Abstract
The photorespiratory Arabidopsis (Arabidopsis thaliana) mutant gld1 (now designated mtkas-1) is deficient in glycine decarboxylase (GDC) activity, but the exact nature of the genetic defect was not known. We have identified the mtkas-1 locus as gene At2g04540, which encodes β-ketoacyl-[acyl carrier protein (ACP)] synthase (mtKAS), a key enzyme of the mitochondrial fatty acid synthetic system. One of its major products, octanoyl-ACP, is regarded as essential for the intramitochondrial lipoylation of several proteins including the H-protein subunit of GDC and the dihydrolipoamide acyltransferase (E2) subunits of two other essential multienzyme complexes, pyruvate dehydrogenase and α-ketoglutarate dehydrogenase. This view is in conflict with the fact that the mtkas-1 mutant and two allelic T-DNA knockout mutants grow well under nonphotorespiratory conditions. Although on a very low level, the mutants show residual lipoylation of H protein, indicating that the mutation does not lead to a full functional knockout of GDC. Lipoylation of the pyruvate dehydrogenase and α-ketoglutarate dehydrogenase E2 subunits is distinctly less reduced than that of H protein in leaves and remains unaffected from the mtKAS knockout in roots. These data suggest that mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of lipoate biosynthesis in leaves and may occur independently of this pathway in roots.
Research on the photorespiratory pathway has triggered progress in many other fields of plant research and helped in the establishment of Arabidopsis (Arabidopsis thaliana) as an important model organism (Somerville, 2001). This high-throughput pathway of plant primary metabolism extends over three cellular compartments, chloroplasts, peroxisomes, and mitochondria, and enables plants to live in an oxygen-containing atmosphere (Osmond, 1981). The biological importance and function of this pathway becomes apparent from the fact that photorespiratory C3 plant mutants grow healthily in air enriched with 1% CO2, but are unable to survive in normal air. This specific feature has been exploited for the selection of chemically induced mutants with defects in individual enzymatic steps of this pathway (Blackwell et al., 1988; Somerville, 2001). One of these mutants, gld1 (originally named glyD), was attributed to a defective Gly decarboxylase (GDC), a mitochondrial multienzyme complex comprising the four protein subunits P, T, L, and H protein (Somerville and Ogren, 1982). H protein plays a key role as both electron acceptor and aminomethyl carrier when it interacts as a mobile substrate, via its lipoamidyl arm, one after the other with P, T, and L protein (for review, see Douce et al., 2001). Major characteristics of gld1 were impaired growth and high accumulation of Gly in normal air, but normal phenotype and healthy growth in air enriched with 1% CO2. Leaf mitochondria were unable to oxidize Gly, and the Gly-bicarbonate carbon exchange reaction of GDC was undetectable in mitochondrial extracts. This specific reaction solely depends on the combined presence of P and H protein, and the authors hypothesized that gld1 represents a defective P-protein gene (Somerville and Ogren, 1982).
Here, we report on the identification of the gld1 locus (now designated mtkas-1) and show that it resides on gene At2g04540 encoding β-ketoacyl-[acyl carrier protein (ACP)] synthase (mtKAS; EC 2.3.1.41), a key enzyme of mitochondrial fatty acid biosynthesis (Wada et al., 1997; Gueguen et al., 2000; Yasuno et al., 2004). One of the major products of mtKAS, octanoyl-ACP, is required for the lipoylation of essential mitochondrial proteins including the H protein of GDC and the dihydrolipoyl acyltransferase (E2) subunits of the pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (KGDH) multienzyme complexes. Western analysis of leaf mitochondria isolated from allelic mtkas mutants revealed heavily impaired lipoylation of H protein, but distinctly smaller effects on the E2 subunits of PDH and KGDH. In roots, lipoylation of the E2 proteins is not affected at all. These data suggest that mitochondrial protein lipoylation in plants, in contrast to current thinking, does not exclusively depend on the mtKAS pathway of fatty acid biosynthesis.
RESULTS AND DISCUSSION
mtkas-1 Is Defective in the Mitochondrial β-Ketoacyl-[ACP] Synthase, mtKAS
The recessive mutation of mtkas-1 was originally mapped to chromosome II at a distance of about ±40 cM from the er-py visible marker region (Artus et al., 1994), which points to a localization of the mutation at one of the ends of this chromosome. To perform a more precise recombination mapping of the mtkas-1 mutation, we crossed mtkas-1 plants with plants of the Landsberg erecta (Ler-0) ecotype and isolated 415 segregants with a photorespiratory phenotype similar to mtkas-1 in the F2 generation (slower growing and pale green leaves, compare Fig. 1). This mapping population was analyzed using three cleaved amplified polymorphic sequence (CAPS; Konieczny and Ausubel, 1993) markers er (50.64 cM), m246 (11.03 cM), and ve017 (69.14 cM). Marker ve017 was not closely linked to mtkas-1. In contrast, recombination between mtkas-1 and the framework marker m246 occurred with a frequency of only 2.99% (4:134 F2 plants; Fig. 2A). This indicated that the mtkas-1 locus resides on the top arm of chromosome II and is not related to any of the three GDC subunit-encoding genes on this chromosome, which all reside far apart from m246 on the bottom arm of chromosome II (The Arabidopsis Genome Initiative, 2000). Next, we used a simple sequence length polymorphism (Bell and Ecker, 1994) marker (on bacterial artificial chromosome [BAC] F5G3, recombination found in 9:403 F2 plants) and another CAPS marker (on BAC T23O15, recombination found in 7:398 F2 plants). The seven recombinants found with this CAPS marker were reanalyzed for recombination with two additional markers (6:7 recombinants with simple sequence length polymorphism marker on BAC F3L12 and 0:7 recombinants with CAPS marker on BAC F7D11). This allowed mapping of the mtkas-1 locus to a 100 to 140 kb interval on the top arm of chromosome II near to marker m246 and very close to BAC clone F7D11 at approximately 1.6 Mb on the pseudomolecule of chromosome II (Fig. 2A). Inspection of this region revealed no gene that is related to known GDC proteins or other enzymes related to the core C2 cycle.
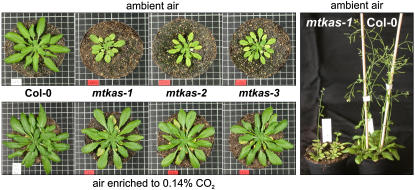
Figure 1.
Wild-type plants in comparison with mtkas-1 plants and two T-DNA insertion lines, mtkas-2 and mtkas-3. Plants were grown for 50 d in normal air (top left section) and in air enriched with 0.14% CO2 (bottom left section), respectively. mtkas-1 plants grow, although very slowly, and even flower under the photorespiratory conditions of ambient air (right section).
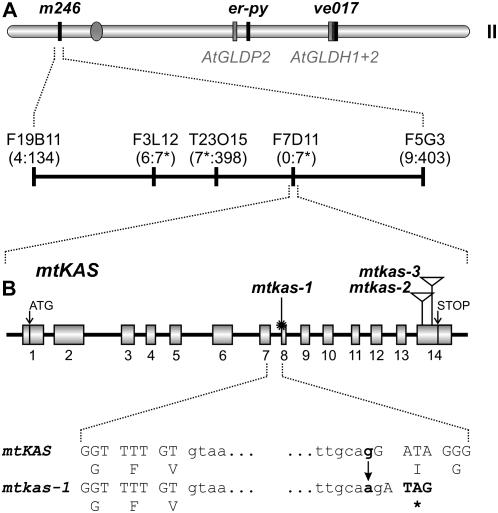
Figure 2.
The GDC-deficient mtkas-1 mutant is defective in mtKAS. A, Mapping of mtkas-1 to a position close to BAC F7D11 on chromosome II. Markers used for recombination mapping are shown by name or the corresponding BAC clone and their positions are indicated by vertical lines. The numbers of recombinants relative to analyzed F2 plants for the respective markers are given in parentheses (for further information see main body of the text). The three GDC subunit-encoding genes present on chromosome II are shown in gray bars and the position of the centromer is indicated by a gray circle. B, Structure of the mtKAS gene and positions of the mutations in the three different alleles, mtkas-1, mtkas-2 (SALK 022295), and mtkas-3 (SALK 087186). The two T-DNA insertions are indicated by triangles. The sequence context of the point mutation (star) at the splice acceptor site of intron 7 is shown in capital (exons) and lowercase letters (intron), respectively. The mutation results in a frameshift and introduces a stop codon with consequential loss of 185 C-terminal amino acids.
To identify the defective gene, we chose to analyze T-DNA insertion lines for a variety of genes in this region. Among these lines, only seedlings of a population of a T-DNA insertion mutant for gene At2g04540 (SALK 022295; mtkas-2) segregated with a phenotype very similar to that of mtkas-1 (Fig. 1). Preliminary analysis of soluble amino acids revealed a highly elevated leaf Gly content, which corresponds with published labeling data for mtkas-1 plants (Somerville and Ogren, 1982). Homozygosity of the T-DNA insertion in this line and in a subsequently isolated allelic T-DNA insertion line (SALK 087186; mtkas-3) was confirmed by PCR with genomic DNA and the positions of insertions determined by sequencing. The two insertions are independent, they reside in the last exon of At2g04540 (Fig. 2B) and, as shown by the absence of corresponding transcripts (Fig. 3), represent knockout alleles for the gene mtKAS. The use of two additional primer combinations for reverse transcriptase (RT)-mediated PCR, covering transcript ranges upstream of the last exon, revealed the presence of aberrant transcripts (data not shown), which is frequently observed with T-DNA insertions in the last exon (for example, Tsunoyama et al., 2004).
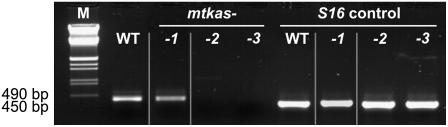
Figure 3.
RT-PCR shows significant amounts of transcripts in leaves of the chemically induced mtkas-1 mutant (-1), while no residual normal mtKAS mRNA is detectable in the homozygous T-DNA knockout plants, mtkas-2 (-2), and mtkas-3 (-3). Wild-type (WT) plants were included as a control, and signals from the 40S ribosomal S16 protein mRNA were used for internal calibration (right). Gray lines enclose lanes (1) that have been sliced in from another part of the same gel.
In contrast to the knockout T-DNA lines mtkas-2 and mtkas-3, significant amounts of mtKAS transcripts are detectable in mtkas-1 plants (Fig. 3). To further support our notion that the mutation in mtkas-1 affects the mtKAS locus, we therefore crossed emasculated plants of the mtkas-1 genotype to homozygous mtkas-2 and mtkas-3 plants. The F1 progenies, which were hybrid hemizygous for both the mtkas-1 and the respective mtkas-2 or mtkas-3 allele, should lack intact mtKAS alleles and the wild-type phenotype should not be restored. Indeed, these hybrid plants showed slow growth with pale-green to yellow leaves in normal air and were indistinguishable from the homozygous parents' photorespiratory phenotype (Supplemental Fig. S1). This lack of complementation provides direct genetic evidence that the photorespiratory phenotype of mtkas-1 is caused by a defect in the single-copy mtKAS gene, At2g04540.
To exactly identify the mutation in the mtKAS gene of mtkas-1 plants, we then used two experimental strategies. First, sequencing of mtkas-1 cDNA obtained by RT-PCR with primers specific for mtKAS revealed a 1-bp deletion, which leads to a frameshift in the open reading frame and a truncated protein. Second, we isolated genomic DNA from mtkas-1 plants and amplified a 3.1 kb fragment by PCR. Sequencing of this genomic fragment revealed a G-to-A transition at the 3′ splice site of the seventh intron, which results in the out splicing of one additional nucleotide (corresponding to the 1-bp deletion in the cDNA) in combination with a premature stop codon (Fig. 2B). Therefore, translation of the mtkas-1 mRNA stops at a position corresponding to Ile-277 with consequential loss of 185 C-terminal amino acids, i.e. about 40% of the native full-length protein. Importantly, none of the analyses to be discussed in the following paragraphs revealed any notable differences between mtkas-1 and the two T-DNA insertion lines, which confirms that all three lines are true knockout mutants.
These four lines of evidence, recombination mapping, phenotype analysis of T-DNA insertion lines, complementation analysis, and sequencing, evidently show that the gld1 (mtkas-1) locus is identical with the single-copy gene encoding the recently predicted (Mekhedov et al., 2000) and identified (Yasuno et al., 2004) Arabidopsis type II mitochondrial fatty acid synthase, mtKAS. Primed with malonyl-ACP, mtKAS catalyzes both the initial condensation step and chain elongation in mitochondrial acyl-ACP biosynthesis and represents the only known source of octanoyl-ACP in plant mitochondria (Yasuno et al., 2004). Elimination of mitochondrial octanoyl-ACP synthesis thus must exert deleterious effects on H protein and GDC, explaining the heavily disturbed photorespiratory metabolism of all three mtkas mutants.
Effects on Photosynthesis and Leaf Amino Acid Content
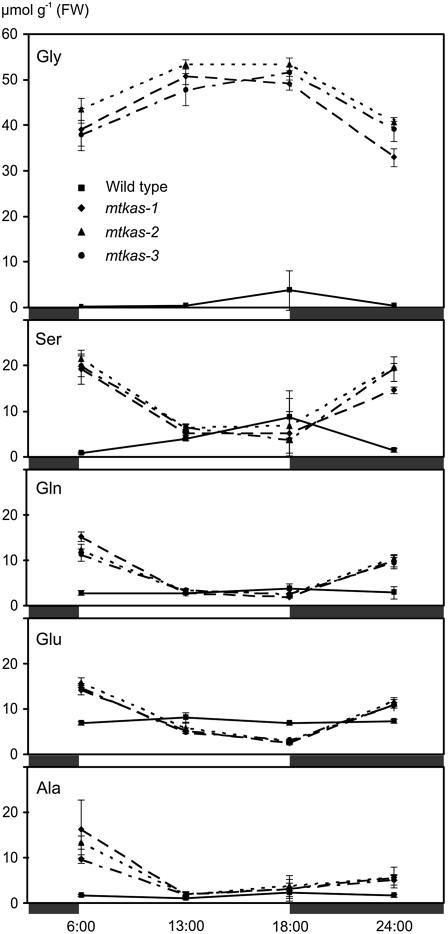
To more exactly determine photosynthetic properties of mtKAS-deficient mutants, we measured the photosynthetic performance of plants grown under elevated CO2. Photosynthetic rates of mtkas-2 plants (7.2 ± 0.2 μmol m−2 s−1) were about 65% of wild-type rates (11.2 ± 0.8 μmol m−2 s−1) at 21% O2 and did not significantly deteriorate during the measurements. We also examined effects of photorespiratory conditions (normal air) on the steady-state levels of soluble amino acids in leaves of all three allelic mtkas mutants over a 24-h cycle (Fig. 4). In comparison with wild-type plants, Gly concentrations were 40- to 50-fold elevated in the mutants during illumination and become reduced by about 20% to 30% during the following dark period. Levels of Ser, Gln, Glu, and Ala changed into the opposite direction, i.e. about wild-type levels were found in the second half of the light phase and increased concentrations during the dark. No or only moderate changes were observed in the content of other amino acids (data not shown).
Figure 4.
Diurnal leaf amino acid profiles confirm heavily disturbed photorespiratory metabolism in the mtkas-1, mtkas-2, and mtkas-3 mutants in comparison with wild-type plants. Light (white boxes) and dark periods (black boxes) are indicated. Symbols and error bars for wild-type (square), mtkas-1 (rhombus), mtkas-2 (triangle), and mtkas-3 (circle) plants grown in normal air represent means ± se from three samples.
Both the recovery of the mtkas mutants by elevated CO2 and these two sets of more quantitative data correspond well with earlier findings (Somerville and Ogren, 1982). Given that mtKAS represents the only known source of octanoyl-ACP synthesis in mitochondria, however, these results are not necessarily to be expected. First, we have recently shown that a total knockout of GDC is lethal even in nonphotorespiratory conditions, indicating that GDC fulfils a nonreplaceable function in vital metabolic processes other than photorespiration (Engel et al., 2007). Second, in addition to H protein, other mitochondrial proteins of strategic importance, mainly the E2 subunits of the α-ketoacid dehydrogenase complexes PDH and KGDH, also require lipoylation for their activity (Mooney et al., 2002; Taylor et al., 2004). Therefore, the observed features of mtkas mutants indicate significant residual activities of all three multienzyme complexes and point to the possibility that mitochondrial protein lipoylation may not be exclusively dependent on mtKAS.
Rates of Gly Decarboxylation Are Very Low in mtkas Mitochondria
To more directly assess effects of mtKAS deficiency on mitochondrial metabolism, we next examined the efficiency of Gly decarboxylation by intact mitochondria (Table I; Supplemental Fig. S2 for details). Good quality of the mitochondrial preparations was indicated by high rates of O2 consumption with malate or NADH and normal respiratory coupling levels (Keech et al., 2005). In accordance with published data for mitochondria isolated from mtkas-1 plants (Somerville and Ogren, 1982), we found that malate-dependent respiratory rates of mitochondria isolated from the mtkas mutants were high and only marginally altered relative to wild type. In contrast, rates with Gly were only about 5% of those measured with wild-type mitochondria. These data strengthen the notion that GDC, at least in photosynthesizing tissues, is very heavily impaired by the deletion of mtKAS.
Table I.
Gly is very poorly respired by leaf mitochondria isolated from mtkas-2 plants, but malate consumption is only marginally affected
Rates were determined in the presence of ADP (state 3) and are expressed as nmol O2 min−1 mg−1 protein. Each value is the mean of six replicates measured with two independent mitochondria preparations for wild-type and mtkas-2 plants (mean ± sd, n = 6 to 9). Respiratory control values (RC) were determined from the state 3 to state 4 transition. An analogous experiment with mtkas-1 mitochondria gave similar results.
| Respiratory Substrate | Wild Type
|
mtkas-2
|
||
|---|---|---|---|---|
| State 3 | RC | State 3 | RC | |
| Malate | 199 ± 57 | 2.9 ± 1.1 | 175 ± 21 | 2.0 ± 0.6 |
| Gly | 253 ± 48 | 1.7 ± 0.2 | 15 ± 5 | 1.4 ± 0.4 |
Activities of PDH and KGDH Are Reduced in mtkas Mitochondria
External pyruvate and α-ketoglutarate gave very low respiratory rates with mitochondria isolated from wild-type Arabidopsis leaves (data not shown), which is typical for leaf mitochondria. We therefore determined PDH and KGDH activities in lysed mitochondria and found clear effects of the mtKAS deletion. PDH activity was reduced to about 40% (0.026 ± 0.05 versus 0.068 ± 0.028 μmol min−1 mg−1 protein, mtkas-2 versus wild-type mitochondria), whereas KGDH was reduced to about 20% (0.024 ± 0.008 versus 0.125 ± 0.047 μmol min−1 mg−1 protein, mtkas-2 versus wild-type mitochondria). From the marginal effects on the growth of the mutants under nonphotorespiratory conditions we infer the presence of an excess of both enzymes in wild-type plants. More importantly, the two enzymes are significantly less reduced in their activities than the mitochondria's capacity to decarboxylate Gly. This finding suggests smaller effects of the deletion of mtKAS on E2-lipoylation and larger effects on the lipoylation of H protein.
mtKAS Is Important But Not Obligatory for Lipoylation of Mitochondrial Proteins
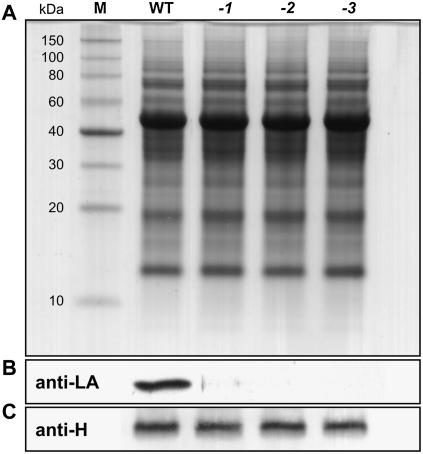
To further examine the possibility that the functional loss of mtKAS might affect subunit lipoylation in the three multienzyme complexes, GDC, PDH, and KGDH, to different extents, we performed western analyses with antisera specific for H protein and lipoic acid (LA), respectively. Lipoylation, within the restrictions of this method, means antigenic reactivity. This definition is important because protein-bound LA can react with the lipid peroxidation product 4-hydroxy-2-nonenal (HNE), which leads to loss of antigenicity of the respective protein domains. It has been shown that H protein is more susceptible to such modification than the E2 subunits of PDH and KGDH (Millar and Leaver, 2000; Taylor et al., 2002). To keep such effects as low as possible, well nourished and in a CO2-enriched atmosphere vigorously growing plants were used for these experiments. In accordance with the heavily impaired capacity of mtkas mitochondria to oxidize Gly (Table I), lipoylated H holoprotein was only detectable in traces in leaf extracts prepared from the three mutants in comparison with wild-type plants (Fig. 5, A and B). Wild-type levels of H apoprotein (Fig. 5C) indicated unaltered turnover of the unlipoylated H apoprotein.
Figure 5.
Lipoylation of H protein depends almost exclusively on a functional mtKAS pathway in leaves. A, Coomassie-stained whole-leaf protein spectra are indistinguishable in comparison of wild-type plants (WT) with mtkas-1 (-1), mtkas-2 (-2), and mtkas-3 (-3) mutants. B, H protein appears almost unlipoylated in the mutants as shown by western analysis and immunodetection of protein-bound LA by a specific antiserum. C, The whole-leaf content of H apoprotein is not reduced in the mutants. An antiserum raised against recombinant H protein was used in this experiment. In all sections, 10 μg soluble leaf proteins of high CO2-grown plants were loaded per slot of a 12% SDS-polyacrylamide gel.
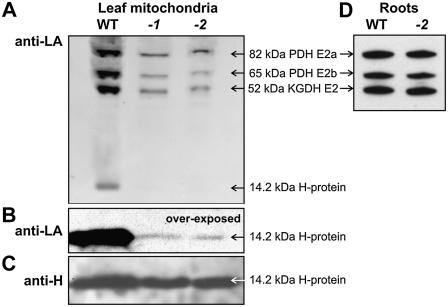
While GDC is an exclusively mitochondrial enzyme, PDH and KGDH are also present in plastids. To obtain additional evidence for H protein and also examine lipoylation of the E2 subunits of PDH and KGDH, we therefore performed similar experiments with mitochondria isolated from wild-type and mutant plants. Again, lipoylation of H protein in the mutant plants was detectable only after strong overexposure of the films (about 1% relative to wild type, Fig. 6, A and B), while the level of H apoprotein remains unchanged (Fig. 6C). In contrast, we observed distinctly higher lipoylation levels for the PDH and KGDH E2 subunits (about 30% relative to wild type, Fig. 6A), which were identified according to antigenicity and apparent molecular masses (Taylor et al., 2004). These percentages, obtained by scanning software, correspond well with the results from Gly-dependent respiration and enzyme activity measurements discussed above.
Figure 6.
Lipoylation of the PDH and KGDH E2 subunits is partially mtKAS dependent in leaves, but mtKAS independent in roots. A, Lipoylation of the mitochondrial E2 subunits of PDH and KGDH is reduced to a much lesser extent than that of H protein. B, Lipoylation of H protein in mitochondrial extracts is detectable only after strong overexposure of the immunoblots. C, The content of unlipoylated H protein remains unchanged. D, Lipoylation of the PDH and KGDH E2 subunits is independent on mtKAS in roots. For these experiments, 3 μg mitochondrial protein (A–C) or 5 μg of root soluble protein (D) of high CO2-grown wild-type (WT), mtkas-1 (-1), and mtkas-2 (-2) plants were loaded per slot of a 12% SDS-polyacrylamide gel. A picture of the complete gel from D is available in the online version of this article as Supplemental Figure S4.
By matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) of peptides obtained from H-protein bands excised from a Coomassie-stained gel run in parallel, signals for the respective lipoylated H-protein peptide were detected for wild-type mitochondria, but below detection limit in mitochondria isolated from mtkas-1 and mtkas-2 plants. In the mutants, only signals corresponding to the respective unlipoylated peptide, but not to HNE-modified lipoylated peptides, could be detected (Supplemental Fig. S3). It shall be noted that modifications can reduce the signal intensity of peptides. While we cannot entirely exclude the possible presence of a small fraction of HNE-modified peptides in our preparations, limited bias caused by HNE would not weaken but even strengthen our argumentation.
H protein of GDC is the dominant lipoylated protein in photosynthesizing plant cells and occurs in large amounts in green leaf mitochondria, representing the major sink for octanoyl chains in these organelles (Wada et al., 1997; Gueguen et al., 2000; Taylor et al., 2004). On the other hand, roots contain only very small amounts of GDC (Douce et al., 2001) and do not require high levels of lipoate biosynthesis. It was therefore tempting to speculate that the degree of E2 lipoylation might be even higher in nonphotosynthetic cells. To test this hypothesis, we finally examined E2 subunit lipoylation in protein extracts prepared from roots (Fig. 6D). Notably, no difference in lipoylation of the PDH and KGDH E2 subunits could be observed, indicating organ-specific effects of the mtKAS knockout. This interesting finding clearly deserves further investigation; however, in the context of this article, it fully supports our notion of an mtKAS-independent route of mitochondrial protein lipoylation.
The existence of an mtKAS-independent pathway for mitochondrial octanoyl-ACP biosynthesis is not supported by the literature. Therefore, the import of free or protein-bound lipoate or octanoate from extramitochondrial compartments appears as more likely. Plastidal de novo synthesis and export of fatty acids (Koo et al., 2004) including lipoate (Yasuno and Wada, 2002) would be one possible source. Alternatively, and possibly more likely, octanoyl chains could be supplied from the β-oxidation of longer-chain fatty acids in peroxisomes (Graham and Eastmond, 2002; Rylott et al., 2006). Lipoate synthase has been identified in plant mitochondria (Yasuno and Wada, 1998); however, such a scavenging pathway would also require lipoyl-protein ligase (LPLA). While such enzymes were unknown in plants until very recently, a bacterial-type bifunctional LPLA of yet unknown subcellular location has been identified in rice (Oryza sativa; Kang et al., 2007).
CONCLUSION
In this report, we identify the genetic lesion in the GDC-deficient mtkas-1 (gld1) mutant (Somerville and Ogren, 1982) and find that the mutation blocks the only known pathway for the de novo synthesis of octanoyl-ACP in mitochondria. This prevents lipoylation of the H-protein subunit of GDC in leaves to a very high degree and explains the heavily disturbed photorespiratory metabolism of this mutant. However, small amounts of lipoylated H protein were found in leaves of three independent mtkas mutants. We conclude that lipoylation of H protein, although only to a very small part in leaves, can occur independently from mtKAS-catalyzed octanoyl-ACP synthesis. This conclusion receives strong support from our analysis of the E2 subunits of PDH and KGDH. Deletion of mtKAS exerts much more limited effects on these proteins in leaves and does not show any apparent effect in roots. There is currently no evidence for an mtKAS-independent pathway for mitochondrial octanoyl-ACP synthesis. We therefore regard import of lipoate or octanoate from the cytosol as a more plausible alternative. Possible sources could be the β-oxidation of longer-chain fatty acids in peroxisomes or plastidal lipoate biosynthesis. These hypotheses will need to be substantiated by further experiments including identification and characterization of the involved scavenging enzymes. Likewise, the protein- and organ-specific differences in mitochondrial protein lipoylation revealed by the functional deletion of mtKAS have no obvious explanation at present and require further investigation.
MATERIALS AND METHODS
Plants and Growth Conditions
Arabidopsis (Arabidopsis thaliana) L. Heynh., ecotype Columbia-0, was used as wild-type reference and ecotype Ler-0 was used for recombination mapping. Seeds of the mtkas-1 mutant (CS8012, Columbia ecotype) were obtained from Jitao Zou and T-DNA insertion lines SALK 022295 and SALK 087186 (Alonso et al., 2003) from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). Seeds were incubated at least 48 h at 4°C to break dormancy prior to germination. Seedlings and adult plants were grown in a 4:1 mixture of soil (Type VM, Einheitserdewerk) and vermiculite and watered with modified Hoagland solution. Unless otherwise stated, plants were grown under a 12/12 h light/dark cycle (22°C/18°C) at 100 to 150 μE m−2 s−1 and 1.400 μL L−1 CO2 in controlled environment chambers.
Map-Based Cloning
To map the mtkas-1 locus, homozygous mtkas-1 plants were crossed to wild-type plants of the Ler-0 background. The segregating F2 generation was grown in normal air and screened for the mutant phenotype. A total of 415 individuals of the pale-green and slow-growing mtkas-1 phenotype were selected, and DNA was extracted individually. PCR analysis was performed with primer combinations listed in Supplemental Table S1, and resulting PCR fragments were differentiated according to size or different restriction sites. For the initial mapping steps, framework markers (http://Arabidopsis.info/new_ri_map.html) were used and, to narrow the region of the mtkas-1 locus, further markers were designed using information from the Monsanto LER sequence collection (Jander et al., 2002).
Isolation of T-DNA Mutants for mtKAS
Genomic DNA of T-DNA lines SALK 022295 (mtkas-2) and SALK 087186 (mtkas-3) was subjected to standard PCR (Master Mix, Quiagen) with primers specific for the left (R741, mLB1) or right (R409, SALK-RB1) border, respectively, and gene-specific primers 04540-A1 (R759 for SALK 022295) and mtKAS-2A (R833 for SALK 087186). All primer sequences are shown in Supplemental Table S2. The obtained fragments were directly sequenced to verify the insertion sites. Homozygous plants were identified by PCR with genomic DNA using a combination of two gene-specific primers (R759 and R726 for SALK 022295, R833 and R795 for SALK 087186) encompassing the respective T-DNA insertion. The knockout of mtKAS in homozygous plants of both mutant lines was verified by RT-PCR using 2.5 μg of leaf RNA (Nucleospin RNA plant kit, Macherey-Nagel) for cDNA synthesis (RevertAid cDNA synthesis kit, MBI Fermentas) and the primer combination R795 and R758. Prior to PCR analysis, cDNA amounts were calibrated according to signals from the constitutively expressed At2g09990 gene encoding the 40S ribosomal protein S16 (primers R176 and R177).
Photosynthesis and Leaf Amino Acid Content
Photosynthetic rates were measured with fully expanded leaves of plants grown in air enriched to 5.000 μL L−1 CO2 using a Licor-6400 gas-exchange system (LI-COR). Measurements were performed at a photosynthetic photon flux density of 500 μE m−2 s−1 supplied by an in-built red/blue LED light source, 380 μL L−1 CO2, 21% (v/v) O2, and a leaf temperature of 25°C.
Amino acids were extracted in 1.8 mL 80% (v/v) ethanol from 100 mg leaf material powdered in liquid nitrogen. After centrifugation, the supernatants were dried by lyophilization and redissolved in 8 mm Na2PO4 (pH 6.8) and 0.4% (v/v) tetrahydrofurane. Individual amino acids were quantified after derivatization with o-phthaldialdehyde as described elsewhere (Eisenhut et al., 2006).
Experiments with Isolated Mitochondria and Leaf Extracts
All plants for these experiments were grown at elevated CO2. Mitochondria were isolated from wild-type and mutant leaves according to Keech et al. (2005) and resuspended in a buffer containing 0.3 m Suc, 10 mm TES-KOH, pH 7.5, 10 mm KH2PO4, and 2 mm EDTA.
Respiratory rates were determined from three (wild type), two (mtkas-2), or one (mtkas-1) independent mitochondria preparations using about 100 μg protein per measurement. The Oxygraph oxygen electrode chambers (Hansatech) contained, in a total volume of 1 mL at 25°C, 0.3 m Suc, 10 mm TES (pH 7.5 with KOH), 10 mm KCl, 2 mm MgSO4, 5 mm KH2PO4, 0.1% (w/v) bovine serum albumin, and mitochondria equivalent to about 100 μg protein (Bradford, 1976). Oxygen consumption was followed without substrate (state 1), in the presence of 12.5 mm malate, Gly, pyruvate, or α-ketoglutarate, respectively, or 0.5 mm NADH (state 2), after subsequent addition of 0.1 mm ADP (state 3), and after consumption of all ADP (state 4).
PDH and KGDH activities were measured in triplicate from three independent mitochondrial preparations (n = 9) for each genotype (wild type and mtkas-2). Assays contained, in 1 mL at 25°C, 50 mm TES-KOH, pH 7.5, 2% (v/v) Triton X-100, 3.3 mm MgCl2, 3 mm Cys, 2.5 mm NAD+, 0.2 mm thiamine pyrophosphate, 1.5 mm pyruvate, or α-ketoglutarate, respectively, and about 100 μg mitochondrial protein. The reaction was started with 0.13 mm coenzyme A and followed by measuring A340 (Schuller and Randall, 1989).
Whole organ extracts were prepared by grinding 500 mg leaf material or 2 g washed roots in liquid nitrogen with subsequent extraction in a 4-fold volume of ice-cold extraction buffer (50 mm HEPES-NaOH, pH 7.6, 1 mm sodium EDTA, 5 mm MgCl2, 10 mm NaCl, 100 mm sorbitol, and 1 mm phenylmethanesulfonyl fluoride) followed by centrifugation at 20,000g for 10 min.
Mitochondrial and whole leaf proteins were separated in 12% (w/v) Tricine-SDS-polyacrylamide gels (Schägger, 2006), electrotransferred to a polyvinylidene difluoride membrane, and immunodetected using standard protocols with primary antibodies raised against recombinant H protein (Kopriva et al., 1996) or against LA (Calbiochem/Merck) at 1:2,000 dilution and chemiluminescence detection.
MALDI-TOF MS Peptide Mass Fingerprinting Analysis of H-Protein Lipoylation
Mitochondrial proteins of wild-type and mtkas1-1 plants were separated in a 12% (w/v) Tricine-SDS-polyacrylamide gels and stained with Coomassie. Excised H-protein bands were reduced and alkylated with tributylphosphine and iodoacetamide, respectively, and digested with trypsin. The resulting peptide mixtures were analyzed by MALDI-TOF MS using a Reflex III mass spectrometer (Bruker Daltonik) as described (Fulda et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The gld1 mutant (mtkas-1) cannot be complemented by crossing with the T-DNA knockout lines mtkas-2 or mtkas-3.
Supplemental Figure S2. Examples of original recorder traces during respiratory measurements showing deficiency in Gly oxidation.
Supplemental Figure S3. Mass spectrometric analysis of H-protein lipoylation.
Supplemental Figure S4. Lipoylation of E2 subunits of PDH and KGDH is independent from the mtKAS pathway of lipoate biosynthesis in roots.
Supplemental Table S1. Primers used for the map-based cloning of mtkas-1.
Supplemental Table S2. Primers used for the detection of T-DNA insertions, RT-PCR experiments, and for the amplification and sequencing of the mtKAS coding region of mtkas-1 genomic DNA.
Supplementary Material
Acknowledgments
We gratefully acknowledge the helpful discussions and support received from Dr. Murray Badger as well as the photosynthesis measurements by Dr. Susanne von Caemmerer (Australian National University, Canberra, Australia). We are grateful to Kirsten van den Daele for initial experiments, Klaudia Michl for performing amino acid analyses, and Dr. Martin Hagemann for critical discussions (all at University of Rostock, Germany), and unknown reviewers for helpful suggestions. This work would not have been possible without the mutant lines obtained from the Nottingham Arabidopsis Stock Centre and seeds of the mtkas-1 mutant (CS8012) kindly provided by Dr. Jitao Zou (Plant Biotechnology Institute, Saskatoon, Canada).
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to H.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hermann Bauwe (hermann.bauwe@uni-rostock.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Artus NN, Naito S, Somerville CR (1994) A mutant of Arabidopsis thaliana that defines a new locus for glycine decarboxylation. Plant Cell Physiol 35 879–885 [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144 [DOI] [PubMed] [Google Scholar]
- Blackwell RD, Murray AJS, Lea PJ, Kendall A, Hall NP, Turner JC, Wallsgrove RM (1988) The value of mutants unable to carry out photorespiration. Photosynth Res 16 155–176 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6 167–176 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Kahlon S, Hasse D, Ewald R, Lieman-Hurwitz J, Ogawa T, Ruth W, Bauwe H, Kaplan A, Hagemann M (2006) The plant-like C2 glycolate pathway and the bacterial-like glycerate cycle cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol 142 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu Ü, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H (May 11, 2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under non-photorespiratory conditions. Plant Physiol 144 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Mikkat S, Huang F, Huckauf J, Marin K, Norling B, Hagemann M (2006) Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6 2733–2745 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41 156–181 [DOI] [PubMed] [Google Scholar]
- Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J (2000) Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem 275 5016–5025 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Jeong HK, Lee E, Natarajan S (2007) Characterization of a lipoate-protein ligase A gene of rice (Oryza sativa L.). Gene 393 53–61 [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeström P (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124 403–409 [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4 403–410 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Ohlrogge JB, Pollard M (2004) On the export of fatty acids from the chloroplast. J Biol Chem 279 16101–16110 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Chu CC, Bauwe H (1996) H-protein of the glycine cleavage system in Flaveria: alternative splicing of the pre-mRNA occurs exclusively in advanced C4 species of the genus. Plant J 10 369–373 [DOI] [PubMed] [Google Scholar]
- Mekhedov S, Martínez de Ilárduya O, Ohlrogge J (2000) Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol 122 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Leaver CJ (2000) The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett 481 117–121 [DOI] [PubMed] [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD (2002) The complex fate of α-ketoacids. Annu Rev Plant Biol 53 357–375 [DOI] [PubMed] [Google Scholar]
- Osmond CB (1981) Photorespiration and photoinhibition: some implications for the energetics of photosynthesis. Biochim Biophys Acta 639 77–98 [Google Scholar]
- Rylott EL, Eastmond PJ, Gilday AD, Slocombe SP, Larson TR, Baker A, Graham IA (2006) The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal beta-oxidation is essential for seedling establishment. Plant J 45 930–941 [DOI] [PubMed] [Google Scholar]
- Schägger H (2006) Tricine-SDS-PAGE. Nat Protoc 1 16–22 [DOI] [PubMed] [Google Scholar]
- Schuller KA, Randall DD (1989) Regulation of pea mitochondrial pyruvate dehydrogenase complex—does photorespiratory ammonium influence mitochondrial carbon metabolism. Plant Physiol 89 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR (2001) An early Arabidopsis demonstration resolving a few issues concerning photorespiration. Plant Physiol 125 20–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem J 202 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277 42663–42668 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T (2004) Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA 101 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Shintani D, Ohlrogge J (1997) Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA 94 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, von Wettstein-Knowles P, Wada H (2004) Identification and molecular characterization of the β-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J Biol Chem 279 8242–8251 [DOI] [PubMed] [Google Scholar]
- Yasuno R, Wada H (1998) Biosynthesis of lipoic acid in Arabidopsis: cloning and characterization of the cDNA for lipoic acid synthase. Plant Physiol 118 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, Wada H (2002) The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517 110–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.