Abstract
We have functionally characterized an Arabidopsis (Arabidopsis thaliana) gene AtHSD1 (At5g50600) that encodes a protein with homology to animal 11-β-hydroxysteroid dehydrogenase (HSD). Transgenic Arabidopsis plants overexpressing AtHSD1 (designated AOHSD plants) under the control of the cauliflower mosaic virus 35S promoter showed increased growth and seed yield as well as increased tolerance of saline stress and reduced seed dormancy. In canola (Brassica napus), transgenic plants overexpressing AtHSD1 also outgrew wild-type plants. AOHSD phenotypes were similar to those of plants that overproduced brassinosteroids (BRs) or overexpressed the BR receptor gene BRI1. A loss-of-function hsd mutant produced by RNA interference displayed a semidwarfed phenotype with reduced sensitivity to BRs. In contrast, AOHSD plants were hypersensitive to BRs and exhibited increased catabolism of abscisic acid (ABA). Germination of AOHSD seeds was less sensitive to ABA, while hsd seed was more sensitive to ABA during germination. AtHSD transcription was rapidly induced by BR treatment in wild type and was expressed widely in aerial plant parts, especially vascular tissues. This study demonstrates that AtHSD1 is involved in regulating growth and development in plants and is likely to promote or mediate BR effects. The gene has significant potential for improving growth and yield of canola and other agricultural crops.
Glucocorticoids are a class of animal steroid hormones involved in metabolic, inflammatory, cardiovascular, and behavioral processes as well as response to stress (Tomlinson et al., 2004). The enzyme 11-β-hydroxysteroid dehydrogenase (HSD) is a key regulator of the level of glucocorticoids and catalyzes the interconversion of biologically active glucocorticoid (cortisol in human and corticosterone in rats and mice) and inactive glucocorticoid (cortisone and 11-dehydrocorticosterone; Kallberg et al., 2002; McCormick et al., 2006). Although 11-hydroxy steroids have not (to our knowledge) been identified in plants, genome sequence annotation has identified eight HSD-like genes in Arabidopsis (Arabidopsis thaliana) whose functions have not been fully characterized.
We initially identified canola (Brassica napus) HSD as being highly expressed in nongerminating, abscisic acid (ABA) analog-treated seed relative to germinating seed (Li et al., 2005). It has been established that HSDs are minor components of oil bodies in the oilseed Brassicaceae and in sesame (Sesamum indicum; Jolivet et al., 2004; d'Andrea et al., 2007). More recently, the protein encoded by HSD1 has been shown to exhibit the properties of an NADP-dependent 11β-, 17β-HSD/17β-ketosteroid reductase (d'Andrea et al., 2007). However, their true substrates and physiological role in oil bodies remain unclear.
Brassinosteroids (BRs) are powerful plant hormones involved in vascular differentiation, seed germination, and vegetative growth (Sasse, 2003). They regulate some growth-specific processes including photomorphogenesis and skotomorphogenesis as well as cell expansion in the presence of a potentially growth-limiting cell wall (Clouse, 2002), and BR-deficient mutants display extreme dwarf phenotypes (Choe et al., 1999; Noguchi et al., 1999). BR biosynthetic pathways have been characterized and many of the genes and proteins involved in the known pathways have been identified (Fujioka and Yokota, 2003).
The use of Arabidopsis mutants that are insensitive to BR resulted in the identification of several components of the BR signaling pathway. The BR receptor BR-INSENSITIVE1 (BRI1) encodes a plasma membrane localized Leu-rich repeat kinase with an extracellular domain that binds brassinolide (BL), the most physiologically active BR. Binding of BL to BRI1 results in phosphorylation of the kinase domain that activates the BRI1 protein leading to BR responses (Friedrichsen et al., 2000; Wang et al., 2001). BRI1 is expressed in various tissues and cells involved in cell elongation (Caño-Delgado et al., 2004). Recent studies have revealed another Leu-rich repeat receptor-like kinase BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) that interacts with BRI1 and regulates BR signaling (Li et al., 2002; Nam and Li, 2002).
Loss-of-function genetic screens have identified additional BR signaling components downstream of BRI1 and BAK1. These include bri2, a negative regulator of the BR pathway, which displays bri1-like phenotypes including dwarfism and BR-insensitive and ABA-hypersensitive responses (Li et al., 2001). Both bri1 ems suppressor1-D (bes1) and brassinazole resistant1-1D can rescue the phenotypes of weak bri1 mutants, and are insensitive to the BR biosynthesis inhibitor brassinazole (BRZ; Asami et al., 2000).
Evidence has accumulated of cross talk between BRs and other hormones including auxin, ABA, jasmonic acid, and ethylene (Krishna, 2003; Sasse, 2003). Of these, the antagonistic interaction between ABA and BR has been explored in a couple of systems at the level of gene expression (Sasse, 2003). The opposing actions of the two hormones can be rationalized insofar as many of the overall effects of both hormones are opposite. In general, BR effects lead to increased growth whereas ABA effects result in slower growth. However, there is an important exception to these opposing effects since both ABA and BR enhance abiotic stress tolerance (Krishna, 2003). Therefore the interaction between these two hormones is complex: at times synergistic and at other times antagonistic. In this context, it is of interest to see if the effects of BRs include causing changes in ABA metabolism.
In this study, we report on an HSD-like gene from Arabidopsis referred to as AtHSD1. Plants overexpressing AtHSD1 constitutively expressed BR response genes and displayed similar phenotypes to those overproducing BRs or BRI1.
RESULTS
Structural Analysis of AtHSD1
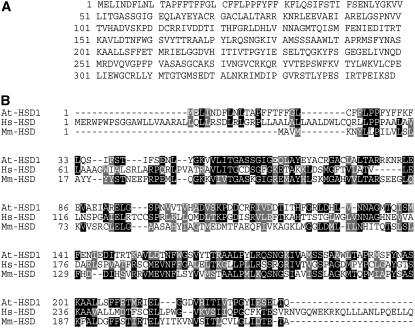
We studied an Arabidopsis gene that is annotated by the Munich Information Centre for Protein Sequences Arabidopsis Genome Database as a homolog of human and animal 11β-HSD, an enzyme that plays an important role in steroid metabolism (Tomlinson et al., 2004; McCormick et al., 2006). We provisionally refer to this gene as AtHSD1. A BLASTN search against the Arabidopsis genome database showed that there are two copies of AtHSD1 in the genome (Arabidopsis Genome Initiative locus At5g50600 and At5g50700, respectively). The AtHSD1 protein belongs to the short chain dehydrogenase/reductase family most of which are known to be NAD- or NADP-dependent oxidoreductases (Kallberg et al., 2002). The AtHSD1 gene consists of six exons and five introns and encodes a protein of 389 amino acid residues (Fig. 1A) with a calculated molecular mass of 39.0 kD and an pI of 6.15. Structural analysis revealed the presence of an N-terminal transmembrane region (amino acids 7–29). The sequence of the N-terminal region of the AtHSD1 protein to amino acid 233 shows high homology to human and mouse HSD proteins (Fig. 1B), whereas the C-terminal sequence (amino acids 234–349) diverges from that of the animal HSDs. In total, there are two identical copies of AtHSD1 and six other homologs (AtHSD2–7): At3g47350, At3g47360, At5g50590, At4g10020, At5g50770, and At5g50690 that, respectively, show 46%, 45%, 49%, 45%, 56%, and 46% sequence identity with AtHSD1.
Figure 1.
Sequence analysis of AtHSD1. A, AtHSD1 amino acid sequence. B, Sequence alignments of Arabidopsis HSD-like (AtHSD1), human HSD (Hs-HSD), and mouse HSD (Ms-HSD) amino acid sequences.
Phenotypes Resulting from Constitutive Overexpression of AtHSD1 Are Similar to Those Produced by Overproducing BRs or BRI Genes
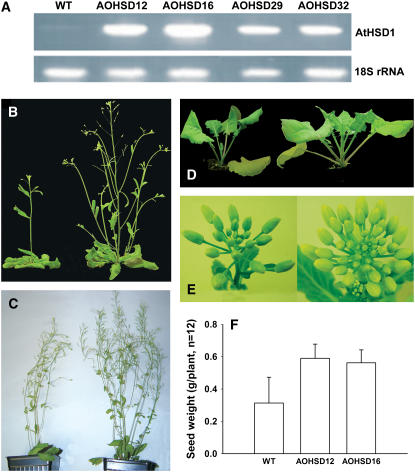
The AtHSD1 cDNA was fused to the cauliflower mosaic virus 35S promoter to drive high levels of gene expression in vegetative tissues. After transformation and selection, 42 kanamycin-resistant T1 transgenic plants were obtained. Reverse transcription (RT)-PCR analysis confirmed higher AtHSD1 mRNA level in T3 plants of four transgenic lines overexpressing AtHSD (AOHSD lines) relative to wild type (Fig. 2A).
Figure 2.
Expression of AtHSD1 and visible phenotypes in wild-type, AOHSD, and BOHSD plants. A, RT-PCR analysis of AtHSD1 transcript levels in Arabidopsis wild-type and overexpression lines AOHSD12, 16, 29, and 32. B, Arabidopsis plants after 1 month: AOHSD (right) and wild type (left). C, Arabidopsis plants after 2 months: AOHSD (right) and wild type (left). D, Canola plants after 1 month: wild type (left) and BOHSD (right). E, Comparison of 6-week-old wild-type (left) and BOHSD2 (right) inflorescences. F, Seed weights per plant of wild type and AOHSD16, n = 12.
The effects of AtHSD1 overexpression on growth of AOHSD plants were monitored during plant development. The number of rosette leaves, flowering time, and flower morphology were not significantly different between wild-type and transgenic plants. However, as shown in Figure 2, B and C, the AOHSD lines outgrew the wild type and mature AOHSD plants were reproducibly approximately 20% taller (43.0 ± 1.4 cm) than wild-type plants (35.8 ± 1.0 cm). Stem diameter was consistently larger in transgenics than in wild type, and the number of branches and siliques were also greater in transgenics than in wild-type plants (Fig. 2C), leading to a doubling of seed weight per plant in two transgenic lines relative to wild type (Fig. 2F). The higher seed yield was due to an increased number of siliques per plant and seed size was not significantly increased (data not shown). The roots of AOHSD plants grown on Murashige and Skoog medium for 7 d were 23% longer (4.8 ± 0.5 cm) than those of wild-type plants (3.9 ± 0.4 cm). Similarly, plants overexpressing AtHSD1 in canola (BOHSD plants) displayed increased growth from the seedling stage to flowering (Fig. 2, D and E). Increased stem diameter was also observed in BOHSD plants relative to wild type. In three transgenic lines (T1 generation), average diameters after 68 d of plant growth (measured just above soil level) were 113%, 127%, and 152% of stems in untransformed controls (average diameter of controls was 0.82 cm).
Overall, these results are similar to the phenotypes produced by overexpression of DWARF4, which encodes a steroid 22α hydroxylase involved in BR biosynthesis (Choe et al., 2001) and overexpression of BRI1 (Nam and Li, 2002). Similar phenotypes were produced by treating untransformed Arabidopsis plants with exogenous BL, whereas treatment with exogenous gibberellin (GA) produced elongated internodes, leading to a quite different leggy morphology (data not shown) similar to that produced by transgenic GA overproduction (Coles et al., 1999).
Seed Dormancy Is Reduced in Overexpression Lines
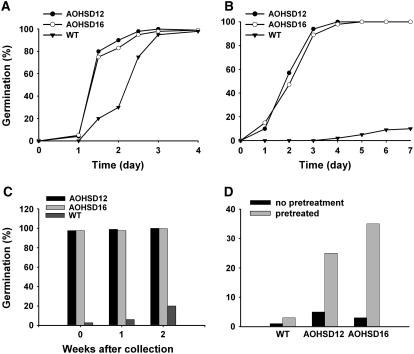
Arabidopsis seeds exhibit primary dormancy (Bentsink and Koornneef, 2002), which means that mature seeds are unable to germinate under the appropriate, permissive environmental conditions. To test whether AtHSD1 overexpression affected seed dormancy, we compared the germination of seeds under different conditions. Overall, transgenic seeds exhibited reduced dormancy compared with the wild type. In the absence of stratification at 4°C, the germination of AOHSD seeds was faster than that of wild type, at 2 d after imbibition reaching 80% to 90% compared to 30% germination of wild type (Fig. 3A). Primary dormancy of wild-type seeds is highest at harvest and decreases during storage (Fig. 3C). The difference between transgenic and wild-type seeds was even more striking when freshly harvested seeds were employed, reaching more than 95% germination in AOHSD seeds compared to less than 10% of wild-type seeds (Fig. 3, B and C). However, the germination of freshly harvested seeds is dependent on light and when seeds were incubated in darkness, almost no germination occurred either in wild-type or transgenic seeds. A 4-d cold treatment (stratification) eliminated primary dormancy in wild-type seeds and the requirement for light. A brief (1-d) cold treatment revealed that transgenic seeds are more sensitive to stratification than wild-type seeds. The cold treatment made little difference to germination of wild-type seeds but significantly increased germination in both transgenic lines (Fig. 3D). Steber and McCourt (2001) reported that BR promotes seed germination and is needed for normal germination in Arabidopsis. Overexpression of AtHSD1 promoted seed germination, possibly by enhancing the effects of endogenous BRs or by leading to elevated BR concentrations.
Figure 3.
Reduced dormancy in AOHSD plants. A, Germination time course for seeds of two AOHSD lines and wild type stored for 4 weeks at room temperature without stratification at 4°C. Germination was scored after 7 d of incubation. B, Germination time course for freshly harvested seed of two AOHSD lines and wild type. Germination was scored after 7 d incubation. C, Germination of seeds from two AOHSD lines and wild type after varying times of storage. Germination was scored after 7 d incubation. D, Germination of two AOHSD lines and wild-type freshly harvested seeds in darkness with or without a 1 d pretreatment at 4°C. Germination was scored after 7 d of incubation.
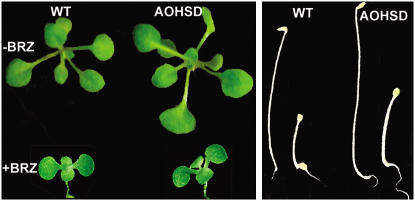
BRZ blocks BR biosynthesis at the C-22 hydroxylation step (Asami et al., 2000) and was used to test whether AtHSD1 effects require BRs. Figure 4 showed that both AOHSD and wild-type seedlings were sensitive to BRZ under both light and dark. Therefore, AtHSD1 effects depend on the presence of BRs and AtHSD1 overexpression appears to increase the sensitivity of plants to BRs.
Figure 4.
Transgenic plants expressing P35S:AtHSD1 are sensitive to the BR biosynthesis inhibitor BRZ under light (left) and dark (right). Three-week-old, light-grown seedlings of wild type and transgenics on Murashige and Skoog medium with or without 1 μm BRZ, and 4-d-old dark-grown seedlings of wild type and transgene with (right) or without (left) 1 μm BRZ.
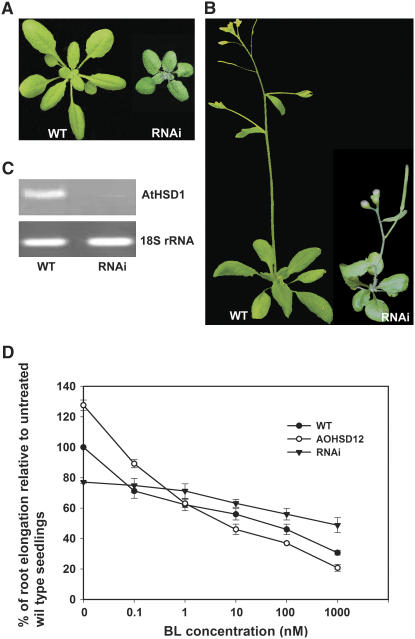
Loss-of-Function Plants Show Semidwarfed Phenotype and Insensitivity to BL
There were no apparent phenotypes in AtHSD1 T-DNA knockout lines, probably because phenotypic effects were masked by gene redundancy. To produce plants with reduced AtHSD gene expression, an AtHSD1 RNA interference (RNAi) transformation was conducted. Six out of 15 transgenic lines displayed a semidwarf phenotype with slightly darker green, wider leaves, and shorter petioles than wild-type plants and typical examples are shown in Figure 5, A and B. Semiquantatitive PCR showed AtHSD1 expression was substantially reduced in a representative RNAi plant relative to wild type (Fig. 5C), and transcripts of other gene family members were also reduced in RNAi plants (data not shown). The phenotypes produced by the RNAi lines are similar to those exhibited by bes1, which contains a defective transcription factor mediating BR-regulated gene expressions (Yin et al., 2005). The responsiveness of various genotypes to exogenously applied BR is shown in Figure 5D. A representative AOHSD line exhibited enhanced BR sensitivity relative to wild type, whereas a typical AtHSD1 RNAi line (hsd) exhibited reduced BR sensitivity compared with wild-type plants. For example, although primary roots of untreated AOHSD plants were longer than those of wild-type plants, they were shorter than wild-type plants at BL concentrations of 10 nm and above. Therefore, expression of AtHSD is directly related to the responsiveness of plants to BR.
Figure 5.
A representative plant expressing AtHSD1 RNAi (hsd) shows a semidwarfed phenotype and reduced sensitivity to BRs. A, Three-week-old wild-type and hsd plants grown in soil. B, Five-week-old wild-type and hsd plants grown in soil. C, RT-PCR analysis of AtHSD1 expression in wild-type and hsd plants. D, Root length grown on media containing different BL concentration. Each measurement is the average of 15 roots. The measurements were taken 7 d after germination.
AtHSD Gene Expression Affects Sensitivity to ABA and ABA Metabolism
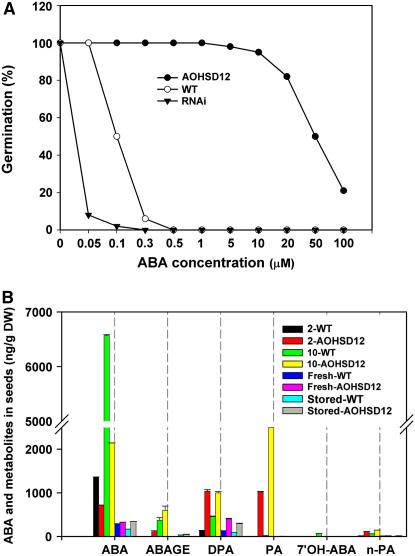
ABA and BRs have been shown to act antagonistically (Mandava, 1988). Therefore, to determine the relationship between AtHSD gene expression and ABA, we measured the germination of seeds on media containing increasing concentrations of (+)-ABA. The results indicated that AOHSD seeds have greatly reduced sensitivity to ABA in germination compared with wild-type Columbia (Col). Wild-type seed germination was almost completely inhibited at 0.5 μm (+)-ABA, whereas transgenic seed germination was not totally blocked even at 100 μm (+)-ABA (Fig. 6A). After germination, AOHSD seedlings grew well on medium containing up to 10 μm (+)-ABA, whereas wild-type Col seedlings failed to grow on medium containing 0.5 μm (+)-ABA (data not shown). Seeds from a typical hsd (RNAi) line were more sensitive to ABA than wild-type seeds; hsd seed germination was strongly inhibited at 0.05 μm (+)-ABA, whereas wild-type seed germination reached 100% (Fig. 6A). The germination of both the BR biosynthetic mutant det2-1 and the BR-insensitive mutant bri1-1 is more sensitive to ABA than the wild type (Steber and McCourt, 2001).
Figure 6.
Effects of (+)-ABA treatments on germination of Arabidopsis seeds and ABA contents under different conditions. A, Comparison of germination in Col wild-type (○), AOHSD (•), and hsd (RNAi; ▾) seeds. Percent germination was determined from a minimum of 120 seeds. B, Endogenous ABA and its metabolites in AOHSD and wild-type seeds under different conditions. Fresh, Freshly isolated seed; Stored, seed stored for 4 weeks at room temperature; 2-, stored seed treated with 2 μm (+)-ABA for 2 d; 10-, stored seed treated with 10 μm (+)-ABA for 2 d; ABA-GE, ABA-Glc ester; DPA, dihydrophaseic acid; 7′-OH-ABA, 7′-hydroxy ABA; n-PA, neo-PA.
To investigate the relationship between ABA metabolism and the ABA insensitivity of AOHSD lines, we profiled ABA and its metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry (Feurtado et al., 2004). There are several pathways of ABA catabolism involving either hydroxylation or conjugation. In most cases, the predominant pathway of ABA catabolism is via hydroxylation at the 8′ position to yield phaseic acid (PA), which is then reduced to dihydrophaseic acid (Zaharia et al., 2005).
As expected (Fig. 6B), the ABA content of freshly harvested wild-type seeds (292 ng/g dry weight) is more than that of seeds stored 4 weeks at room temperature (168 ng/g dry weight), consistent with increased germination after storage (Fig. 3C). The ABA contents of both freshly harvested and stored seeds of AOHSD lines (326 and 344 ng/g dry weight, respectively) are a little higher than in wild-type seeds. After 2 d treatment of stored wild-type seeds with 2 and 10 μm (+)-ABA, the tissue ABA levels (1,360 and 6,570 ng/g dry weight, respectively) were much higher than in untreated wild-type seeds (168 ng/g dry weight). In contrast, the ABA levels in AOHSD seeds were lower after ABA treatments (717 ng/g dry weight in the 2 μm treatment and 2,140 ng/g dry weight in the 10 μm ABA treatment) than in wild-type seeds, but the levels of all ABA metabolites were higher than in wild-type seeds (Fig. 6B). For example, PA only significantly accumulated in ABA-treated AOHSD seed. The elevated presence of ABA catabolites and reduced ABA content in AOHSD seed relative to wild type are consistent with increased flux through the ABA metabolic pathway.
Wild-type seeds did not germinate after ABA treatment (Fig. 6A) or if they were untreated but freshly harvested (Fig. 3B). However, AOHSD seeds can germinate and grow well despite the presence of high concentrations of applied ABA (Fig. 6A). The results described above suggest that AOHSD seeds have a higher capacity for ABA metabolism that is manifested in greater catabolism of exogenously applied ABA. However, despite increased ABA catabolism in AOHSD seeds, it is noteworthy that ABA nonetheless accumulates to high levels (e.g. to 2,140 ng/g dry weight in AOHSD seeds treated with 10 μm ABA) but the seeds nonetheless germinate readily. Therefore, the transgenic seeds demonstrate markedly reduced sensitivity to ABA.
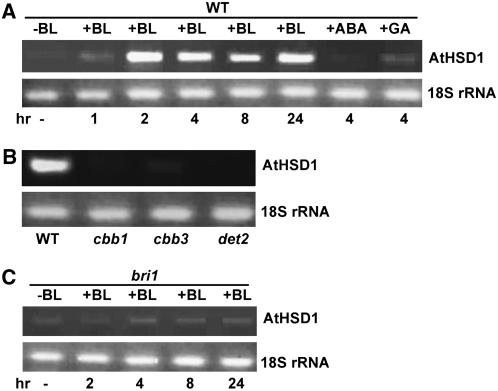
AtHSD1 Is Induced by BR
To determine if AtHSD1 is responsive to BL, the most active BR, a time course of BL treatment was performed. Induction of AtHSD1 occurred rapidly with a maximum around 2 h. Treatment with other plant hormones, such as ABA and GA3, did not significantly induce gene expression (Fig. 7A). On the other hand, AtHSD1 expression was severely reduced in all BR-deficient mutants tested, such as cbb1, cbb3, and det2 (Fig. 7B), suggesting that AtHSD1 was induced specifically by BRs during plant growth. To further test the genetic relationship between AtHSD1 and BR perception, we treated bri1 seedlings with BL. The results showed that the induction of AtHSD1 was reduced in bri1 (Fig. 7C), suggesting that AtHSD1 transcription is dependent on BR perception.
Figure 7.
AtHSD1 expression is induced by BL. A, BL-induced accumulation of AtHSD1 mRNA in wild-type plants. B, Low expression of AtHSD1 in BR-deficient mutants cbb1, cbb3, and det2. C, BL-induced accumulation of AtHSD1 mRNA is reduced in the bri1 background.
Differential Expression of Genes in Overexpression Plants
To examine the effects of AtHSD1 gene expression on other genes, expression profiling was conducted using Arabidopsis cDNA microarrays. Comparisons were made between 4-week-old seedlings of wild-type (Col) and AOHSD lines (Table I) and the experiments were repeated four times. Statistical analysis using Significant Analysis of Microarrays (SAM) showed 127 genes to be significantly differentially expressed by more than 2-fold, including 38 up-regulated genes and 89 down-regulated. Among the 38 induced genes, there were several with functions similar to those of known BL-induced genes encoding putative cell elongation or expansion-associated proteins such as pectinesterase and xyloglucan fucosyltransferase. BRs are known to increase expression of many genes involved in cell wall biosynthesis and modification, consistent with their effects on increasing cell expansion and division (Haubrick and Assmann, 2006). Other growth-related genes included elongation factor 1B as well as auxin or GA induced genes (Goda et al., 2002). The auxin-responsive gene SHY2/IAA3 (At1g04240) regulates auxin responses and such genes are known to also mediate BR responses (Nakamura et al., 2006). There were seven up-regulated genes involved in photosynthesis, which is also consistent with the increased growth of the plants. In addition, a Pro-rich family protein (At4g38770) playing an active role in plant defense reactions (Fowler et al., 1999) was also contained in this group. Some of the up-regulated genes were involved in lipid metabolism and other conversions of primary metabolism. The results suggest that the transgenic plant phenotype of increased growth is consistent with the observed changes in gene expression.
Table I.
Genes up-regulated in AOHSD transgenic plants relative to wild type
| Locus | Annotation | GenBank Accession |
|---|---|---|
| At1g12080 | Expressed protein | AY140106 |
| At5g13420 | Transaldolase, putative | NM_121345 |
| At2g45180 | Protease inhibitor/seed storage/lipid transfer protein family | NM_129813 |
| At2g42520 | DEAD box RNA helicase | NM_001035807 |
| At4g30960 | CBL-interacting protein | AF436831 |
| At2g34430 | Chlorophyll A-B binding protein/LHCII type I | AF339687 |
| At3g47470 | Chlorophyll A-B binding protein 4, LHCI type III CAB-4 | AF325012 |
| At5g54270 | Chlorophyll A-B binding protein/LHCII type III (LHCB3) | AF361858 |
| At1g29910 | Chlorophyll A-B binding protein 2, LHCII type I CAB-2 | AY065165 |
| At5g54280 | Myosin heavy chain | NM_124808 |
| At3g08640 | Alphavirus core protein family | AY081327 |
| At2g30820 | Expressed protein | NM_128635 |
| At3g56510 | TBP-binding protein | NM_115509 |
| At2g34420 | Chlorophyll A-B binding protein/LHCII type I | AF419587 |
| At3g33530 | Transducin family protein | NM_114071 |
| At4g27440 | Protochlorophyllide reductase B | AY081465 |
| At1g15820 | Chlorophyll A-B binding protein, chloroplast (LHCB6) | AF332425 |
| At5g02160 | Expressed protein | AF325034 |
| At2g45010 | Expressed protein | AF327424 |
| At1g45201 | Lipase class 3 family protein | NM_202246 |
| At5g05070 | Chlorophyll A-B binding protein/LHCII type II (LHCB2.2) | NM_120589 |
| At4g38770 | Proline-rich family protein | AY092992 |
| At4g39660 | Alanine-glyoxylate aminotransferase, putative | NM_120126 |
| At5g35735 | Auxin-induced protein | AW784033 |
| At4g24050 | Short-chain dehydrogenase/reductase (SDR) family protein | AF439829 |
| At4g35160 | O-methyltransferase family 2 protein | AY099803 |
| At1g03400 | Ethylene synthesis regulatory protein E8, putative | AY133559 |
| At1g48690 | Auxin-responsive GH3 | NM_103764 |
| At1g04240 | Auxin-responsive protein | AF332393 |
| At5g04960 | Pectinesterase family protein | NM_120578 |
| At3g16770 | AP2 domain-containing protein | AY142562 |
| At1g74420 | Xyloglucan fucosyltransferase, putative (FUT3) | AF417473 |
| At5g19510 | Elongation factor 1B | AF360304 |
| At5g50700 | Short-chain dehydrogenase/reductase (SDR) family protein | AF446888 |
| At2g14900 | GA-regulated protein 1 (GASA1) | AW985337 |
| At2g40890 | Cytochrome P450 98A3, putative | NM_180006 |
| At1g20620 | Catalase 3 (SEN2) | NM_001035995 |
| At5g19770 | Tubulin α-3/α-5 chain (TUA3) | NM_121982 |
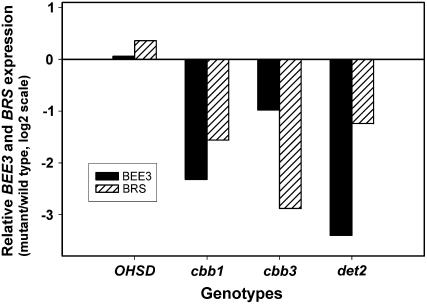
The BR ENHANCED EXPRESSION3 (BEE3) gene encodes a basic helix-loop-helix transcription factor that is induced by BR and is a positive regulator of BR responses. The bri1 SUPPRESSOR gene (BRS) encodes a secreted Ser carboxypeptidase that acts as a negative regulator of BR action, although its true mode of action is unknown (Haubrick and Assmann, 2006). Expression of both of these genes is significantly reduced in genotypes with defects in BR synthesis or responses (Fig. 8). However, expression of these genes is slightly increased in AOHSD plants and this result, together with the increased expression of BR-related genes in Table I, suggests elevated BR signaling and responses in plants overexpressing HSD.
Figure 8.
Expression of BEE3 and BRS in various genotypes. Real-time PCR was used to compare the expression of BEE3 and BRS in OHSD plants and in mutants defective in BR biosynthesis or signaling.
AtHSD1 Expression Is Tissue Specific
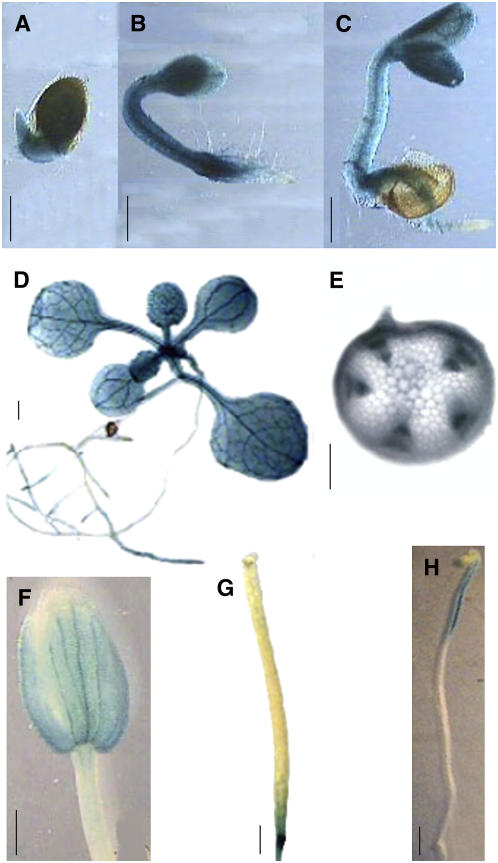
An AtHSD1 promoter-reporter gene fusion was employed to study the tissue specificity of AtHSD1 expression. The promoter region of AtHSD1 (approximately 1.5 kb) was isolated from Arabidopsis genomic DNA and fused to the GUS protein coding sequence (PHSD1:GUS). Histochemical analysis of GUS activity in transgenic plants harboring PHSD1:GUS showed a high level of expression in the above-ground parts of seedlings, and weak expression in root tissues in both light and darkness (Figs. 9, A–D and 8H). PHSD1:GUS was strongly expressed in vascular tissues (Fig. 9E) and this is consistent with the known involvement of BRs in vascular differentiation (Sasse, 2003). The vascular localization of expression may be related to our observation (noted earlier) that stem diameter was consistently higher in transgenic plants than in wild type. GUS activity was also observed in the bud and silique pedicels (Fig. 9, F and G).
Figure 9.
Expression of PHSD1:GUS in various tissues of transgenic plants. GUS activity was detected in germinating seeds at 1 (A), 2 (B), and 4 d (C) poststratification, Roots, cotyledons, and apical meristem (D), vascular tissues (E), petal (F), base of silique (G), and seedling growing under darkness (H).
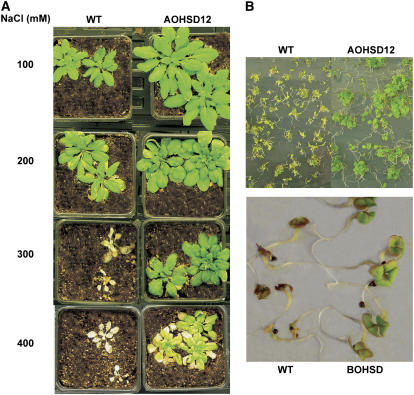
Increased Stress Tolerance of AOHSD and BOHSD Plants
The abiotic stress tolerance of plants overexpressing P35S:HSD were assessed by their ability to tolerate a saline growth medium. Wild-type Arabidopsis plants died after application of 300 mm NaCl, whereas transgenic plants were healthy and unbleached at 400 mm (Fig. 10A), although their rate of growth was reduced relative to untreated material. The effect of salt stress on seedling vigor was also assessed. Both wild-type and transgenic seeds of both Arabidopsis and canola were germinated on medium containing 100 mm NaCl and after 2 weeks a significant number of wild-type seedlings had died, whereas only a few of the transgenic seedlings failed to grow. Furthermore, wild-type seedlings were stunted and discolored, whereas transgenic seeds appeared healthy (Fig. 10B).
Figure 10.
Effect of salt stress on survival of plants. A, Two-week-old plants were flooded once a week for 4 weeks with 100, 200, 300, and 400 mm of NaCl. B, Seedlings of Arabidopsis and canola were germinated and grown for 2 weeks on agar medium supplemented with 100 mm NaCl.
DISCUSSION
We have characterized the functional effects of a gene with homology to animal 11β-HSD. Although 11-hydroxysteroids are important in regulating growth and development in animals, they have not been found in plants so the possible enzymatic functions of plant HSDs are uncertain. Enzyme assays of AtHSD have demonstrated that it possesses 11β- and 17β-HSD activity (d'Andrea et al., 2007) but its true substrate(s) is/are unknown. Our data suggest that AtHSD produces BR-like effects such as increased growth, branching, and flower production as well as thicker stems and increased stress tolerance. In addition, expression of the native HSD gene was induced by BL. Although GAs are also associated with rapid growth and increased height (e.g. Coles et al., 1999), we observed that exogenous application of GA produced a more elongated type of growth than that produced in AOHSD and BOHSD plants or by application of BL to control plants. Furthermore, treatment of plants with GAs or transgenic overproduction of GAs has not, to our knowledge, been associated with increased stress tolerance. On the other hand, the increased ABA catabolism and reduced sensitivity to ABA that we measured in AOHSD plants cannot account for the increased growth observed in OHSD plants and for their enhanced stress tolerance. Typically, ABA-deficient and ABA-insensitive mutants grow no better than wild type and are susceptible to water loss (e.g. Leon-Kloosterziel et al., 1996). Conversely, although ABA-hypersensitive mutants are more tolerant of drought stress, they typically grow slowly (e.g. Pei et al., 1998).
However, the role of HSD in relation to BR action has not been established. One possibility is that AtHSD is responsible for catalyzing a step in the biosynthesis of these hormones. However, the possibility that AtHSD functions in BR signaling cannot be excluded. For example, the fact that OHSD plants are hypersensitive to BR and that hsd (RNAi) plants are relatively insensitive to BR (Fig. 5) is more consistent with a role in mediating responses to BR than in controlling levels of BR.
Previous reports have suggested a reciprocal relationship between BR and ABA. The experiments reported here also provide examples of this phenomenon. AOHSD plants are hypersensitive to BR and insensitive to ABA and conversely, RNAi plants are insensitive to BR. We also show that enhanced BR effects in AOHSD seeds are associated with a greater ability to catabolize exogenous ABA, suggesting either faster ABA metabolic flux or at least a higher capacity for ABA metabolism. The antagonistic relationship between ABA and BR raises the puzzling question of how BR-like effects of HSD can be associated with enhanced stress tolerance (which is promoted by ABA) as observed in this study (Fig. 9) and in previous ones (Krishna, 2003). In this context, we have noted previously that a canola homolog of HSD was highly expressed in nongerminating, ABA analog-treated seed relative to germinating seed (Li et al., 2005), which is consistent with its known role as a component of oil bodies (Jolivet et al., 2004; d'Andrea et al., 2007) that are degraded during germination. However, these facts are difficult to reconcile with our observation that overexpression of AtHSD promotes germination. Indeed, there is no apparent relationship between the involvement of HSD in oil bodies and its role in promoting BR-mediated vegetative growth.
The interaction between ABA and BR is clearly complex: partially antagonistic and partially synergistic. A full understanding of the role that HSD plays in this hormonal interaction will require further experiments to define its mode of action. For example, analysis of the effects of HSD expression on active BR levels will be required to characterize the relationship of HSD to BR biosynthesis. Genetic experiments to determine whether HSD overexpression complements BR-deficient and/or BR-insensitive mutants are also required. However, irrespective of its precise mode of action, AtHSD is clearly an important contributor to plant growth and development and provides the potential for producing increased yield and stress tolerance in crop plants.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild type, and Arabidopsis transgenic plants were transformed with Agrobacterium tumefaciens GV3101 using the floral dip method (Clough and Bent, 1998). Canola (Brassica napus) was transformed by the method of Cardoza and Stewart (2003). To examine the growth of seedlings, Arabidopsis and canola seeds were surface sterilized with 10% bleach solution for 5 min and washed three times with sterile water. The sterilized seeds were sown in 9 cm petri dishes on Murashige and Skoog medium containing 1% (w/v) agar. Ten days after germination, seedlings with similar sizes were transferred to soil and grown in a growth chamber under long-day growth conditions (16-h light followed by 8-h darkness) at 20°C ± 2°C. To measure germination, sterilized seeds were incubated on Murashige and Skoog medium under continuous light at 23°C. Germination was scored after 7 d of incubation unless otherwise noted.
The cbb1 and cbb3 mutant seeds were kindly provided by Dr. Carsten Müssig, and the det2 mutant seeds by Dr. Jianming Li. bri1 (cbb2, CS292) mutant seeds were obtained from the Arabidopsis Biological Resource Center. For the mRNA analysis following various hormone treatments, 4-week-old wild-type or mutant plants were sprayed with 1 μm BL (OlChemIm Ltd., http://www.olchemim.cz/INDEX_e.HTM), 100 μm (+)-ABA, or 5 μm GA (GA4+7), respectively, and incubated for varying times. For (+)-ABA dose response curves, seeds (>120) were germinated and grown in light on Murashige and Skoog medium containing varying concentrations of (+)-ABA and germination was scored after 7 d. The seeds were treated for 2 d on Murashige and Skoog medium containing 2 and 10 μm (+)-ABA, respectively, then collected, washed, and frozen in liquid nitrogen.
Isolation of AtHSD1 cDNA and Constructs
A genome database search resulted in the identification of a genomic sequence encoding a protein that shares high homology with human and animal HSD, especially in the N-terminal region. We named this gene AtHSD1. The coding region of AtHSD1 gene was generated by PCR amplification of plasmid Uvi51 containing full coding sequence of this gene, the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGAGTTGATAAACGACTTTCTC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATCCGACTTGATTTCTGGAGT-3′ (attB sites for recombination cloning are shown in bold, and the sequence corresponding to AtHSD1 is underlined) were used for PCR. For generating AtHSD1 overexpression lines, the PCR product was introduced into the binary vector pK7WG2 (Karimi et al., 2002). The insert in the construct was sequenced to confirm the orientation and sequence and then transformed to Arabidopsis and canola. For AtHSD1 RNAi, an AtHSD1 full-length cDNA was amplified and cloned into pK7GWIWG2(I) in antisense/sense (Karimi et al., 2002).
Construction and Expression of Promoter-Reporter Gene
The region of AtHSD1 from −1,534 to +75 bp was generated by PCR amplification of genomic DNA using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCAATGGAACCGAAAGCCTAA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAGGCAAGAAGAAGCAGAGACC-3′ (attB sites for recombination cloning are shown in bold, and the sequence corresponding to AtHSD1 is underlined). The resultant 1,609-bp fragment was subcloned into binary vector pMDC163 (Curtis and Grossniklaus, 2003), and the resulting PHSD1:GUS construct was transferred into Arabidopsis (Col).
For GUS staining, seedlings were immersed in 50 mm sodium phosphate buffer (pH 7.2) with 1 mm 5-bromo-4-chloro-3indolyl-d-GlcUA and incubated at 37°C for 12 h. Chlorophyll was extracted by passing through increasing concentrations of ethanol. Nineteen independent T2 transgenic lines were analyzed and commonly observed staining patterns were recorded.
RT-PCR Analysis
Total RNA (2.5 μg) from each sample, treated with RNase-free DNase (Promega), was used for reverse transcriptase reactions. First-strand cDNA was synthesized with random hexamers using a SuperScript first-strand synthesis system according to the manufacturer's instructions (Invitrogen Life Technologies), and QuantumRNA 18S internal standards (Ambion) were used as a positive control for quantification of the relative amounts of cDNA. One microliter of RT reaction mixture was used as a template in a 20 μL PCR. For AtHSD amplification, the primers HSD1-1 (5′-TGCCAAGTCGATAGTGAACG-3′) and -2 (5′-CAGTAACCGACAACCCCACT-3′) were used for PCR. For amplification of BRS1, the primers BRS1-1 (5′-CCAACCAAAAGTGGCATTCT-3′) and BRS-2 (5′-TGTTGATACGAAACGGTCCA-3′) were employed. For amplification of BEE3, the primers BEE3-1 (5′-CGACGAGGGAAAATAAACGA-3′) and BEE3-2 (5′-CATGGATTCCACAGCATCAG-3′) were employed. The amplification conditions were 94°C (30 s), 56°C (30 s), and 72°C (30 s) and 28 cycles. RT-PCR was repeated twice.
HPLC-Mass Spectrometry Analyses
HPLC was performed using a Waters 2695 separation module (Waters). The extraction and purification of ABA and its metabolites, HPLC conditions, addition of internal standards, mass spectrometry, and quantification of endogenous levels of compounds were performed as described by Feurtado et al. (2004). Each analysis was performed in triplicate.
Microarray Analysis of Gene Expression Profiling in AOHSD Plants
Four-week-old seedlings of AOHSD and wild-type plants were harvested and frozen quickly in liquid nitrogen. Total RNA was extracted using RNeasy mini kits (Qiagen). Each total RNA sample (50 μg) was converted to cDNA and labeled using the Cyscribe Post-Labeling kit (Amersham Bioscience, RPN5660) following the manufacturer's instructions. The CyScribe GFX Purification kit (Amersham Bioscience, RPN5660X) was used to purify the fluorescently labeled cDNA probe by removing free nucleotides and unincorporated CyDye molecules. Arabidopsis 12 K cDNA microarrays (average length approximately 300 bases) from the Keck Foundation Biotechnology Resource Laboratory at Yale University School of Medicine were used. Hybridized slides were scanned sequentially for Cy3- and Cy5-labeled probes with a ScanArray 4000 laser scanner at a resolution of 10 Am. The experiments were repeated four times. Image analysis and signal quantification were performed using Quantarray (GSI Lumonics). Clones showing a signal value <800 in both Cy3 and Cy5 channels were eliminated from the analysis. The average of the resulting total Cy5 and Cy3 signals were used to calculate the ratios that were used for normalization. Data storage, preliminary data processing, and Lowess normalization were performed with the Bioarray Software Environment (Saal et al., 2002). Background-subtracted clone signals were used to calculate Cy5/Cy3 ratios. For each gene, the statistical significance of differences in expression between the control and AOHSD plants was calculated using SAM (P value ≤ 0.005; Tusher et al., 2001). SAM is a statistical technique for finding significant genes in a set of microarray experiments. SAM computes a statistic for each gene, measuring the strength of the relationship between gene expression and the response variable. It uses repeated permutations of the data to determine if the expression of any genes are significantly related to the response. The threshold for significance is determined by a tuning parameter delta, chosen by the user based on the false-positive rate. Users also have an option to choose a fold-change parameter, to ensure that called genes change at least a prespecified amount. Genes with a ratio of transgenic to wild-type plants of greater than 2.0 or less than 0.5 were selected.
Salt Stress Tolerance
The salt sensitivity of plants was evaluated by growth in pots in a controlled environment chamber. After 2 weeks of growth under normal conditions, plants were flooded once a week with solutions containing varying NaCl concentrations.
Acknowledgments
We thank Andrew Ross and Steve Ambrose for liquid chromatography-mass spectrometry analyses, Li Forseille for assistance with some of the canola experiments, and Suzanne Abrams for helpful discussions. In addition, we thank one of the anonymous reviewers for drawing our attention to some relevant publications.
This work was supported by the Genome and Health initiative of the National Research Council of Canada and by Genome Canada and Genome Prairie under the program “Enhancing canola through genomics.” This article is National Research Council of Canada number 48416.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Adrian J. Cutler (adrian.cutler@nrc-cnrc.gc.ca).
References
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M (2002) Seed dormancy and germination. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0050, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng J-C, Nam KH, Li J, Chory J (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 5341–5351 [DOI] [PubMed] [Google Scholar]
- Cardoza V, Stewart CN (2003) Increased Agrobacterium-mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyls segment explants. Plant Cell Rep 21 599–604 [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al (1999) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26 573–582 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002) Brassinosteroids. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0009, www.aspb.org/publications/arabidopsis/
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17 547–556 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462– 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Andrea S, Canonge M, Beopoulos A, Jolivet P, Hartmann MA, Miquel M, Lepiniec L, Chardot T (2007) At5g50600 encodes a member of the short-chain dehydrogenase reductase superfamily with 11β- and 17β-hydroxysteroid dehydrogenase activities associated with Arabidopsis thaliana seed oil bodies. Biochimie 89 222–229 [DOI] [PubMed] [Google Scholar]
- Feurtado JA, Ambrose SJ, Cutler AJ, Ross AR, Abrams SR, Kermode AR (2004) Dormancy termination of western white pine (Pinus monticola Dougl. Ex D. Don) seeds is associated with changes in abscisic acid metabolism. Planta 218 630–639 [DOI] [PubMed] [Google Scholar]
- Fowler TJ, Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol 121 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Huter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54 137–164 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubrick LL, Assmann SM (2006) Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ 29 446–457 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Roux E, d'Andrea S, Davanture M, Negroni L, Zivy M, Chardot T (2004) Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 42 501–509 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Opperman U, Jornvall H, Persson B (2002) Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci 11 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Krishna P (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22 289–297 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Li F, Wu XZ, Tsang E, Cutler AJ (2005) Transcriptional profiling of imbibed Brassica napus seed. Genomics 86 718–730 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222 [DOI] [PubMed] [Google Scholar]
- Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39 23–52 [Google Scholar]
- McCormick KL, Wang XD, Mick GJ (2006) Evidence that the 11 β-hydroxysteroid dehydrogenase (11β-HSD1) is regulated by pentose pathway flux: studies in rat adipocytes and microsomes. J Biol Chem 281 341–347 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S (2006) Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J 45 193–205 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid singaling. Cell 110 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290 [DOI] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg Å, Peterson C (2002) BioArray software environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol 3 SOFTWARE0003 [DOI] [PMC free article] [PubMed]
- Sasse JM (2003) Physiological actions of brassinosteroids: an update. J Plant Growth Regul 22 276–288 [DOI] [PubMed] [Google Scholar]
- Steber CM, McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JW, Walker EA, Bujalska IJ, Draper N, Lavery GG, Cooper MS, Hewison M, Stewart PM (2004) 11β-Hydroxysteroid dehydrogenase type 1: a tissue-specific regulator of glucocorticoid response. Endocr Rev 25 831–866 [DOI] [PubMed] [Google Scholar]
- Tusher V, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to transcriptional responses to ionizing radiation. Proc Natl Acad Sci USA 98 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410 380–383 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259 [DOI] [PubMed] [Google Scholar]
- Zaharia LI, Walker-Simmons MK, Rodriguez CN, Abrams SR (2005) Chemistry of abscisic acid catabolites and analogs. J Plant Growth Regul 24 274–284 [Google Scholar]