Abstract
DNA polymerase β (pol β) is a 39-kDa protein that functions in DNA repair processes in mammalian cells. As a first step toward understanding mechanisms of polymerase fidelity, we developed a genetic method to identify mammalian pol β mutator mutants. This screen takes advantage of a microbial genetics assay and the ability of rat pol β to substitute for Escherichia coli DNA polymerase I in DNA replication in vivo. Using this screen, we identified 13 candidate pol β mutator mutants. Three of the candidate mutator mutants were further characterized in vivo and shown to confer an increased spontaneous mutation frequency over that of wild-type pol β to our bacterial strain. Purification and subsequent analysis of one of our putative mutator proteins, the pol β-14 protein, showed that it possesses intrinsic mutator activity in four different assays that measure the fidelity of DNA synthesis. Therefore, residue 265, which is altered in pol β-14 and another of our mutant proteins, pol β-166, is probably critical for accurate DNA synthesis by pol β. Thus, our genetic method of screening for pol β mutator mutants is useful in identifying active mammalian DNA polymerase mutants that encode enzymes that catalyze DNA synthesis with altered fidelity compared with the wild-type pol β enzyme.
DNA polymerases catalyze the template-directed incorporation of a dNMP into a growing DNA primer and function in essential cellular processes, including DNA replication, DNA repair, and recombination (1). While catalyzing the synthesis of DNA, polymerases commit errors that may become fixed as mutations during subsequent rounds of replication. DNA polymerases are postulated to be a significant source of mutations in cells, some of which may result in tumorigenesis or tumor progression (2). Therefore, it is important to elucidate the mechanisms polymerases employ to synthesize DNA accurately.
Studies of mutational spectra with purified proteins (3–5) and kinetic analysis (6, 7) of wild-type (WT) DNA polymerases reveal that these enzymes actively discriminate between the correct and the incorrect dNTP. Furthermore, different polymerases seem to employ different mechanisms of dNTP discrimination (4, 7). For example, polymerases may discriminate between the correct and the incorrect dNTP at the initial binding step, at a step after initial binding occurs during a conformational change, during chemical formation of the nascent phosphodiester bond, or when the polymerase attempts to add the next dNMP to the primer terminus (7).
Characterization of mutant DNA polymerases indicates that specific amino acid residues of DNA polymerases are critical for accurate DNA replication. Many amino acid side chains of prokaryotic and bacteriophage DNA polymerases, such as Escherichia coli DNA polymerase III and T4, which most likely function in the fidelity of DNA synthesis, have been identified by mutation, through the use of genetic screens and selections (8–10). Because there have been few genetic screens for rapidly identifying mutant mammalian or viral DNA polymerases, little information is available regarding amino acid side chains of mammalian polymerases which affect the fidelity of DNA synthesis. Current methods for isolating functionally significant mammalian DNA polymerase mutants require residues of mammalian polymerases to be altered individually, based upon evolutionary conservation or structural information (11–13). However, many site-directed mutations result in inactive enzymes that cannot be used to probe function (14).
To identify amino acid side chains that function in mammalian DNA polymerase β (pol β) fidelity, we recently developed a genetic system to isolate rat pol β mutator mutants. pol β is a mammalian DNA polymerase that most likely functions in base excision repair (15, 16). Correlation of structure and function of this polymerase is now possible because the crystal structure of this enzyme complexed with DNA and a dideoxycytidine residue was solved recently (17). When produced in E. coli, pol β is an active enzyme which can functionally replace DNA Polymerase I (pol I) in DNA repair, lagging strand replication, and plasmid replication (18–20). This complementation system was used to obtain a large collection of functionally significant pol β mutants that display altered phenotypes in vivo (19, 21). The present study describes a new method for identifying pol β mutants that confer a mutator phenotype to an E. coli bacterial strain; we demonstrate that a mutator mutant isolated using this method encodes an intrinsically error-prone pol β enzyme. The mutant pol β protein carries a single amino acid residue alteration in a region of the pol β structure not predicted a priori to affect polymerase fidelity. Thus, our method permits rapid genetic identification of active mammalian pol β mutants that encode enzymes with altered fidelities of DNA synthesis.
MATERIALS AND METHODS
Bacterial Strains and Media.
Strain SC18–12 is derived from E. coli B/r and has the genotype recA718 polA12 uvrA155 trpE65 lon-11 sulA1 (22). The SC18–12 strain was used in screening of a cDNA library of pol β mutants (19). DH5αMCR has the genotype mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 (lacZYA-argF)U169 deoR recA1 endA1 phoA supE44 thi-1 gyrA96 relA1 and was used in cloning experiments. NR9099, MC1061, and CSH50 F′ are strains used in the M13 fidelity assays and have genotypes as described (23). The strain used to detect mutations in the Herpes simplex virus thymidine kinase gene (HSV-tk) was strain FT334 and has the genotype recA13 upp tdk (24).
ET medium was E salts (25) supplemented with 0.4% glucose and 20 μg/ml Trp. Eglu medium is ET without Trp. Transformants were selected on Luria–Bertani agar (26) supplemented with 30 μg/ml chloramphenicol (Cam) and 12 μg/ml tetracycline. Nutrient broth was prepared according to the manufacturer’s directions (Difco). Minimal agar and soft agar used in M13 assays was as described by Bebenek and Kunkel (23). HSV-tk selection medium is described by Eckert and Drinkwater (24).
Generation of Rat pol β Mutants.
Mutants were generated using two different methods. The first method involved treatment of the rat pol β cDNA (supplied by S. H. Wilson, National Institute of Environmental Health and Safety, Research Triangle Park, NC) with nitrous acid as described (19) and transformation of this library into the SC18–12 strain to yield approximately 10,000 transformants (19).
The second method we employed to create mutants was to amplify the rat pol β cDNA using PCR under mutagenic conditions as described (27). This resulted in mutation of an area of the pol β gene which is hypothesized to be near to the dNTP binding site. These 89 bp of the rat pol β cDNA lie between the single ClaI and SphI sites within the rat pol β cDNA and correspond to amino acid residues 255–285 of the pol β protein (19). To mutate this section, we used the 577+ primer which has the sequence 5′-GCTGGATCCTGAGTACATCGC-3′ and the 1046− primer which has the sequence 5′-CGCTCCGGTCCTTGGGTTC-3′. The PCR conditions were 94°C for 1 min, 60°C for 1 min, 72°C for 1 min for a total of 30 cycles. After the PCR, the ClaI–SphI 89-bp pol β fragment was purified by agarose gel electrophoresis and ligated into the vector pβ-lacZα, which had been digested with ClaI and SphI. The pβ-lacZα vector is pβL (19), carrying the lacZα fragment from the pCRII vector (Invitrogen) in place of the 89-bp ClaI–SphI pol β fragment. This library of pol β mutants was transformed into the DH5αMCR strain by electroporation, and approximately 7000 transformants exhibiting no β-galactosidase activity were pooled. DNA was prepared from this pool and used to transform the SC18–12 strain. These transformants were screened for mutator activity in the Trp+ reversion assay as described below.
Trp+ Reversion Assay to Detect pol β Mutator Mutants.
The 15 pol β mutants that were previously identified in a functional complementation assay, pol β-5 to -20 (21), and the mutants generated by PCR-mediated mutagenesis were analyzed in the Trp+ reversion assay. Individual SC18–12 transformants were picked into 2 ml of Luria–Bertani broth containing 1 mM isopropyl β-d-thiogalactopyranoside, and were incubated in 24-well microtiter plates (Falcon) with aeration for 16–24 hr at 37°C. After centrifugation, the cells were resuspended in saline, and an aliquot of the cells was spread onto Eglu agar. The plates were incubated at 30°C for 3 days, and Trp+ revertants were counted. Transformants which produced 10 or more Trp+ revertants were judged to be candidate mutator mutants. In this assay, the SC18–12 strain carrying the WT pol β cDNA (pol β-WT) usually produces 0–1 Trp+ revertants.
Confirmation of a Mutator Phenotype.
The phenotype of each of the candidate mutators was confirmed as described (21). The spontaneous mutation frequencies were calculated using the method of Vaccaro and Siegel (28).
Purification of the pol β-WT and -14 Proteins.
The rat pol β cDNA was subcloned into pPR977 (New England Biolabs) to generate pMBP-β. This construct carries the pol β cDNA fused at its 5′ end to the maltose binding protein gene with a linker between the two that encodes a thrombin cleavage site. Upon induction of expression with isopropyl β-d-thiogalactopyranoside, a fusion protein of approximately 85 kDa is produced.
pol β protein was purified according to the manufacturer’s directions (New England Biolabs) with the following modifications. Cells containing both pMBP-β and pLysS (Novagen), a plasmid which expresses lysozyme, were grown as described (New England Biolabs) and lysed by one cycle of freezing and thawing. After affinity purification of the fusion protein, the protein was cleaved with thrombin in 50 mM Tris·Cl pH 7.4/5 mM CaCl2/100 mM NaCl/0.5 mM EDTA/0.011 μg of thrombin (Sigma) per μg of fusion protein for 45 min at 21°C in a total volume of 1 ml. After cleavage, the reaction mix was loaded onto a 1-ml single-stranded DNA cellulose (Sigma) column. The maltose binding protein was present in the void volume, and the pol β protein remained bound to the column and was eluted with 10 volumes of column buffer containing 1 M NaCl and 15% glycerol; pol β was present in the first few 1-ml fractions. One liter of cells yielded approximately 1 mg of pol β protein. The amount of protein was quantitated by the method of Bradford (29). The amino-terminal sequence of the 39-kDa band (determined by the Keck Center for Biotechnology at Yale University) was Gly-Ser-Met-Ser-Lys-Arg-Lys-Ala-Pro-Gln-Glu; the Met residue is followed by eight amino acid residues that are identical to the pol β protein, thus identifying the 39-kDa band as pol β. The Gly and Ser residues just before the Met are derived from the maltose binding protein fusion peptide and remain after cleavage with thrombin. The pol β-14 protein was purified exactly as the pol β-WT protein (data not shown). The protein preparations were at least 95% homogeneous based on Coomassie blue staining.
Steady-State Kinetic Measurements.
Standard methods of kinetic analysis were employed, and linearity was observed as a function of time for initial rate determinations (30, 31). Activated DNA was prepared by the method of Spanos and coworkers (32). The concentration of WT enzyme used was 10 nM, and the concentration of pol β-14 enzyme was 106 nM. Km and kcat parameters were derived from Hanes–Woolf plots using the enzyme kinetics computer program (Trinity Software). For measurements made with activated DNA, each value was obtained from at least two independent experiments and is reported with a standard error. For measurements made with the substrate employed in the kinetic fidelity assay, data are presented as the averages of at least four independent experiments.
M13mp2-Based Reversion Assays.
The fidelity of pol β-WT and -14 was measured using two M13mp2-derived templates, each of which contain a 390-bp gap opposite the lacZα gene and were constructed as described (23). All bacteriophages and their corresponding strains were obtained from Thomas Kunkel (National Institute of Environmental Health and Safety, Research Triangle Park, NC). For gap-filling DNA synthesis, 0.1 pmol of gapped DNA was incubated with 20 pmol of rat pol β-WT or 200 pmol of pol β-14 in 30 μl of buffer containing 50 mM Tris·Cl (pH 8.4), 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 200 μg/ml BSA, 500 μM dNTPs, and [α-32P]dTTP (800 Ci/mmol; 1 Ci = 37 GBq) at 37°C for 1 hr at 37°C, and the reactions were terminated by the addition of EDTA to a final concentration of 15 mM. An aliquot of each reaction was analyzed on a 0.8% agarose gel containing ethidium bromide to be certain that the gap was filled completely (23). In each case, the reaction products comigrated with a double-stranded nicked molecular size standard, indicating that the gap had been filled to completion within our limits of detection (≥90%). Aliquots of each reaction were also transfected into the MC1061 strain, and the polymerase-induced mutation frequency was calculated from the ratios of mutant (blue) and nonmutant (colorless) plaques on CHS50 F′ indicator E. coli as described (23).
HSV-tk Forward Mutation Assay.
This assay was used to assess all possible errors committed by each polymerase and can detect large deletion or addition errors, multiple mutations, and complex mutations in addition to single-base substitution and frameshift mutations within a variety of sequence contexts. The assay focuses on polymerase-mediated errors in the 5′ region of the HSV-tk gene target, which contains the sequence encoding the 32-aa ATP-binding site of the thymidine kinase enzyme. DNA templates were created by hybridization of single-stranded HSV-tk containing DNA to a 20-mer oligonucleotide at a 1:1 molar ratio, such that DNA synthesis is initiated 34 nt downstream of the ATP-binding site. The polymerase reactions contained 2 pmol of template DNA, 50 mM Tris·Cl (pH 8.5), 50 mM NaCl, 1 mM DTT, 1 mM dNTPs, 200 μg/ml BSA, and either 20 pmol of pol β-WT or 300 pmol of Pol β-14 enzyme. All reactions were incubated at 37°C for 1 hr and terminated by the addition of EDTA to 15 mM. The extent of DNA synthesis was determined in a parallel reaction, supplemented with 5 μCi of [α-32P]dCTP (3000 Ci/mmol). Reaction products were separated on an 8% denaturing polyacrylamide gel, and autoradiography was used to determine if DNA synthesis had proceeded past the 5′ end of the mutational target. The DNA reaction products were digested with MluI and EcoRV restriction enzymes, and 203-bp DNA fragments were purified and analyzed for HSV-tk-inactivating mutations (unpublished work). Briefly, the fragments were hybridized to a gapped duplex molecule, and the resulting heteroduplex plasmid molecules were used to transform recA upp tdk E. coli strain FT334 (24). Selection of the bacteria with 50 μg/ml Cam ensures exclusive analysis of mutations derived from the DNA strand produced during in vitro synthesis. HSV-tk mutant plasmids are selected by plating the bacteria in the presence of 5′-fluoro-2′-deoxyuridine. The resulting HSV-tk mutant frequency is calculated as the number of colonies resistant to both 5′-fluoro-2′-deoxyuridine and Cam divided by the total number of Cam-resistant colonies.
Kinetic Gel Fidelity Assay.
The mispair formation efficiencies of purified pol β proteins in vitro were quantified kinetically using a gel fidelity assay (33, 34). 5′-32P-end-labeled 20-mer (35) and nonlabeled 46-mer oligodeoxyribonucleotides (HPLC-purified from Operon Technologies, Alameda, CA) were hybridized under conditions yielding >90% DNA•DNA duplex (36) to form the primer•template:
[5′-32P]GCAGGAAAGCGAGGGTATCC
GTCCTCGTCCTTTCGCTCCCATAGGGTGTTTCAGGT-
CGCATGGTAT-5′
The efficiencies of incorporation of each dNMP opposite template G21 (underlined) were determined in polymerization reactions (10 or 20 μl) containing pol β-WT or pol β-14, 20 nM primer•template, and a single dNTP at 0–2500 μM in 50 mM Tris•HCl, pH 8.0 (22°C)/10 mM MgCl2/2 mM DTT/20 mM NaCl/20 mM KCl/2.5% glycerol/0.2 mg/ml BSA. After preincubation of reaction mixtures for 5 min at 37°C in the absence of dNTPs, polymerizations were initiated by the addition of a single dNTP, and incubations were continued for an additional 3–15 min until termination by mixing with an equal volume of 0.5 M EDTA. Primers (20-mers) and extension products (≥21-mers) were separated by PAGE (8 M urea/16% polyacrylamide gel) and then quantified using a phosphorimager and imagequant software (Molecular Dynamics). Vmax/Km values for correct (dCTP) and incorrect (dATP, dGTP, and dTTP) nucleotides were determined from the initial slopes of Michaelis–Menten plots (33), and the frequencies of nucleotide misincorporation (f) were calculated as described (33, 34). Enzyme concentrations and incubation times were adjusted to optimize detection of misincorporation products, while ensuring that all reactions were conducted in the steady-state within the linear range of substrate utilization (<25% primer extension). Relatively high concentrations of pol β-14 were required (6 and 60 nM for correct and incorrect dNTPs, respectively) compared with pol β-WT (0.5 and 5 nM, respectively) due to the lower specific activity of the mutant protein. As expected in steady-state, Vmax values were directly proportional to enzyme concentration; appropriate adjustments were made in the calculations for misincorporation frequencies.
RESULTS
Screen to Identify pol β Mutator Mutants in E. coli.
We developed a genetic screen to identify mammalian pol β mutator mutants. Our screen is based upon the fact that pol β is able to substitute for E. coli pol I in DNA replication and repair in the SC18–12 strain (18). Briefly, pol β increases the rate of joining of Okazaki fragments on the lagging strand during DNA replication, and it confers methylmethane sulfonate resistance to this otherwise methylmethane sulfonate-sensitive strain, suggesting that it also substitutes for pol I in base excision repair. By using this genetic screen we identified 13 candidate pol β mutator mutants. Three of the mutator mutants were characterized further in vivo and were shown to confer a mutator phenotype to the SC18–12 strain. One mutant protein, pol β-14, was purified to relative homogeneity and was shown to possess intrinsic mutator activity in vitro.
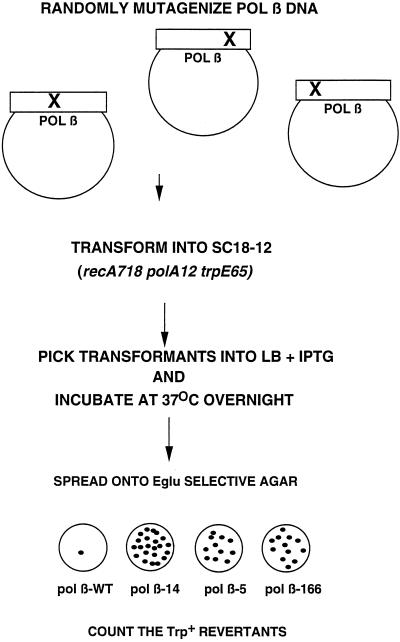
To identify putative pol β mutator mutants, we made use of the fact that the SC18–12 strain possesses a chromosomal mutation of the trpE gene, referred to as the trpE65 allele. The trpE65 allele contains an ochre mutation at an unidentified location within the trpE structural gene (37), resulting in the inability of the SC18–12 strain to synthesize anthranilate synthetase, an enzyme needed by the cell to produce Trp. Therefore, this strain must be maintained on medium containing a source of Trp. A detectable Trp+ phenotype can be produced by five out of six possible base substitution errors which may occur by either direct reversion at the site of the TAA nonsense codon or at the anticodon loop of one of three tRNAs to generate a suppressor mutation, resulting in insertion of either Ser, Tyr, or Gln across from the nonsense codon during protein synthesis (38). We developed a screen which scores for the number of Trp+ revertants produced by a single transformant, as diagrammed in Fig. 1. By employing this screen, we identified 13 pol β transformants that yielded increased numbers of Trp+ revertants. We detected mutants which produced between 10 and 60 Trp+ revertants in the screen. Two of these candidate mutators were identified in a previous screen as mutants which were partially able to substitute for E. coli pol I in DNA repair (21), and 11 of the candidate mutator mutants were from the pool of 800 transformants generated by PCR-mediated mutagenesis.
Figure 1.
Protocol to detect pol β mutator mutants carrying a randomly mutagenized pol β cDNA are picked into Luria–Bertani broth containing 1 mM isopropyl β-d-thiogalactopyranoside and grown in 24-well microtiter plates overnight at 37°C. Aliquots of each well are spread onto Eglu agar and incubated for 3 days at 30°C. In this assay the pol β-WT strain produces 0–1 Trp+ revertants and mutator mutants such as pol β-14, -5, and -166 produce greater than 10 Trp+ revertants.
The pol β Mutants Confer a Mutator Phenotype to the SC18–12 E. coli Strain.
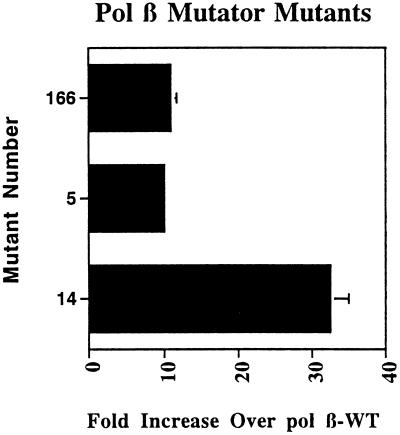
To confirm that the candidate mutants we identified in the screen possessed true mutator phenotypes, we compared the spontaneous mutation frequencies of three of these mutants with that of the pol β-WT strain. As shown in Fig. 2, the pol β-166 and -5 mutants have spontaneous mutation frequencies approximately 10-fold higher than the pol β-WT strain, whereas the pol β-14 mutant has a spontaneous mutation frequency that is 30-fold higher than the pol β-WT strain (21). Analysis of the types of Trp+ mutations resulting from the pol β-5 and -14 mutants, by cross-streaking with T4 phages possessing nonsense mutations in essential genes (38) shows that pol β-5, -14, and -WT produce nearly equal numbers of true revertants and suppressor tRNA mutants. Among the tRNA mutants, our data show that the pol β-14 and -5 mutants produce predominantly transversion mutations in tRNA genes while substituting for pol I of E. coli. Twenty-eight of 35 suppressor mutations generated by pol β-14 were GC to TA transversions, and 51 of 58 suppressor mutations produced by pol β-5 were GC to TA transversions, whereas 26 of 27 suppressor mutations generated by pol β-WT were GC to AT transition mutations. The pol β-14 mutation alters Tyr-265 of the protein to Cys, and the pol β-5 mutation changes Pro-312 to Ser (21). To identify the mutation present within the pol β-166 cDNA, we determined its DNA sequence as described (21) and found that it was also mutated at position 265, resulting in a Tyr-265 → His amino acid change. Both the Pro-312 and Tyr-265 amino acid residues are located in the carboxyl terminus of the pol β protein, exterior to the DNA binding cleft. Preliminary analysis of the 10 remaining mutants indicates that they are also mutators and characterization of these mutants is ongoing (unpublished data). Each of these mutants contains a single mutation resulting in an amino acid alteration different from those of the pol β-14, -166, and -5 proteins.
Figure 2.
pol β mutator mutants. The -fold increase over pol β-WT represents the spontaneous mutation frequency of the mutant divided by that of the pol β-WT. Each bar represents data averaged from three independent experiments. The mutant number refers to the pol β mutant cDNA which is present on the pβL vector and contained within the SC18–12 strain. Error bars represent standard deviations.
pol β-14 Has Intrinsic Mutator Activity in Vitro.
The in vivo data demonstrate that the pol β-5, -14, and -166 mutants confer a mutator phenotype to the SC18–12 E. coli strain. One interpretation of these data is that the mutator phenotypes are caused in a direct manner, by a mutator polymerase committing errors as it substitutes for pol I in DNA replication. To test this hypothesis, we purified recombinant pol β-WT and -14 proteins (data not shown) and compared their fidelities of DNA synthesis in four separate in vitro assays; the results are shown in Table 1.
Table 1.
Fidelities of purified pol β-WT and pol β-14 in vitro
| Type of error | Assay | Error frequency, ×10−4
|
Increase, -fold | |
|---|---|---|---|---|
| pol β-WT | pol β-14 | |||
| Base substitution | TGA reversion | 1.9 ± 0.10 | 87 ± 19 | 46 |
| G:N kinetics | 9.8 ± 6.0 | 830 ± 460 | 85 | |
| Frameshift | TTTTT reversion | 1.9 ± 0.7 | 470 ± 20 | 240 |
| All | tk loss of function | 21 ± 0.5 | 540 ± 0.7 | 25 |
The TGA target is in a modified version of the lacZα gene and refers to the opal codon reversion assay which detects base substitution mutations. The TTTTT target is in a modified version of the lacZα gene and refers to the frameshift reversion assay which detects predominantly frameshift mutations. For M13mp2-derived assays with pol β-WT, at least 85,000 plaques were scored; with pol β-14, at least 15,000 plaques were scored. At least 30,000 colonies were analyzed for each HSV-tk experiment. Data represent the averages of two independent experiments and are presented with standard errors. The values for the kinetic assay are the average nucleotide misinsertion frequencies (f) of the three G:N mispairs examined. Initial evaluation of the polymerase activities of these recombinant proteins on activated calf thymus DNA showed that the Km (dTTP) for pol β-14, 1.9 ± 0.3 μM, is quite similar to that of pol β-WT (1.2 ± 0.2 μM). In contrast, the mutant protein exhibited a Km (activated DNA) of 80 ± 6.5 μM and kcat of 0.0015 sec−1, which are significantly lower than those of the pol β-WT protein (362 ± 15 μM and 0.03 sec−1, respectively). Our recombinant pol β-WT protein and an independent preparation lacking the extra amino acids at the amino terminus (supplied by Akio Matsukage, Aichi Cancer Center Research Institute, Nagoya, Japan) yielded essentially identical values in these kinetic assays (data not shown). Initial evaluation of polymerase activities on the DNA substrate employed in the kinetic fidelity assay showed similar trends as that observed with activated DNA. The Km (app) was 190 ± 45 μM for pol β-WT and 83 ± 27 μM for pol β-14. The kcat for WT using this DNA substrate was 2.6 sec−1, and for pol β-14 in the kcat was 0.055 sec−1.
First, we used an opal codon reversion assay to detect eight possible base substitution mutations that can be committed by a polymerase at three adjacent template positions (39). In this assay, the pol β-14 protein committed errors with a frequency about 50 times higher than that of the WT protein.
Second, a kinetic assay (33, 34) was used to quantify the relative rates of nucleotide misincorporation catalyzed by the pol β-14 and -WT proteins opposite a single template G residue in a synthetic 5′-32P-end-labeled 20-mer primer•46-mer template. As shown in Table 1, the pol β-14 protein was on average 85 times more error-prone than the pol β-WT protein for the three G:N mispairs examined at this template site. The increase in misincorporation rate by pol β-14 was similar for G:A (76 times), G:G (86 times), and G:T (100 times; data not shown). Thus, the increased base substitution error rate of pol β-14 is due, at least in part, to increased insertion of the wrong nucleotide.
Third, to quantify frameshift fidelity, we employed a reversion assay that detects minus-one base pair frameshifts at a TTTTT sequence or at 36 other sites located elsewhere in the lacZα gene (40). We obtained a spontaneous mutation frequency of 470 × 10−4 for the pol β-14 protein in this frameshift reversion assay, which is 240-fold higher than that of the WT enzyme, as shown in Table 1. This demonstrates that the pol β-14 protein commits frameshift errors at a much higher frequency than the WT protein.
Finally, we used a forward mutation assay to assess all possible errors committed by each protein (unpublished work). The HSV-tk assay detects large addition or deletion errors and multiple mutations, in addition to frameshift and base substitution mutations. In this forward mutation assay, the pol β-14 protein commits errors at a frequency of 540 × 10−4, which is approximately 25-fold more frequent than errors committed by WT protein. Together, these in vitro fidelity assays show that pol β-14 catalyzes the synthesis of DNA with lower fidelity than the WT enzyme.
DISCUSSION
A Genetic Screen to Identify DNA pol β Mutator Mutants.
We have designed a new screen to identify mammalian pol β mutator mutants. This screen is based upon our original discovery that rat pol β is able to substitute for E. coli pol I in DNA replication and repair in the SC18–12 E. coli strain. The SC18–12 strain carries the trpE65 allele, rendering this strain incapable of growth in the absence of Trp. Trp+ revertants result from base substitution mutations at either the ochre codon within the trpE gene or at one of four tRNA anticodon loop sequences (38). We reasoned that a pol β mutator enzyme which substituted for pol I in lagging strand replication in the SC18–12 strain would commit errors, resulting in a Trp+ phenotype, and allow us to identify E. coli strains carrying such mutators. Using this assay, we identified 13 candidate pol β mutator mutants; these mutators produced an increased number of Trp+ revertants compared with SC18–12 E. coli harboring the pol β-WT cDNA.
We confirmed that three of our mutants, the pol β-14, -5, and -166 mutants, confer an increased spontaneous mutation frequency over that of pol β-WT to the SC18–12 strain. In addition, we demonstrated that one of these, the pol β-14 mutant, encodes an intrinsically error-prone polymerase as determined in four different assays which measure the accuracy of DNA synthesis in vitro. Therefore, we conclude that our screen is able to identify mutants which encode pol β enzymes that catalyze the synthesis of DNA with less accuracy than the wild-type protein.
Tyr-265 Is Important for pol β Fidelity.
Amino acid residue Tyr-265 of the pol β protein was altered in two of our mutator mutants. The pol β-166 protein contains His at position 265, whereas the pol β-14 protein has Cys at this position. The fact that position 265 is found to be altered in two mutator mutants out of a total 13 identified suggests that residue 265 plays an important role in fidelity. Amino acid residue 265 is part of the carboxyl terminus of the pol β protein. Some amino acid residues of this domain directly interact with the DNA template strand or with the dNTP substrate; this is not the case for Tyr-265. In the crystal structure of pol β, residue 265 is located in helix M in a small hydrophobic pocket exterior to the DNA binding cleft, a position that suggests that Tyr-265 could participate in intramolecular interactions which maintain the overall structure of the pol β protein (17). Alteration of residue 265 might disrupt the structure of the pol β enzyme in the vicinity of helix M, causing a change in the overall geometry of the active site of the enzyme, and result in a polymerase which synthesizes DNA less accurately than the WT protein. Alternatively, Tyr-265 may interact with other residues of pol β to facilitate a conformational change during the catalytic cycle; conformational changes of this nature have been hypothesized to play a role in DNA polymerase fidelity (7). This hypothesis is substantiated by recent evidence suggesting the existence of a hinge region within the pol β enzyme which is lined with hydrophobic residues, including Tyr-265 (41). Pelletier et al. proposed that this hinge region is responsible for movement of the carboxy terminus from an open conformation to a closed conformation which might represent a rate-limiting step in replication fidelity (41). Further biochemical and biophysical characterization of the pol β-5, -14, and -166 proteins is necessary to understand the contribution of Tyr-265 and Pro-312 to pol β fidelity.
Benefits of a Genetic Screen for Mammalian pol β Mutator Mutants.
Molecular structure–function studies combined with kinetic analysis of mutant proteins have the potential to yield specific information concerning the molecular mechanism of accurate DNA polymerization. The purpose of these studies is to identify amino acid residues of a DNA polymerase that affect the fidelity of DNA synthesis and to determine their precise function.
Most structure–function studies of DNA polymerases have focused on identifying residues of these enzymes that are necessary for catalysis. The major strategy employed is to alter single amino acid residues of the protein using site-directed mutagenesis and to determine the resultant effects on catalysis. These methods are used to change amino acid residues believed to be located near to the active site of the enzyme or which are thought to contact DNA or dNTP based upon hypotheses generated from the enzyme’s crystal structure. Because a three-dimensional structure is not available for the majority of mammalian polymerases, amino acid residues conserved among a family of polymerases are chosen for alteration. The resulting mutant enzymes are then purified and studied in vitro to determine if they are active proteins with intrinsic mutator activity. These strategies have been successful in some cases (13, 42, 43). However, the primary goal of the majority of polymerase structure–function studies is to identify catalytic residues of the protein. Catalytic residues might not necessarily function in polymerase accuracy (30). Moreover, mutation of a polymerase residue which functions in catalysis is likely to result in an inactive protein, making it difficult to characterize these mutant proteins in vitro (12, 14, 30, 44).
Another strategy to reveal amino acid residues of a DNA polymerase which affect fidelity is to employ a genetic screen. Genetic screens and selections have been used with good results to identify mutator and antimutator mutants of several DNA polymerases, including T4 and DNA polymerase III of E. coli (8–10). Our screen designed to detect pol β mutator mutants has three advantages over traditional methods of isolation and characterization of mammalian polymerase mutator proteins. First, this screen has the potential to identify large numbers of polymerase mutator mutants rapidly. Our initial results with pol β suggest that pol β mutator mutants are rare; in this study, they appeared at a frequency of about 1 out of 100 screened, not taking into account the frequency of false positives. Thus, the ability to screen large numbers of pol β mutants is important to identify each amino acid residue of the protein that affects fidelity. Second, this screen facilitates mutagenesis studies that examine relatively large domains of the protein without a priori assumptions about the role of individual amino acids in fidelity. This is important because alteration of Tyr-265 would not have been predicted to result in an enzyme with mutator activity on the basis of homology with other polymerases or by an examination of the structure of the pol β ternary complex. Therefore, our genetic screen complements the site-directed mutagenesis approach. Third, our screen identifies pol β mutants that produce catalytically active proteins which commit errors at higher frequencies than pol β-WT. Because the screen is performed under conditions that preclude growth of the SC18–12 strain in the absence of functional pol β protein, mutants that are isolated must encode an active pol β protein. This greatly facilitates subsequent biochemical and biophysical studies of fidelity mechanisms. Therefore, we envision that a genetic screen will be a very useful tool in the identification of pol β mutants for further study.
Acknowledgments
This paper is dedicated to Dr. Lawrence A. Loeb on the occasion of his 60th birthday. We thank Dr. Thomas A. Kunkel for the M13mp2 constructs, Dr. Akio Matsukage for his gift of rat pol β protein, Dr. Samuel Wilson for the rat pol β cDNA clone, and Dr. Richard Bockrath for T4 nonsense phage mutants and advice. We also thank Jessica Kosa for discussions which focused our thoughts concerning the structure of the pol β enzyme. This research was supported by American Cancer Society Research Investigation Grant NP-930 (to J.B.S.). J.B.S. is also a recipient of an American Cancer Society Junior Faculty Research Award.
Footnotes
Abbreviations: WT, wild type; pol β, DNA polymerase β; pol I, DNA polymerase I; pol β-WT, WT pol β cDNA.
References
- 1.Kornberg A, Baker T A. DNA Replication. 2nd Ed. New York: Freeman; 1992. pp. 113–164. [Google Scholar]
- 2.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 3.Loeb L A, Kunkel T A. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel T A, Bebenek K. Biochim Biophys Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 5.Schaaper R M. Genetics. 1993;134:1031–1038. doi: 10.1093/genetics/134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echols H, Goodman M F. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 7.Johnson K A. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 8.Hall R M, Brammar W J. Mol Gen Genet. 1973;121:271–276. [Google Scholar]
- 9.Oller A R, Fijalkowska I J, Schaaper R M. Mutat Res. 1993;292:175–185. doi: 10.1016/0165-1161(93)90145-p. [DOI] [PubMed] [Google Scholar]
- 10.Reha-Krantz L J. J Mol Biol. 1988;202:711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- 11.Copeland W C, Dong Q, Wang T S. Methods Enzymol. 1995;262:294–303. doi: 10.1016/0076-6879(95)62025-7. [DOI] [PubMed] [Google Scholar]
- 12.Menge K L, Hostomsky Z, Nodes B R, Hudson G O, Rahmati S, Moomaw E W, Almassy R J, Hostomska Z. Biochemistry. 1995;34:15934–15942. doi: 10.1021/bi00049a008. [DOI] [PubMed] [Google Scholar]
- 13.Beard W A, Osheroff W P, Prasad R, Sawaya M R, Jaju M, Wood T G, Kraut J, Kunkel T A, Wilson S H. J Biol Chem. 1996;271:12141–12144. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 14.Date T, Yamamoto S, Tanihara K, Nishimoto Y, Matsukage A. Biochemistry. 1991;30:5286–5292. doi: 10.1021/bi00235a023. [DOI] [PubMed] [Google Scholar]
- 15.Clairmont C, Sweasy J B. J Bacteriol. 1996;178:656–661. doi: 10.1128/jb.178.3.656-661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobol R W, Horton J K, Kuhn R, Gu H, Singhal R K, Prasad R, Rajewsky K, Wilson S H. Nature (London) 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 17.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 18.Sweasy J B, Loeb L A. J Biol Chem. 1991;267:1407–1410. [PubMed] [Google Scholar]
- 19.Sweasy J B, Loeb L A. Proc Natl Acad Sci USA. 1993;90:4626–4630. doi: 10.1073/pnas.90.10.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweasy J B, Chen M Z, Loeb L A. J Bacteriol. 1995;177:2923–2925. doi: 10.1128/jb.177.10.2923-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweasy J B, Yoon M S. Mol Gen Genet. 1995;248:217–224. doi: 10.1007/BF02190803. [DOI] [PubMed] [Google Scholar]
- 22.Witkin E M, Roegner-Maniscalco V. J Bacteriol. 1992;174:4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bebenek K, Kunkel T A. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 24.Eckert K A, Drinkwater N R. Mutat Res. 1987;178:1–10. doi: 10.1016/0027-5107(87)90079-0. [DOI] [PubMed] [Google Scholar]
- 25.Vogel H L, Bonner D M. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 26.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 27.Rice G C, Goeddel D V, Cachianes G, Woronicz J, Chen E Y, Williams S R, Leung D W. Proc Natl Acad Sci USA. 1992;89:5467–5471. doi: 10.1073/pnas.89.12.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaccaro K, Siegel E C. Mol Gen Genet. 1975;141:251–262. doi: 10.1007/BF00341803. [DOI] [PubMed] [Google Scholar]
- 29.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Polesky A, Steitz T A, Grindley N D F, Joyce C A. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 31.Tanababe K, Bohn E W, Wilson S H. Biochemistry. 1979;18:3401–3406. doi: 10.1021/bi00582a029. [DOI] [PubMed] [Google Scholar]
- 32.Spanos A, Sedgwick S G, Yarronton G T, Hubscher U, Banks G R. Nucleic Acids Res. 1981;9:1825–1840. doi: 10.1093/nar/9.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boosalis M S, Mosbaugh D W, Hamatake R, Sugino A, Kunkel T A, Goodman M F. J Biol Chem. 1989;264:11360–11366. [PubMed] [Google Scholar]
- 34.Creighton S, Bloom L B, Goodman M F. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 36.Ji X, Klarmann G J, Preston B D. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 37.Witkin E M. Genetics. 1963;48:916. [Google Scholar]
- 38.Bockrath R, Mosbaugh P. Mol Gen Genet. 1986;204:457–462. doi: 10.1007/BF00331024. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel T A, Soni A. J Biol Chem. 1988;263:4450–4459. [PubMed] [Google Scholar]
- 40.Bebenek K, Joyce C M, Fitzgerald M P, Kunkel T A. J Biol Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 41.Pelletier H, Sawaya M R, Wolfe W, Wilson S H, Kraut J. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 42.Copeland W C, Lam N K, Wang T S. J Biol Chem. 1993;268:11041–11049. [PubMed] [Google Scholar]
- 43.Dong Q, Copeland W C, Wang T S. J Biol Chem. 1993;268:24163–24164. [PubMed] [Google Scholar]
- 44.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]