Abstract
The robustness of eight common food web properties is examined with respect to web size. We show that the current controversy concerning the scale dependence or scale invariance of these properties can be resolved by accounting for scaling constraints introduced by webs of very small size. We demonstrate statistically that the most robust way to view these properties is not to lump webs of all sizes, but to divide them into two distinct categories. For the present data set, small webs containing 12 or fewer species exhibit scale dependence, and larger webs containing more than 12 species exhibit scale invariance.
Keywords: scale invariance, community structure, connectance
The search for invariant properties has a long tradition in the natural sciences. In food web ecology, some food web properties (e.g., the fractions of top, intermediate, and basal species; the number of links per species; and rigid circuits) have been shown to be scale-invariant: they remain constant across webs of different kinds and sizes (1–5). These scaling properties fostered the formulation of interesting hypotheses on how food webs are assembled and how they operate (6–14). However, together with criticisms of the data used to demonstrate these properties (15, 16), recently compiled food webs have cast doubt on the validity of the scale-invariant laws (17–22). One recent study in particular heightened the controversy by proposing that most food web properties are scale-dependent (23). Interestingly, this analysis relied on a previously published data set (5) for which the properties were found to be scale-invariant. This discrepancy is due to the addition of small webs (with <10 species) to the new analysis, which were discarded from the original study because of worries about trivial biases that would affect these properties in small webs. Thus, the current controversy can be resolved by developing criteria for whether it is more correct to lump webs of all sizes for the analysis, or to separate them into two size classes.
Our objective here will be to analyze systematically a suite of competing models describing the relationship between food web properties and web size. We shall present quantitative evidence that the most robust model is a piecewise linear regression, indicating that food web properties are bounded to different scaling regions (24, 25) that should not be combined. This is an important result that both strengthens our understanding of the organization of food webs and provides a resolution to the controversy concerning the scaling behavior of food web properties.
The following properties will be considered: the link density (L/S, the total number of links divided by the number of species); the fraction of top (%T; having no predator), intermediate (%I; being predator and prey), and basal (%B; autotroph and detritus) species; and the fraction of links between top and intermediate (%TI), top and basal (%TB), intermediate and intermediate (%II), and intermediate and basal species (%IB). The properties are regressed against the number of species S in the webs. The suite of models that we consider is as follows: (i) linear regression, (ii) least squares fit to the power function, (iii) exponential asymptotic regression, (iv–vi) second- to fourth-order polynomial regression, (vii) discontinuous piecewise linear regression, and (viii) continuous piecewise linear regression. The properties are regressed against the number of species S in the webs.
We used the original dataset of Sugihara et al. (5), because it spans food webs from the very small (2 species) to the large (83 species) and because the original authors provided taxonomic refinement at the level of individual species in most cases. For the latter reason, we used biological species and not trophic species in our analyses (a trophic species is the union of all species sharing the same predators and prey). The criteria used for web selection and potential sources of taxonomic and sampling bias in this collection can be found in Schoenly et al. (ref. 26, pp. 623–626). In this data set, small webs do not represent highly aggregated or incomplete webs, as suggested by some authors (15, 16); most of them are small because they come from small habitats and are time-specific.
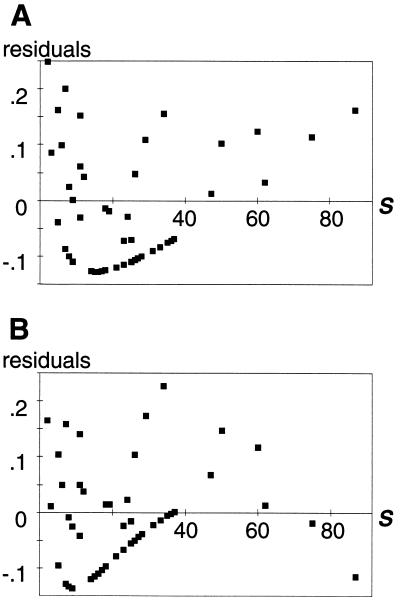
As noted elsewhere (23), simple linear regressions give non-zero slopes for many of the properties, indicating dependence on web size. As seen in Table 1, however, the fits are very poor (as indicated by the adjusted coefficients of determination Radj2), and, more importantly, the residuals often exhibit a U-shaped pattern (Fig. 1A and Table 1), as well as heteroscedasticity (i.e., the variance increases with web size; see Fig. 2 and Table 1). These systematic errors indicate that a linear model is not a robust description of the relationship. Fitting a power function (Y = a·Sb, with Y being the property, S the number of species, and a and b fitted constants) provides better results for most properties, but it does not remove the structure in the residuals. The exponential asymptotic model (Y = a + b·e−c·S, with a, b, and c being fitted constants) performs well for all properties but the fraction of top species. It is the most appropriate model for the fraction of links between top and intermediate species. However, for five of the properties, the residuals show evidence of heteroscedasticity. The second-order polynomial is the most suitable model for the fraction of links between intermediate and intermediate species and performs well for all but two properties. Nonetheless, for six of the properties, the residuals continue to exhibit strong structure (Fig. 1B and Table 1), indicating that second-order polynomial models are inappropriate here. In general, third-order polynomials do not represent significant improvements over second-order polynomials and are not considered further. However, fitting fourth-order polynomials provides significant improvements over the second-order polynomials for six of the properties and removes the higher-order structure in the residuals. Unfortunately, for four properties, heteroscedasticity still remains, indicating that these various polynomial models are inadequate descriptions of the data.
Table 1.
Performance and systematic fit of seven models for the eight food web properties
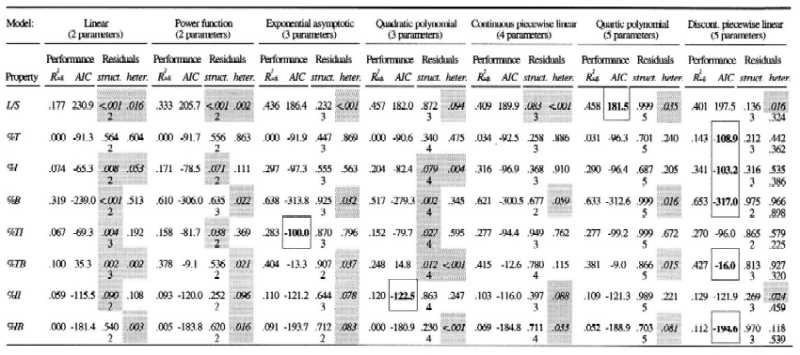
The models investigated are: (i) a linear regression, (ii) a power function, (iii) an exponential asymptotic model, (iv) a second-order polynomial regression, (v) a continuous piecewise linear regression with a breakpoint at c = 12, (vi) a 4th-order polynomial regression, and (vii) a discontinuous piecewise linear regression with a breakpoint at c = 12. The performance of a model is expressed by the adjusted coefficient of determination (Radj2; here, we prefer Radj2 over its unadjusted form, which does not take into account the number of parameters in the model). To determine which model is the most appropriate, given its number of parameters, we computed the Akaike Information Criterion (ref. 27; AIC). The model that minimizes the AIC is considered to be the best one (indicated by boldface type and frame). A systematic error is evaluated in two ways, first by fitting polynomial regressions to the residuals to determine if higher order structure (struct.) is present (see Fig. 1). The P values associated with the F test for the polynomial regressions are given here, together with the order of the polynomial regression applied to the residuals (we tried up to seventh order polynomials; the one giving the lowest P value is shown here). Significant P values (shaded) indicate the presence of systematic pattern in the residuals. Second, we evaluated the presence of heteroscedasticity (heter.; see Fig. 3) by fitting a linear regression to the absolute values of the residuals (28). The P values associated with the F test are given here. A poor fit to the linear regression indicates the absence of heteroscedasticity. For the piecewise model, the first P value is for webs with 12 species or less, and the second is for larger webs. As an example, the discontinuous piecewise linear model explains 40.1% of the variance in the link density property L/S, and gives an AIC of 197.5; a cubic regression of the residuals on S gives a P value for the F test of 0.136; a regression of the absolute values of the residuals on S gives a P value of 0.016 for the webs with 12 species or less and a P value of 0.324 for the webs with >12 species.
Figure 1.
Examples of model fit and systematic error. Residuals for the fraction of basal species using (A) a linear regression and (B) a second-order polynomial regression. The residuals exhibit patterns that are captured by fitting higher-order polynomial to them (see Table 1).
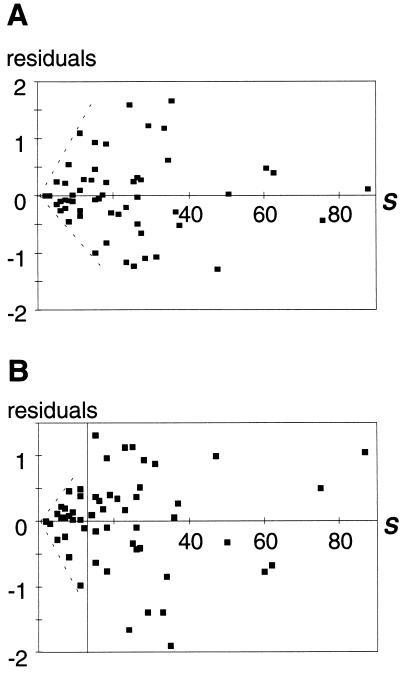
Figure 2.
Residuals for the link density property for the fourth-order polynomial regression (A) and for the discontinuous piecewise linear regression (B). Heteroscedasticity can be demonstrated by fitting a linear regression to the absolute values of the residuals (see Table 1). The residuals display heteroscedasticity for all webs in A and for the small webs in B. A line separates the large webs (n = 36) from the small webs (n = 24) in B. Only the residuals of large webs with the piecewise model exhibit homogeneous variances.
We now show that these problems in model fit or systematic error can be resolved most simply with a model that assumes the existence of two distinct scaling regions. The simplest model of this kind is a piecewise linear regression. For each food web property, we use a model of the following form:
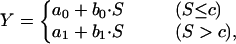 |
1 |
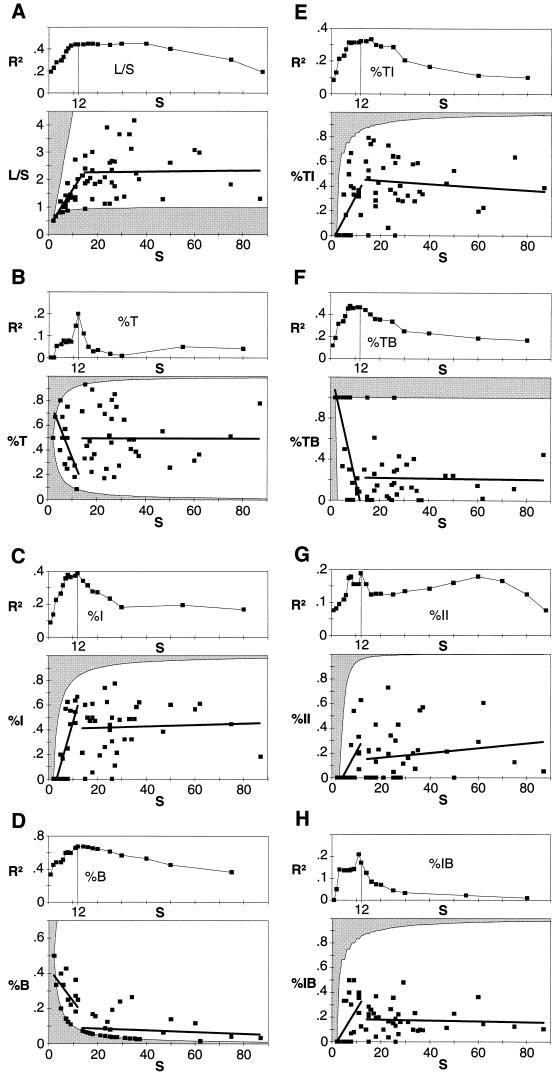
with a0, b0, a1, b1, and c being fitted constants. Of particular interest, the parameter c indicates the position of the breakpoint—that is, the natural boundary in web size separating the scaling regions of small and large webs. To find that boundary, a trial and error analysis with different values of c is done, and the performance (as expressed by the coefficient of determination R2) of the piecewise model is evaluated (first plots of Fig. 3). For most of the properties, the maximal value of R2 is obtained at or near c = 12 (the maximum is at c = 30 for the link density, but the value of R2 is very similar to that at c = 12). For economy, we shall adopt the value of 12 for the parameter c for all the piecewise regressions of the food web properties (we do not claim that 12 is a universal limit; rather it is a concordant characteristic for this data set). As can be seen in the Table 1, for five out of the eight properties, the discontinuous piecewise model is the most appropriate model, and it performs close to the best models for the remaining three properties.
Figure 3.
Data and discontinuous linear piecewise regression models of the food web properties for the 60 original food webs of Sugihara et al. (5). For each property, the first plot gives the value of the coefficient of determination, R2, for different positions of the breakpoints (the constant c in Eq. 1). The second plot presents the data and the regression lines for a breakpoint at S = 12 (S is the number of species). The upper and lower limits of the properties, given that there are no loops of the kind where species A eats species A or species A eats species B and species B eats species A are indicated by the shaded region. The properties, the coefficients of the piecewise linear regressions (from Eq. 1), and the results of t tests for the slopes b0 and b1 are as follows. (A) link density, L/S: a0 = 0.211, b0 = 0.140 (t = 6.49, P < 0.0001), a1 = 2.216, b1 = 0.0015 (t = 0.180, P = 0.858); (B) fraction of top species, %T: a0 = 0.785, b0 = −0.048 (t = −4.75, P = 0.0001), a1 = 0.496, b1 = 0 (t = −0.004, P = 0.996); (C) fraction of intermediate species, %I: a0 = −0.216, b0 = 0.067 (t = 5.35, P < 0.0001), a1 = 0.408, b1 = 0.0005 (t = 0.267, P = 0.791); (D) fraction of basal species, %B: a0 = 0.430, b0 = −0.019 (t = −2.69, P = 0.0134), a1 = 0.096, b1 = −0.0005 (t = −0.865, P = 0.393); (E) fraction of links between top and intermediate species, %TI: a0 = −0.064, b0 = 0.038 (t = 2.92, P = 0.0080), a1 = 0.474, b1 = −0.0013 (t = −0.635, P = 0.530); (F) fraction of links between top and basal species, %TB: a0 = 1.285, b0 = −0.107 (t = −4.79, P < 0.0001), a1 = 0.218, b1 = −0.0002 (t = −0.088, P = 0.931); (G) fraction of links between intermediate and intermediate species, %II: a0 = −0.167, b0 = 0.037 (t = 3.73, P = 0.0012), a1 = 0.121, b1 = 0.0019 (t = 0.956, P = 0.346); (H) fraction of links between intermediate and basal species, %IB: a0 = 0.055, b0 = 0.032 (t = 2.76, P = 0.0115), a1 = 0.186, b1 = −0.0004 (t = −0.351, P = 0.728).
Finally, we tried the continuous form of the piecewise linear model, where the intercept a1 in Eq. 1 is replaced by a0 + b0·c, thereby eliminating one parameter. As seen from Fig. 3, this model may provide a good fit as, for all properties except the fraction of top species (Fig. 3B), the extremities of the first lines come close to the starting points of the second ones. However, low values of the AIC indicate that this model is not the most appropriate and, moreover, the residuals show higher-order structure for the L/S property, and heterogeneous variance for the L/S, %B, %II, and %IB properties (Table 1).
The discontinuous piecewise linear model is the best performer among the suite of models examined. Of greater importance, this piecewise model removes awkward structure in the residuals, which is present in all the other models considered. This indicates that a simple linear model with a single breakpoint is a more satisfactory description of the various scaling relationships examined here. The link density property is one possible exception for which a polynomial model appears to perform slightly better. However, the residuals for this property exhibit heteroscedasticity (Fig. 2A) and, moreover, these polynomial models lack motivation. With the discontinuous piecewise model, while smaller webs show the same heteroscedasticity, the larger webs, having >12 species, are homoscedastic (Fig. 2B and Table 1). The heteroscedasticity of small webs is an artifact that will be discussed below.
The piecewise model shows clearly that all of the properties behave differently on either side of the boundary (second plots of Fig. 3). While small webs containing 12 or fewer species exhibit strong scale dependence, larger webs are statistically scale-invariant (see the legend of Fig. 3 for the values of t tests performed on the slopes). The changes in slope between small and large webs are highly significant for all properties (F test for difference between regression coefficients; the smallest difference is for the fraction of links between intermediate and intermediate species: F[1,31] = 28.04, P <.00001).
The scale-dependent behavior of small webs follows from a simple feature of such small systems: the rarity of cannibalistic loops and of loops of the kind where species A eats species B and species B eats species A. For example, a web with two species will have a link density, a fraction of top species and a fraction of basal species of 0.5. There are no degrees of freedom for any of these properties in such a small web. Indeed, for small webs in general, some of the properties are constrained to narrow upper and lower limits. As shown by the shaded regions in the plots of Fig. 3, these limits canalize strongly the L/S and %B properties (Fig. 3 A and D), forcing them to behave in a scale-dependent way. For the link density property, this is clearly the mechanism behind the heteroscedasticity seen in small webs. These limits also impose constraints on the %I, %TI, and %II properties (Fig. 3 C, E, and G), but to a lesser extent. For the other properties, the scale dependence of small webs may still reflect these boundary constraints, since %T + %I + %B = 1 and %TI + %TB + %II + %IB = 1. A property forced to exhibit scale dependence will in turn affect the behavior of its sister properties. These findings are in accord with earlier results (5) that warned against the trivial biases of small webs. This is not to say that the study of small webs is of no interest. They are certainly worth being studied for their own merit, but not with regard to scale invariance.
In the present analysis, we used biological species and not trophic species. The use of trophic species is aimed at alleviating the problem of variable taxonomic resolution among webs, thus rendering their comparison less dependent of this bias. We performed the analysis of the discontinuous piecewise model with the trophic versions of the webs to see if this lumping procedure could alter our conclusions. The first effect of this lumping process is to shrink the range of species numbers in the data set, from 2–83 to 2–54. It also displaces the position of the breakpoint (c in Eq. 1) from 12 to 7. Qualitatively, the results with trophic species are similar to those with biological species, that is, small webs are statistically scale-dependent (except for %T and %II), while webs with more than seven species are statistically scale-invariant. There is one notable exception: the link density property shows scale dependence for large webs. The generality and biological significance of this contrasting result needs further investigation.
The scale invariance revealed here for large webs gives support to earlier claims for the existence of scaling laws (1–5) and to the models predicting these patterns (6–14). The least robust property appears to be the link density (29). First, for this data set, scale invariance is revealed only with biological species. Second, the pattern found here for the link density property (scale dependence for small webs and scale invariance for large webs) is predicted by the random web/link sample bias model of Kenny and Loehle (30), which generates random webs with sampling error on the trophic links. Third, the scale invariance of the link density is likely not to be upheld in some circumstances (13, 29), such as in webs that are dominated by nonselective generalists. For example, Havens (21) studied 50 webs representing pelagic communities of small lakes and ponds and found that link density increases 4-fold over a range of 10–74 species. Havens proposed that such a scale-dependent relationship is likely to appear in webs such as his, which have a predominance of indiscriminate herbivores. This characteristic is most likely to exist in aquatic communities, where filter feeders are common (20, 21, 31, 32) and where predation by fishes is often size-dependent rather than species-dependent (33, 34). Thus, the behavior of the link density property as a function of species richness may well depend upon the dominant feeding mode in the system investigated; it is not likely to be universally constant or dependent on food web size. Finally, it is worth noting that, for the link density as well as for the other properties, the proportion of the variance explained by the piecewise model remains low (Table 1). Noise may account for a portion of this unexplained variance, but other factors may also be important for our understanding of the variability of these properties in different webs. Much work is needed here.
Insofar as we have shown that small webs exhibit different scaling properties than large webs and that this distinction can be objectively quantified, it is incorrect to lump them to support or criticize the generality of food web properties. This problem is less acute in recently compiled food webs (16, 17, 19, 20, 35–37), which are larger and more finely resolved. These are thoroughly and specifically compiled for the study of trophic interactions. With more such studies, it should be possible to provide greater resolution to the phenomenon of scale invariance and its exceptions.
Acknowledgments
We are grateful to Debal Deb, Paul Dixon, Karl Havens, Cleridy Lennert, Neo Martinez, Claudia Pahl-Wostl, Peter Perkins, Gary Polis, Dave Raffaelli, Kenneth Schoenly, Philip Warren, Kirk Winemiller, and the anonymous reviewers for their constructive suggestions. The use of the continuous piecewise linear and exponential asymptotic models was proposed by anonymous reviewers. L.-F.B. was supported by the Swiss National Science Foundation and the Ciba–Geigy Jubilee Foundation, and G.S. was supported by Office of Naval Research Grants N00014-95-1-0034 and N00014-92-J-4068, and by endowment income from the John Dove Isaacs Chair in Natural Philosophy.
References
- 1.Briand F, Cohen J E. Nature (London) 1984;307:264–266. [Google Scholar]
- 2.Cohen J E, Briand F. Proc Natl Acad Sci USA. 1984;81:4105–4109. doi: 10.1073/pnas.81.13.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J E, Newman C M. Proc R Soc London Ser B. 1985;224:421–448. [Google Scholar]
- 4.Briand F, Cohen J E. Science. 1987;238:956–960. doi: 10.1126/science.3672136. [DOI] [PubMed] [Google Scholar]
- 5.Sugihara G, Schoenly K, Trombla A. Science. 1989;245:48–52. doi: 10.1126/science.2740915. [DOI] [PubMed] [Google Scholar]
- 6.Yodzis P. J Theor Biol. 1981;92:103–117. [Google Scholar]
- 7.Sugihara, G. (1982) Ph.D. thesis (Princeton University, Princeton).
- 8.Sugihara G. In: Population Biology: Proceedings of Symposia in Applied Mathematics. Levin S A, editor. Providence, RI: Am. Math. Soc.; 1984. pp. 83–101. [Google Scholar]
- 9.Sugihara G. In: Mathematical Topics in Population Biology, Morphogenesis, and Neurosciences: Proceedings of an International Symposium Held in Kyoto, November 10–15, 1985. Teramoto E, Yamaguchi M, editors. Berlin: Springer; 1987. pp. 53–59. [Google Scholar]
- 10.Pimm S L. Food Webs. London: Chapman & Hall; 1982. [Google Scholar]
- 11.Pimm S L. The Balance of Nature? Chicago: Univ. of Chicago Press; 1991. [Google Scholar]
- 12.Lawton J H, Warren P H. Trends Ecol Evol. 1988;3:242–245. doi: 10.1016/0169-5347(88)90167-X. [DOI] [PubMed] [Google Scholar]
- 13.Cohen J E, Briand F, Newman C N. Community Food Webs: Data and Theory. Berlin: Springer; 1990. [Google Scholar]
- 14.Drake J A. J Theor Biol. 1990;147:213–233. [Google Scholar]
- 15.Paine R T. Ecology. 1988;69:1648–1654. [Google Scholar]
- 16.Polis G A. Am Nat. 1991;138:123–155. [Google Scholar]
- 17.Winemiller K O. Am Nat. 1989;134:960–968. [Google Scholar]
- 18.Warren P H. Am Nat. 1990;136:689–700. [Google Scholar]
- 19.Martinez N D. Ecol Monogr. 1991;61:367–392. [Google Scholar]
- 20.Deb D. Oikos. 1995;72:245–262. [Google Scholar]
- 21.Havens K E. Science. 1992;257:1107–1109. doi: 10.1126/science.257.5073.1107. [DOI] [PubMed] [Google Scholar]
- 22.Martinez N D. Am Nat. 1992;139:1208–1218. [Google Scholar]
- 23.Martinez N D. Am Nat. 1994;144:935–953. [Google Scholar]
- 24.Sugihara G, May R M. Trends Ecol Evol. 1990;5:79–86. doi: 10.1016/0169-5347(90)90235-6. [DOI] [PubMed] [Google Scholar]
- 25.Hastings H, Sugihara G. Fractals: A User’s Guide for the Natural Sciences. Oxford: Oxford Univ. Press; 1993. [Google Scholar]
- 26.Schoenly K, Beaver R A, Heumier T A. Am Nat. 1991;137:597–638. [Google Scholar]
- 27.Sakamoto Y, Ishiguro M, Kitagawa G. Akaike Information Criterion Statistics. Tokyo: KTK Scientific Publishers; 1986. [Google Scholar]
- 28.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice–Hall; 1984. [Google Scholar]
- 29.Pimm S L, Lawton J H, Cohen J E. Nature (London) 1991;350:669–674. [Google Scholar]
- 30.Kenny D, Loehle C. Ecology. 1991;72:1794–1799. [Google Scholar]
- 31.Havens K E. Oikos. 1993;68:117–124. [Google Scholar]
- 32.Cohen J E. Philos Trans R Soc London B. 1994;343:57–69. [Google Scholar]
- 33.Brooks J L, Dodson S I. Science. 1965;150:28–35. doi: 10.1126/science.150.3692.28. [DOI] [PubMed] [Google Scholar]
- 34.Zaret T M. Predation and Freshwater Communities. New Haven, CT: Yale Univ. Press; 1980. [Google Scholar]
- 35.Warren P H. Oikos. 1989;55:299–311. [Google Scholar]
- 36.Hall S J, Raffaelli D. J Anim Ecol. 1991;60:823–842. [Google Scholar]
- 37.Goldwasser L, Roughgarden J. Ecology. 1993;74:1216–1233. [Google Scholar]