteins) were identified in the various washing and extraction procedures (Table II), including the V-PPase (homologs to AtAVP-3), the integral V0-ATPase subunit a3, and the putative plasma membrane intrinsic protein 3a (homologs to AtPIP2.7). Moreover, only 27 membrane proteins (26.5%) were detected in three of the five treatments. Less than the half of all membrane proteins (46 of 102 proteins) were identified in at least two procedures (Table II).
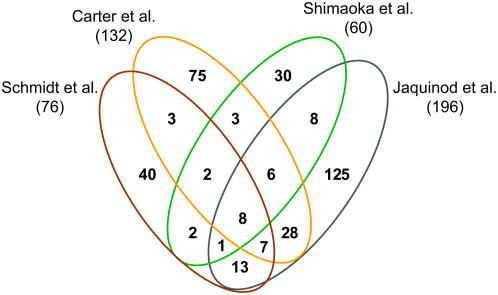
Figure 5 shows the cross correlation of identified cauliflower membrane proteins in our study with the three largest Arabidopsis proteomic approaches of the vacuolar membrane (Carter et al., 2004; Shimaoka et al., 2004; Jaquinod et al., 2007). Well-known proteins of the chloroplast, mitochondria, plasma membrane, and the ER proteins AtACA1, calnexin 1, and AtRER1A were excluded from the analysis. Of 76 vacuolar and putative vacuolar membrane proteins 40 proteins (52%) were exclusively identified in our study, suggesting a higher relative abundance of these proteins in cauliflower buds vacuoles compared to Arabidopsis leaf mesophyll and cell culture vacuoles. Interestingly, when the corresponding Arabidopsis genes for the newly discovered vacuolar and putative vacuolar membrane proteins were analyzed for their expression pattern by GENEVESTIGATOR (www.genevestigator.ethz.ch), a very large part of them was mainly expressed in young, developing parts (elongation zone, epidermal atrichoblast, root hair zone, shoot apex) of the plant (Supplemental Fig. S1). This result confirms the interest to perform vacuolar proteomics in different tissues and at different developmental stages.
Figure 5.
Overlap between different proteomic analyses of the tonoplast. The Venn diagram compares membrane proteins identified in cauliflower with membrane proteins identified in the three largest Arabidopsis tonoplast proteomic analyses (Carter et al., 2004; Shimaoka et al., 2004; Jaquinod et al., 2007). Numbers in parentheses indicate the total number of identified proteins with at least one transmembrane domain in the particular study, excluding contaminating proteins of the proteomic analyses. The prediction of TMDs by TMHMM was considered for membrane proteins identified by Carter et al. (2004).
We detected 11 out of 13 subunits (Sze et al., 2002). Subunit VHA-a is abundant in all five different fractions, whereas only subunits VHA-c″ and VHA-e were not identified. VHA-c″ occurs with low abundance and VHA-e is a very small protein (approximately 8 kD) and, therefore, probably migrates too fast for detection in the 10% SDS-PAGE as used under our conditions. Until now, no tonoplast proteome is available containing these two subunits (Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004; Endler et al., 2006; Jaquinod et al., 2007). Calculating the total number of peptides from all subunits of the V-ATPase we detected 1,029 peptides. Compared to the total number of peptides from the nonvacuolar ATPases, only 146 peptides from four subunits of the F-ATPase (FHA-β, FHA-ɛ, FHA-γ, and FHA-D) and 31 peptides from the P-ATPase were identified (see Supplemental Table S1). To conclude, 85.3% of peptides (1,029 of 1,206) of all ATPases are of vacuolar origin. This comparison probably reflects much more the enrichment of the vacuolar membrane in our fraction than the total proportion of proteins. It is also interesting to note that it has been reported that in young tissues the V-PPase is the predominant pump (Maeshima, 2001). Despite the fact that our proteomic study is not quantitative, the large number of peptides corresponding to this proton pump indicates that this may also be true for cauliflower buds.
To get a hint on the reliability of identification and for the vacuolar localization we made GFP fusion constructs for five unknown proteins (see Fig. 4). Four of them were targeted to the tonoplast, the fifth to the plasma membrane. The first of the novel tonoplast proteins, At1g16390, has similarities to organic cation transporters like At1g79360 with 11 TMDs. Organic cation transporters influence the plasma concentration of many endogenous molecules and of an even wider array of xenobiotic compounds. They are expressed in many tissues and play a critical role in fundamental cellular processes (Bednarczyk et al., 2003). However, their function in plants has not been investigated.
The second novel tonoplast protein with nine TMDs, At5g45370, belongs to the nodulin N21 family. The closest homologs are At4g19185 and At3g45870 with 74% and 66% similarity, respectively. At5g45370 belongs to the drug/metabolite transporter superfamily (Jack et al., 2001), however, until now, no member of the family has been characterized.
Our third tonoplast candidate, At1g06470, is an integral membrane protein with eight predicted TMDs. The closest homologs, At2g25520 and At5g25400, both with 28% similarity, belong to a group of phosphate translocators that function as antiport systems using inorganic phosphate and phosphorylated C3, C5, or C6 compounds as counter substrates (Flügge, 1999). Preliminary tests investigating whether this gene could code for the vacuolar phosphate transporter indicated that the corresponding deletion mutants were not affected in vacuolar phosphate and sugar contents (data not shown).
The fourth tonoplast membrane protein, At4g28770, is a putative senescence-associated protein with three TMDs. There is just one close homolog, At2g20230, exhibiting a similarity of 75%. During senescence the vacuole has an important role in degradation of proteins and possibly also in temporary storage of sugars and amino acids. Furthermore, many compounds stored in the vacuole are retranslocated to young, growing tissues or seeds during senescence. The increase of the corresponding transcript level during senescence may indicate that this membrane protein may be related to such a process.
We are aware that from the localization of five proteins we cannot directly extrapolate the number of tonoplast proteins in our preparation. However, if we assume that around 80% of the novel identified proteins containing at least two membrane spanning domains are vacuolar 27 so far unknown vacuolar membrane proteins have to be postulated to exist.
In conclusion, despite the fact that the vacuole contains less than 1% of the cellular protein, a large number of proteins can be identified when large amounts of highly purified vacuoles are used in combination with the proteomic approach. This will allow for more detailed investigation into the role of vacuolar transporters in storage and homeostasis. Vacuoles are likely to fulfill functionally differing roles in various plants and tissues. Using the vacuolar proteomic approach will therefore not only help to identify new vacuolar proteins but allow us to elucidate the functional roles of vacuoles in a given plant and tissue.
MATERIALS AND METHODS
General
Restriction enzymes were purchased from New England Biolabs. Oligonucleotides were obtained from Mycrosynth. DNA sequencing was conducted by the University of Zürich. Unless otherwise indicated, all chemicals were purchased from Sigma. Escherichia coli strain XL-1 Blue (Stratagene) was generally employed for DNA cloning procedures. All cloning procedures were conducted using standard methods previously described by Sambrook et al. (1989).
Plant Material and Growth Conditions
Cauliflower (Brassica oleracea) buds were obtained at commercial fresh produce outlets. Arabidopsis (Arabidopsis thaliana) cells were grown in Murashige and Skoog medium as previously described (May and Leaver, 1993). The cell cultures were maintained at 25°C in an orbital shaker (150 rpm) under normal illumination. Cells were collected 7 d after subculture (Millar et al., 2001).
Tonoplast Preparation
Intact vacuoles and tonoplast fragments from cauliflower buds were prepared as previously reported (Leigh et al., 1979; Marty and Branton, 1980). Briefly, 750 g of fresh tissue was sliced three times in 500 mL of ice-cold collection medium (1 m sorbitol, 5 mm EDTA, 4 mm dithiothreitol [DTT], 50 mm Tris-HCl, pH 7.6) using a motor-driven tissue slicer. All subsequent manipulation was done at 4°C. The filtrate was pooled and centrifuged at 8,000g for 15 min. The sediments (crude vacuole homogenate) containing intact vacuoles, nuclei, mitochondria, plastids, and tissue fragments were resuspended in 2.5 mL of 15% (w/v) Nycodenz (Axis-Shield) in isolation medium (1.5 m sorbitol, 1 mm EDTA, 4 mm DTT, 10 mm Tris-HCI, pH 7.6) and pooled before filtration through one layer of Miracloth (Chicopee Mills, Inc.) prewetted with 15% (w/v) Nycodenz in isolation medium. The filtrate (16 mL) was divided into four tubes and overlaid first with 5 mL of isolation medium containing 8% (w/v) Nycodenz and 4 mL of isolation medium lacking Nycodenz. The discontinuous gradients were centrifuged for 45 min at 100,000g in a slowly accelerated and very slowly decelerated swing-out rotor. After centrifugation the vacuoles were found at the interface of the 0% and 8% Nycodenz layers. Vacuoles were lysed by hypotonic treatment in 10 mL of 10 mm Tris-HCI, pH 7.6. The vacuole membranes were sedimented at 140,000 g for 30 min. The sediments of vacuole membrane were extensively drained and the purity of the final fraction was assessed using marker enzymes (Marty-Mazars et al., 1995). The purified fraction was directly solubilized for electrophoresis or resuspended in the appropriate elution solution (see subfractionation of tonoplast proteins) for extraction.
Subfractionation of Tonoplast Proteins
To remove most of the soluble proteins, the purified vacuole membrane fractions were individually treated. The tonoplast fraction (0.2 mg) was washed either by resuspending them in (1) 80% acetone; (2) in 50 mm MOPS/NaOH pH 7.8, 1 mm DTT containing 0.1 m NaOH (final pH approximately 10); (3) in 20 mm HEPES-KOH pH 7.2 containing 0.3 m KI. After 30 min on ice, the mix was centrifuged (17,600g, 15 min, 4°C) to separate two fractions: the supernatant containing membrane associated proteins released by the respective treatment and the pellet containing the integral membrane proteins. The pelleted proteins were suspended in 30 μL Laemmli buffer (Laemmli, 1970) and analyzed by SDS-PAGE. In an additional approach, hydrophobic tonoplast proteins were extracted from purified vacuolar fraction using a C/M (5:4, v/v) mixture (Marty-Mazars et al., 1995; Marmagne et al., 2004). The tonoplast proteins (1 mg) were resuspended in 200 μL of 50 mm MOPS/NaOH pH 7.8, 1 mm DTT (C/M), or 20 mm sodium phosphate buffer pH 6 (C/M pH 6) and 1.8 mL cold C/M (5:4, v/v) was slowly added. The resulting mixtures were stored for 15 min on ice before centrifugation (4°C) for 20 min at 17,600g. The organic phase, which contains the hydrophobic proteins soluble in C/M was used for further protein analyses. C/M-soluble proteins were dried under nitrogen and the pellet proteins were then suspended in 30 μL Laemmli buffer (Laemmli, 1970) and analyzed by SDS-PAGE.
Protein Determination
Protein concentrations were determined by the dye-binding method of Bradford (1976) using bovine serum albumin as a standard.
Western Blotting
Proteins (10 μg) were separated using SDS-PAGE (Laemmli, 1970) with 10% acrylamide gels. Western blotting was carried out using antibodies for γ-TIP, α-subunit of the chloroplastic ATPase (ATP-α), luminal binding protein (BiP), and plasma membrane aquaporin (PAQ1). Secondary antibody (antirabbit, coupled to horseradish peroxidase, Promega) was diluted 1:25,000 in Tris-buffered KI (0.2 m Tris-HCl pH 7.5, 0.5 m NaCl). Horseradish peroxidase activity was detected using a Chemiluminescence Blotting Substrate kit (SuperSignal Chemiluminescent Working Solution, Pierce) according to manufacturer's instructions.
Marker Enzyme Assays
Cytochrome c oxidase (CCO; EC 1.10.2.2), antimycin A-insensitive NADH-cytochrome c reductase (CCR; EC 1.6.99.3), and latent inosine diphosphatase (IDPase; EC 3.6.1.6) were assayed according to standard procedures (Green, 1983; Lord, 1983; Moore and Proudlove, 1983). The H+-transporting two-sector ATPase (F-ATPase; EC 3.6.3.14), the plasma membrane ATPase (P-ATPase; EC 3.6.1.35), and V-ATPase (EC 3.5.1.3) were measured as described by Ratajczak et al. (1999).
SDS-PAGE
The buffer system of Laemmli (1970) was used for SDS-PAGE. Slab gels were 1 mm thick and consisted of a 10% (w/v) acrylamide resolving gel and a 4% (w/v) acrylamide stacking gel. Polypeptides in the resolving gel were stained with Coomassie Brilliant Blue (Fairbanks et al., 1971). For each protein fraction the gels were cut into four to 10 slices (see Table II), and each gel segment was subjected to trypsin in gel digest as described (Shevchenko et al., 1996).
Mass Spectrometric Protein Identification
Tryptic peptides of each fraction were resuspended in 5 μL of 5% acetonitrile and 0.2% formic acid (v/v) in water and loaded on laboratory-made silica capillary columns (i.d. of 75 μm, length of 9 cm; BGB Analytik AG) packed with C18 reversed-phase material (Magic C18 resins; 5 μm, 200-Å pore; Michrom BioResources). The peptide mixture was separated and eluted by a gradient from 5% to 65% acetonitrile over 2 h followed by an increase up to 80% during an additional 15 min. The flow rate at the tip of the column was adjusted to approximately 200 nL/min. LC was coupled on-line to an LCQDeca XP ion trap mass spectrometer (Thermo Finnigan) equipped with a nanospray ionization source. Mass analysis was performed with a spray voltage of 2.0 to 2.5 kV and one MS full scan followed by three data-dependent MS/MS scans of the three most intense parent ions. The dynamic exclusion function was enabled to allow two measurements of the same parent ion during 1 min followed by exclusion for 1 min.
Analysis and Interpretation of MS Data
The SEQUEST software (Thermo Finnigan) was used to search the Arabidopsis protein database (www.ncbi.nlm.nih.gov/, 3701, 01.12.2004). Arabidopsis and cauliflower are closely related species facilitating the detection of homologous proteins (e.g. 95% identity for aquaporins; Barrieu et al., 1998). Furthermore, this database was chosen because it is well annotated and has a size that is compatible with the statistical tools used for the evaluation of our proteomics data (see below). Data files were created by the SEQUEST software for every MS/MS scan with a total ion count of at least 5 × 104, minimal peak count of 35, and a precursor ion mass in the range of 300 to 2,000 mass-to-charge ratio. Data were searched against the database restricted to tryptic peptides without modifications (except for carboxyamidomethylated Cyss 57.0513 and oxidized Mets 15.9994), allowing a parent mass error tolerance of 2 D and daughter ion error tolerance of 0.8 D.
To assess false-positive identification rates statistically, we performed Peptide and Protein-Prophet data analyses (Keller et al., 2002). These statistical models allow for the assessment of false-positive identification rates and we chose a cutoff of 0.9 for accepting protein identity, suggesting a false-positive identification rate of 10% (see Table II; Supplemental Table S1). Supplemental Table S1 lists all identified membrane proteins including the Atg number, the total and unique number of peptides assigned to the protein, the protein probability calculated by Protein-Prophet, the peptide sequences, the sequence coverage in percent, and the mass differences of the theoretical and experimental masses observed for the precursor ions. In most instances, we identified the same protein from different biochemical fractions further substantiating the reliability of the identification (see Table II). In some cases where we accepted proteins with a lower Protein-Prophet score (but with an acceptable SEQUEST score [see above]) we performed additional experiments to demonstrate their vacuolar localization.
To further validate the identification procedures, we cloned the respective genes and analyzed the localization of these proteins by transient GFP localization assays (see Fig. 4; Table II). For the SEQUEST-based data interpretation we proceeded as follows: We accepted cross-correlation scores (Xcorr) of at least 2.5 for doubly, and 3.5 for triply charged ions. For peptide identifications with a dCN (normalized difference in correlation score, giving the differences between the front ranking and the following possible hit) lower than 0.1, the spectra of lower ranking hits were also examined. Identifications with a dCN of 0.0 resulting from different members of protein families, isoforms, or redundant database entries that could not be distinguished by the identified peptides are given in Table II.
GENEVESTIGATOR
The expression pattern of novel vacuolar membrane proteins and putative vacuolar membrane proteins identified by MS was analyzed using the Meta-Analyzer of GENEVESTIGATOR (Zimmermann et al., 2004). Signal intensity levels of all genes were checked using the digital northern tool. Genes showing almost always an absent call were excluded from the analysis.
Isolation of RNA
Plant material that was flash frozen in liquid nitrogen and stored at −80°C was ground in liquid nitrogen and transferred into a sterile 2 mL Eppendorf tube. RNA from different tissues (leaves, roots, flowers, stems, or protoplasts) of Arabidopsis was extracted using an RNA isolation kit, according to the manufacturer's instructions (RNeasy for plant material, Qiagen). Following extraction of total RNA, a DNase treatment (DNA-free, Ambion) was used to eliminate contamination with genomic DNA conditions. Quantification and purity of RNA were determined spectrophotometrically according to the method described by Sambrook et al. (1989). The integrity of RNA samples was determined by formaldehyde agarose gel electrophoresis using a 1.2% gel prestained with ethidium bromide (Lehrach et al., 1977).
Plasmid Construction
To construct GFP fusion proteins we amplified, via PCR, the entire cDNA of At1g06470, At1g16390, At1g17840, At4g28770, and At5g45370, using the Expand High FidelityPLUS PCR system (Roche) and either cDNA generated from Arabidopsis tissues (first-strand cDNA synthesis kit, Amersham Pharmacia Biotech) or RIKEN Arabidopsis full-length cDNA clones (Seki et al., 1998, 2002), respectively, as templates. The primers we used are listed in Table III. The obtained DNA fragments were cleaved with XbaI and XhoI (see Table III) and inserted in frame in the vector pGFP2 (Kost et al., 1998) leading to C-terminal fusions to GFP.
Table III.
Primer used for cloning into pGFP2
| Primer Name and Specificity | Sequence |
|---|---|
| At1g16390 | |
| 2_XbaI, 5′ | 5′-TCTAGAATGGCTGACTCAACTCGG-3′ |
| 2_XhoI, 3′ | 5′-CTCGAGACCAATAAATTGTCTTTTTGCC-3′ |
| At1g17840 | |
| 3_XbaI, 5′ | 5′-TCTAGAATGGAGATCGACGCAAGCAGGC-3′ |
| 3_XbaI, 3′ | 5′-CTCGAGCCATCTGCGAGCTCCATC-3′ |
| At4g28770 | |
| 16_XbaI, 5′ | 5′-GCTCTAGAATGCGACACCATTGTTGCCATGTC-3′ |
| 16_XhoI, 3′ | 5′-CCGCTCGAGAGCTGATGGAGTATGACTCTG-3′ |
| At1g06470 | |
| 23_XbaI, 5′ | 5′-TCTAGAATGGAGCAAAGAGTGCAGCTC-3′ |
| 23_XhoI, 3′ | 5′-CTCGAGGGGACTATTTTCTTGATCATCC-3′ |
| At5g45370 | |
| 31_XbaI, 5′ | 5′-TCTAGAATGGCAGCTCCAGCAATCC-3′ |
| 31_XhoI, 3′ | 5′-CTCGAGTTTACCAGTCTCATCTCTG-3′ |
Transformation of Arabidopsis Cell Culture or Onion Epidermal Cells and Confocal Laser Scanning Microscopy
Cells from an Arabidopsis cell suspension culture (Millar et al., 2001) were transiently transformed with purified plasmid DNA using Qiagen columns according to the manufacturer's protocol (Qiagen). The GFP fusion constructs were introduced into Arabidopsis protoplasts (Frangne et al., 2002) by a polyethylene glycol-mediated transformation procedure (Jin et al., 2001). Alternatively, transient expression of the fusion constructs was carried out after biolistic transformation into onion (Allium cepa) epidermal cells using the Helium Biolistic particle delivery system (Bio-Rad; Takeuchi et al., 1992).
Expression of the GFP fusion proteins was monitored 2 and 3 d after transformation by confocal laser scanning mircroscopy using a Leica DM IRE2 microscope with a 63× Plan-Apochromat oil immersion objective for protoplasts or with a 20× Plan-Apochromat glycerol immersion objective for onion cells coupled to Leica TCS SP2 spectral confocal and multiphoton microscope (Leica Microsystems). Fluorescing cells were imaged at an excitation wavelength of 488 nm and the emission signal was recovered between 495 and 530 nm for GFP fusion, and excitation of 543 nm and emission signal recovered between 555 and 620 nm for DsRed fusion. Images were processed with the Leica confocal software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression pattern of newly identified vacuolar and putative vacuolar membrane proteins was investigated using Meta-Analyzer of GENEVESTIGATOR.
Supplemental Table S1. Table lists soluble and membrane proteins identified in different subfractions.
Supplementary Material
Acknowledgments
We thank Dr. Katrin Czempinsky (University of Potsdam, Germany) for providing us with the KCO1∷DsRed fusion construct, Prof. Maeshima (University Nagoya, Japan) for the antibodies, Dr. Helene Barbier-Brygoo and Dr. Anne Marmagne (Centre National de la Recherche Scientifique Gif-sur-Yvette, France) for advice in C/M extraction, Dr. Christian Panse and Dr. Jonas Grossmann (Functional Genomics Center Zurich) for help with data acquisition, and Shaun W. Peters (University of Zurich) for critical reading of the manuscript.
This work was supported by the Plant Science Center Zurich-Basel (Graduate Research Fellowship) and the project Novel Ion Channels in Plants (grant no. EU HPRN–CT–00245).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Enrico Martinoia (enrico.martinoia@botinst.unizh.ch).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 [DOI] [PubMed] [Google Scholar]
- Barrieu F, Thomas D, Marty-Mazars D, Charbonnier M, Marty F (1998) Tonoplast intrinsic proteins from cauliflower (Brassica oleracea L. var. botrytis): immunological analysis, cDNA cloning and evidence for expression in meristematic tissues. Planta 204 335–344 [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarczyk D, Ekins S, Wikel JH, Wright SH (2003) Influence of molecular structure on substrate binding to the human organic cation transporter, hOCT1. Mol Pharmacol 63 489–498 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) Rapid and quantitative method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–252 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi K (2003) The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporter. Plant Cell 15 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czempinski K, Frachisse JM, Maurel C, Barbier-Brygoo H, Mueller-Roeber B (2002) Vacuolar membrane localization of the Arabidopsis “two-pore” K+ channel KCO1. Plant J 29 809–820 [DOI] [PubMed] [Google Scholar]
- De Angeli A, Monachello D, Ephritikhine G, Franchisse JM, Thomine S, Gambale F, Barbier-Brygoo H (2006) The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442 939–942 [DOI] [PubMed] [Google Scholar]
- Dozolme P, Marty-Mazars D, Clémencet MC, Marty F (1995) Monoclonal antibody TeM 106 reacts with a tonoplast intrinsic protein of 106 kDa from Brassica oleracea L. J Cell Sci 108 1509–1517 [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerlich V, Linka N, Reinhold T, Hurth MA, Traub M, Martinoia E, Neuhaus HE (2003) The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc Natl Acad Sci USA 100 11122–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks G, Steck TL, Wallach DFH (1971) Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10 2606–2617 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 5 325–345 [DOI] [PubMed] [Google Scholar]
- Flügge UI (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50 27–45 [DOI] [PubMed] [Google Scholar]
- Frangne N, Eggmann T, Koblischke C, Weissenböck G, Martinoia E, Klein M (2002) Flavone glucoside uptake into barley and Arabidopsis cell culture vacuoles-energization occurs by H+ and by ABC type mechanisms. Plant Physiol 128 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Frangne N, Gomes E, Martinoia E, Palmgren MG (2000) The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast1. Plant Physiol 124 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JR (1983) The Golgi apparatus. In JL Hall, AL Moore, eds, Isolation of Membranes and Organelles from Plant Cells. Academic Press, London, pp 119–134
- Hirschi K (2001) Vacuolar H+/Ca2+ transport: who's directing the traffic? Trends Plant Sci 6 100–104 [DOI] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ (2000) Expression of Arabidopsis CAX2 in tobacco: altered metal accumulation and increased manganese tolerance. Plant Physiol 124 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CG, Hao YJ, Honda C, Kita M, Moriguchi T (2003) Putative PIP1 genes isolated from apple: expression analyses during fruit development and under osmotic stress. J Exp Bot 54 2193–2194 [DOI] [PubMed] [Google Scholar]
- Hurth MA, Suh SJ, Kretzschmar T, Geis T, Bregante M, Gambale F, Martinoia E, Neuhaus HE (2005) Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol 137 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack DL, Yang NM, Saier MH Jr (2001) The drug/metabolite transporter superfamily. Eur J Biochem 268 3620–3639 [DOI] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J (2007) A proteomic dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-golgi network to the central vacuole in Arabidopsis. Plant Cell 13 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16 393–401 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, Hames C, Alcon C, Neumann D, Vansuyt G, Curie C, Schroder A, Kramer U, et al (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boedker H (1977) RNA molecular weight determinations by gel electrophoresis under denaturing conditions; a critical re-examination. Biochemistry 16 4743–4751 [DOI] [PubMed] [Google Scholar]
- Leigh RA, Branton D, Marty F (1979) Methods for the isolation of intact vacuoles and fragments of tonoplast. In E Reid, ed, Plant Organelles, Methodological Surveys (B) Biochemistry, Vol 9. E. Harwood Ltd., Chichester, England, pp 69–80
- Lord JM (1983) Endoplasmic reticulum and ribosomes. In JL Hall, AL Moore, eds, Isolation of Membranes and Organelles from Plant Cells. Academic Press, London, pp 119–134
- Luu DT, Maurel C (2005) Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell Environ 28 85–96 [Google Scholar]
- Maeshima M (2001) Tonoplast transporters: organization and function. Annu Rev Plant Physiol Plant Mol Biol 52 469–497 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics 3 675–691 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet B, Forestier C, Kolukisaoglu U, Mueller-Roeber B, Schulz B (2002) Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214 345–355 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58 83–102 [DOI] [PubMed] [Google Scholar]
- Marty F (1999) Plant vacuoles. Plant Cell 11 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F, Branton D (1980) Analytical characterization of beetroot vacuole membrane. J Cell Biol 87 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty-Mazars D, Clémencet MC, Dozolme P, Marty F (1995) Antibodies to the tonoplast from the storage parenchyma cells of beetroot recognize a major intrinsic protein related to TIPs. Eur J Cell Biol 66 106–118 [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Moore AL, Proudlove MO (1983) Mitochondria and sub-mitochondrial particles. In JL Hall, AL Moore, eds, Isolation of Membranes and Organelles from Plant Cells. Academic Press, London, pp 153–184
- Peiter E, Maathuis F, Mills L, Knight H, Pelloux J, Hetherington AM, Sanders D (2005) The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434 404–408 [DOI] [PubMed] [Google Scholar]
- Ratajczak R, Hinz G, Robinson DG (1999) Localization of pyrophosphatase in membranes of cauliflower inflorescence cells. Planta 208 205–211 [DOI] [PubMed] [Google Scholar]
- Rea P, Li ZS, Lu YP, Drozdowicz YM, Martinoia E (1998) From vacuolar GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49 727–760 [DOI] [PubMed] [Google Scholar]
- Rea P, Sanders D (1987) Tonoplast energization: two H+ pumps, one membrane. Physiol Plant 71 131–141 [Google Scholar]
- Reisen D, Marty F, Leborgne-Castel N (2005) New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol 5 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. In C Nolan, ed, A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York
- Sazuka T, Keta S, Shiratake K, Yamaki S, Shibata D (2004) A proteomic approach to identification of transmembrane proteins and membrane-anchored proteins of Arabidopsis thaliana by peptide sequencing. DNA Res 11 101–113 [DOI] [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J 15 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68 850–858 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K, Maeshima M, Yokota A, Tomizawa K, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Sze H, Schumacher K, Muller ML, Padmanaban S, Taiz L (2002) A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci 7 157–161 [DOI] [PubMed] [Google Scholar]
- Szponarski W, Sommerer N, Boyer JC, Rossignol M, Gibart R (2004) Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics 4 397–406 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Dotson M, Keen NT (1992) Plant transformation: a simple particle bombardment device based on flowing helium. Plant Mol Biol 18 835–839 [DOI] [PubMed] [Google Scholar]
- Theodoulou FL (2000) Plant ABC transporters. Biochim Biophys Acta 1465 79–103 [DOI] [PubMed] [Google Scholar]
- Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34 685–695 [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt tolerant tomato plants accumulate salt in the foliage but not in the fruits. Nat Biotechnol 19 765–768 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.