Abstract
The dimer of bovine pancreatic ribonuclease A (RNase A) discovered by Crestfield, Stein, and Moore in 1962 has been crystallized and its structure determined and refined to a 2.1-Å resolution. The dimer is 3D domain-swapped. The N-terminal helix (residues 1–15) of each subunit is swapped into the major domain (residues 23–124) of the other subunit. The dimer of bull seminal ribonuclease (BS-RNase) is also known to be domain-swapped, but the relationship of the subunits within the two dimers is strikingly different. In the RNase A dimer, the 3-stranded beta sheets of the two subunits are hydrogen-bonded at their edges to form a continuous 6-stranded sheet across the dimer interface; in the BS-RNase dimer, it is instead the two helices that abut. Whereas the BS-RNase dimer has 2-fold molecular symmetry, the two subunits of the RNase A dimer are related by a rotation of ∼160°. Taken together, these structures show that intersubunit adhesion comes mainly from the swapped helical domain binding to the other subunit in the “closed interface” but that the overall architecture of the domain-swapped oligomer depends on the interactions in the second type of interface, the “open interface.” The RNase A dimer crystals take up the dye Congo Red, but the structure of a Congo Red-stained crystal reveals no bound dye molecule. Dimer formation is inhibited by excess amounts of the swapped helical domain. The possible implications for amyloid formation are discussed.
Keywords: protein structure, x-ray crystallography, protein–protein interactions, amyloid
Bovine pancreatic ribonuclease A (RNase A) is a 124-residue enzyme that catalyzes the hydrolysis of RNA with specificity for pyrimidines at the 3′-side of the cleavage site. Studies of RNase A have made great contributions to our understanding of protein structure and function, including early studies of the spontaneous renaturation of proteins (1), amino acid sequence determination (2), synthesis of proteins (3), protein three-dimensional structure (4, 5), and peptide–protein interactions (6). Here, we pursue another aspect of RNase A: its formation of oligomers by 3D domain swapping.
In 1962, Crestfield et al. (7) reported that RNase A forms dimers and higher oligomers after dialysis against 50% acetic acid and lyophilization. The dimer remains active with the same specific activity as the monomer. From studies of chemical modification of active site residues, Crestfield et al. proposed that RNase A forms a dimer by exchanging its N-terminal helix (7), a phenomenon now called 3D domain swapping (8).
In a 3D domain-swapped dimer, two subunits exchange identical “domains.” That is, a structural unit of one subunit takes the place of the identical structural unit of the other subunit and vice versa. The term 3D domain swapping was coined to describe a dimer of diphtheria toxin (8). Since then, more than 10 crystal structures of 3D domain-swapped proteins have been reported (9), including a trimer (10). The interface between domains found both in the monomer and in the domain-swapped oligomer is termed the “closed interface,” and the interface found only in the oligomer is termed the “open interface” (9, 11). These two types of interfaces are illustrated in Fig. 2 later in this paper. 3D domain swapping can endow oligomers with additional properties, e.g., metal binding in the case of CksHs2, a cell cycle regulatory protein in yeast (12). In addition, domain swapping is a possible mechanism for the formation of protein aggregates, which can be favored by mutational and environmental changes (9, 11). The amyloid-like fibrils formed by intact Ig light chains have been suggested by Klafki et al. (13) to form by a mechanism that is essentially domain swapping. Studies of oligomers of the well characterized protein RNase A may therefore illuminate the formation of amyloid and other protein aggregates.
Figure 2.
The structures of three RNase molecules, with red domains all in the same orientation. The hinge loops (residues 16–22) are shown in green. (A) The RNase A monomer (Protein Data Bank code: 1RTB) (28). The helix to be swapped (residues 1–15) is shown in blue; the major domain (residues 23–124) is shown in red. The “closed interface” is that between the blue helix and the red major domain. (B) The BS-RNase domain-swapped dimer (Protein Data Bank code: 1BSR) (2). The two intersubunit disulfide bonds are shown in yellow. The “open interface” here is between the adjacent red and blue helices. (C) The RNase A dimer. Subunit 1 (blue, residues 1–124) and subunit 2 (red, residues 201–324) are related by a ∼160-degree rotation about an axis roughly perpendicular to the page. The hinge loops are in the same orientation as those in Fig. 1. Notice the 6-stranded beta-sheet formed from three strands of each subunit. The open interface here is between the adjacent red and blue strands of the beta-sheet. Notice also that the open interfaces in B and C differ but that the closed interfaces in A, B, and C are the same. Functional unit 1 (see Table 3) consists of the blue swapped helix and the red major domain. The diagrams are made with the program molscript (37).
Bull seminal (BS)-RNase is a 124-residue ribonuclease from bovine seminal plasma that shares 81% sequence identity with RNase A. The structure of BS-RNase shows it is a domain-swapped dimer (14). Domain swapping endows this dimer with additional biological properties, such as allostery (15), and antitumor and immunorepression activity (16, 17), functions not possessed by the monomer and the non-domain-swapped dimer of BS-RNase. The mechanism for these acquired biological properties remains under study (18). Solution and computational studies of BS-RNase and RNase A suggest that the hinge loop (residues 16–22) connecting the swapped domain (residues 1–15) and the major domain (residues 23–124) permits the domain exchange, perhaps with Pro-19 in BS-RNase being important (19–21). The structure of the RNase A monomer was first determined to 2 Å by Kartha et al. in 1967 (4) and later refined to 1.26 Å by Wlodawer et al. (22). Also, the structure of RNase S, the RNase A cleaved by subtilisin, was determined by Wyckoff et al. (5). The structure of the RNase A dimer, however, has gone undetermined until now.
Our rationale for studying binding of Congo Red to the RNase A dimer is the possible relationship of domain swapping to amyloid formation, mentioned above. Congo Red is a clinical probe for amyloid; when it binds to amyloid a green birefringence is given off under polarized light (23). Several models of Congo Red binding to amyloid have been proposed (24, 25), but further structural evidence is needed to understand this interaction. We observed that Congo Red stains crystals of the RNase A dimer, which led us to examine the crystal structure of a potential RNase A–Congo Red complex.
There exists a subtilisin cleavage site between residues 20 and 21 in RNase A. After cleavage, the N-terminal segment, the S-peptide, remains associated with the rest of the protein, the S-protein (6). Low pH is required to separate the S-peptide from the S-protein. Because the swapped helices in the RNase A dimer are contained within the S-peptide, exogenous S-peptide may compete with the formation of the dimer. Studies of this competition may thus illuminate the mechanism of domain swapping.
Our structural study of the RNase A dimer was undertaken to see if it is domain-swapped, and if so, to compare its mode of 3D domain swapping with the BS-RNase dimer. Other goals included studying the mechanism of binding of Congo Red to amyloids and the formation of protein aggregates.
METHODS
Purification of the RNase A Dimer.
The RNase A dimer was formed and isolated as reported by Crestfield et al. (7). RNase A was purchased from Sigma (RNase A III-A). The powder was dissolved in 50% acetic acid at 20 mg/ml and was kept at 4°C overnight. The sample was lyophilized, redissolved in 10 mM phosphate buffer (pH 6.5), 0.1 M sodium sulfate, and loaded on an HPLC size exclusion column. The yield of the dimer was about 20%. The dimer was concentrated using Centriprep concentrators with a Mr cutoff of 10,000 and was adjusted to 15 mg/ml. Some trimer and possibly higher order oligomers were suggested by HPLC chromatography.
Crystallization and Data Collection.
Crystals of the RNase A dimer were grown using the hanging-drop vapor diffusion method. Dimer at a concentration of 15 mg/ml in 10 mM phosphate buffer (pH 6.5), 0.1 M sodium sulfate, and 10 mM ADP was mixed with an equal volume of reservoir solution containing 100 mM phosphate buffer (pH 7.5), 1.6 M ammonium sulfate, 0.6 M NaCl, and 2% PEG 400 and was equilibrated against the reservoir solution at room temperature in 4–6-μl drops. Needle-like microcrystals appeared in 1–2 weeks, and x-ray quality crystals of 0.1 × 0.1 × 0.5 mm3 were obtained by microseeding.
The crystals were transferred to droplets containing the reservoir solution without NaCl, in which the crystals were stable. The 1% Congo Red (Sigma) solution was added to the droplets to a final concentration of 0.1%. After 1 day, the crystals developed a deep red color, showing that the dye is more highly concentrated in the crystals than in the solution. These crystals were then used to collect x-ray diffraction data for the potential Congo Red–RNase A complex.
Initially, cryogenic x-ray diffraction data to a 2.8-Å resolution were collected with a Rigaku R-AXIS IV imaging plate detector. Data to a 2.1-Å resolution were collected with an R-AXIS II imaging plate detector at room temperature with a larger crystal (0.1 × 0.1 × 0.5 mm3). The x-ray source was a Rigaku RU-200 generator running at 50 kV, 100 mA with focusing mirrors. Crystals were rotated about φ, and oscillation images were collected every 1.5°. The data were processed and reduced using the programs denzo and scalepack (26). The statistics of data collection are listed in Table 1.
Table 1.
Statistics of x-ray data collection for crystals of the RNase A dimer
| RNase A dimer | RNase A dimer | Dimer + Congo Red | |
|---|---|---|---|
| Cryogenic protectant | — | 25% glycerol | 25% glycerol |
| Temperature | Room temperature | Cryo | Cryo |
| Wavelength, Å | 1.54 | 1.54 | 1.54 |
| Resolution range, Å | 40–2.1 | 40–2.8 | 40–2.5 |
| Total reflections | 30,428 | 14,830 | 20,706 |
| Unique reflections | 16,601 | 6,766 | 9,588 |
| Completeness, % | 96.0 | 95.9 | 97.7 |
| Rsym*, % | 9.7 | 10.1 | 8.7 |
| Space group | P32 | P32 | P32 |
| Unit cell constants | a = b = 57.0 Å | a = b = 56.3 Å | a = b = 56.2 Å |
| c = 81.4 Å | c = 80.7 Å | c = 80.3 Å |
Rsym = Σhkl Σi |I(hkl)i − 〈I(hkl)〉|/Σhkl Σi 〈I(hkl)i〉.
Structure Determination.
The structure of the RNase A dimer was determined by molecular replacement method using amore (27). The search model was the RNase A monomer (28) without the hinge loop. The BS-RNase dimer (14) failed as a search model, for reasons which the structure now makes clear. Model building was undertaken using all data (no σ cutoff) during 20 rounds of crystallographic refinement in xplor (29). A “round” of refinement is defined as sequential application of positional, simulated annealing and isotropic temperature factor (B factor) refinement, followed by visual inspection of electron density maps coupled with manual model building using the molecular graphics program frodo (30). The solvent molecules were added using program xtal view (version 4.0) (31). Model atom positions of the hinge loops were verified by the examination of conventional and simulated annealing omit maps (32).
Competition with the Dimer Formation by the S-Peptide.
The S-peptide (Sigma) and intact RNase A monomer were mixed at various molar ratios and were subjected to the same procedure as used to prepare dimer (7). The concentration of RNase A was kept the same in all samples. RNase A solution without the S-peptide was used as a control.
RESULTS
Structure Solution and Refinement.
The molecular replacement solution was obtained using 10–3.5 Å data. The model was refined to 2.8 Å using xplor. At 2.8 Å, it was evident that the dimer is domain-swapped, but the hinge loops could not be completely built based on difference electron density maps. The model at 2.8 Å was further refined against the new data to 2.1 Å, and the two hinge loops could be completely built into the electron density map (Fig. 1). The statistics of the model and the refinement are listed in Table 2. The quality of the model was examined using procheck (34), verify3d (35), and errat (36). The Ramachandran plot reveals that 100% of the non-glycine, non-proline residues lie in the additional allowed regions.
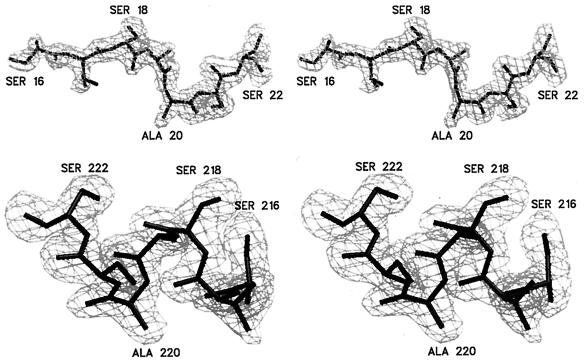
Figure 1.
Stereo view of the electron density superimposed on the model of the hinge loops of the RNase A dimer. These loops link the swapped helix (residues 1–15) to the major domain (residues 23–124). The electron density is a simulated annealing omit Fo–Fc map (32), contoured at 2.5 σ using the graphical program setor (33). Loop 1 (Upper, residues 16–22) is extended, whereas loop 2 (Lower, residues 216–222) forms a helix. These hinge loops are shown with the same orientation as those in Fig. 2C.
Table 2.
The atomic refinement of the RNase A dimer at 2.1 Å resolution
| Resolution range | 10–2.1 Å | No. of reflections | 16,471 |
| R-factor* | 19.2% | No. of protein atoms | 1,902 |
| Free R-factor* | 25.8% | No. of waters | 93 |
| rmsd bonds | 0.01 Å | No. of chloride ions | 2 |
| rmsd angles | 1.56° | No. of sulfate ions | 1 |
| Average atomic displacement parameter 20.1 Å2 | |||
R = Σhkl |F(hkl)o − F(hkl)c|/Σhkl F(hkl)o. rmsd, root-mean-square deviation.
The RNase A Dimer Is an Asymmetric, 3D Domain-Swapped Dimer.
The two RNase A subunits exchange their N-terminal α-helices to form a domain-swapped dimer (Fig. 2C). The two polypeptide chains are labeled as residues 1–124 and residues 201–324, respectively. In the dimer, the active sites (composed of residues His-12, Lys-41, and His-119 in the monomer) are now formed by the residues from two subunits. In other words, one active site contains residues His-212, Lys-41, and His-119, and the other active site contains residues His-12, Lys-241, and His-319. The exchange of the helices does not disrupt the active site, which is consistent with the observation that the RNase A dimer retains full activity (7). The two subunits are related by an approximate 160-degree rotation, as opposed to a 2-fold axis between the two subunits as in BS-RNase (14). The two hinge loops have different conformations. Loop 1 (residues 16–22) is more extended, with higher atomic displacement parameters than loop 2 (residues 216–222) (Figs. 1 and 2C). The two loops have average atomic displacement parameters of 24 Å2 in loop 1 and 14 Å2 in loop 2.
Upon domain swapping, the hinge loops change conformation, but the rest of the molecule does not. Structural comparison by the program align (38) of the main chains of the two “functional units” (residues 1–15 + residues 223–324, and residues 201–215 + residues 23–124) gives a root-mean-square deviation of 0.48 Å. When compared with the RNase A monomer without the hinge loop, both functional units give a root-mean-square deviation of 1.3 Å (Table 3). This means that each half of the RNase A dimer has a nearly identical conformation as the RNase A monomer, except at the hinge loops.
Table 3.
Structural comparison of the main chains of the RNase A monomer, the RNase A dimer, and the BS-RNase dimer, giving the root-mean-square deviation (rmsd, in Å) of the functional units (FU)*
| RNase A-FU† | RNase A-FU1 | RNase A-FU2 | BS-RNase-FU1 | BS-RNase-FU2 | |
|---|---|---|---|---|---|
| RNase A-FU | 0 | 1.3 | 1.3 | 1.1 | 0.6 |
| RNase A-FU1 | 0 | 0.5 | 1.4 | 1.2 | |
| RNase A-FU2 | 0 | 1.5 | 1.3 | ||
| BS-RNase-FU1 | 0 | 0.9 | |||
| BS-RNase-FU2 | 0 |
The functional unit in RNase A is the monomer without the hinge loop (residues 16–22), and in the dimers is the monomer-like unit made up of residues 1–15 and 23–124 but lacking the hinge loop. Notice that all of the functional units have essentially the same structure. The functional unit is essentially the same in all three structures; only the hinge loop is changed, which results in a very different dimeric structure.
The definition of functional units are as follows. RNase A-FU: RNase A monomer functional unit, residues 1–15 + 23–124; RNase A-FU1: RNase A dimer functional unit 1, residues 1–15 + 223–324; RNase A-FU2: RNase A dimer functional unit 2, residues 201–215 + 23–124; BS-RNase-FU1: BS-RNase functional unit 1, residues 1A–15A + 23B–124B; BS-RNase-FU2: BS-RNase functional unit 2, residues 1B–15B + 23A–124A.
The RNase A and BS-RNase Dimers Have Different Orientations of Subunits.
As mentioned above, BS-RNase is a domain-swapped dimer, and it shares 81% sequence identity with RNase A (4 of the 23 sequence differences occur in the hinge loop). Our structure shows that the “functional units” within RNase A and BS-RNase have essentially the same conformation (Table 3). As part of this similarity, they possess a similar “closed interface,” the interface between the swapped domain (residues 1–15) and the other major domain (residues 23–124). The unexpected result is that the orientation of the subunits in the RNase A dimer differs substantially from that of the BS-RNase, with different parts of the protein involved in the “open interface,” the new interface formed in the domain-swapped dimer, not present in the monomer. In the RNase A dimer, the “open interface” is formed between two antiparallel β-strands (residues 97–103 and residues 297–303) (Fig. 2C), with six hydrogen bonds between the strands. However, in the BS-RNase dimer, the “open interface” is formed primarily between two α-helices (residues 24–33) connected by two intersubunit disulfide bonds (Fig. 2B). There is no hydrogen bonding between the two helices (14).
No Congo Red Molecule Is Found in the Structure of Congo Red-Stained Dimer.
The model of the RNase A dimer at 2.1 Å was refined against Congo Red-stained data to 2.5 Å using xplor. No significant peak for Congo Red could be found in the Fo − Fc electron density map. Considering the high level of noise in the Fo − Fc map, we computed a Fo (Congo Red–RNase A complex) − Fo (the RNase A dimer) map. Again, no Congo Red molecule was found. This indicates that the binding of Congo Red to the RNase A dimer is nonspecific.
The S-Peptide Interferes with the Formation of the Dimer.
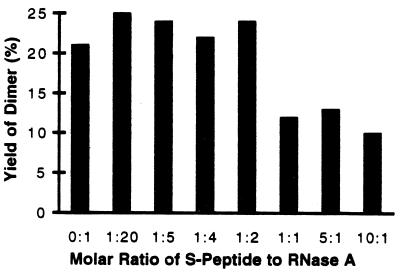
The data shown in Fig. 3 suggest that the S-peptide interferes with the formation of the RNase A dimer; at molar ratios of the S-peptide to RNase A larger than 1, the yield of the dimer decreases significantly. From the law of mass action, we would expect that the S-peptide occupying the binding site of the swapped domain would compete with it. In contrast, a low molar ratio of the S-peptide to RNase A slightly increases the yield of the dimer over no S-peptide (Fig. 3).
Figure 3.
Yield of the RNase A dimer in the presence of varying concentrations of the S-peptide (residues 1–20). Notice that larger concentrations of the S-peptide diminish the yield of dimer, presumably by competing for the domain-swapping site that would otherwise bind the swapped domain (residues 1–15). Notice also in the absence of the S-peptide, the yield of dimer is also diminished slightly. This small difference can be interpreted as shown in Fig. 4.
DISCUSSION
Orientations of Subunits in the RNase A and BS-RNase Dimers.
Our results show that the RNase A dimer is a 3D domain-swapped dimer as is the BS-RNase dimer, but with a distinctly different orientation of subunits than in BS-RNase. Stated differently, the hinge loop of RNase A (with sequence STSAASS) has a different conformation from that of BS-RNase (with sequence GNSPSSS). The hinge loop of BS-RNase forms a turn at the position of Pro-19. In RNase A, residue 19 is an Ala, which allows the hinge loops to be more flexible. Because of the flexibility of the hinge loops and the absence of intersubunit disulfide bond in the RNase A dimer, the two subunits in RNase A dimer adopt different orientations, permitting the formation of six hydrogen bonds at the “open interface,” creating a 6-stranded beta-sheet, extending across the dimer interface. But the formation of this sheet forces an asymmetric relationship between the two subunits. The asymmetric relationship is reflected in the hinge loops, with one hinge loop (residues 216–222) forming a helix, and the other hinge loop (residues 16–22) being extended. In both the RNase A and BS-RNase dimers, much of the intersubunit adhesion arises from the swapped N-terminal helices. In RNase A, an additional favorable binding energy may come from the six new hydrogen bonds in the “open interface.” This additional binding energy is not accessible to the BS-RNase dimer because its intersubunit disulfide bonds hold the subunits in an orientation that prevents these hydrogen bonds from forming.
As mentioned above, BS-RNase acquires additional biological properties when it is domain-swapped (15–17). The replacement of A19P, E28L, K31C, and S32C on RNase A led to domain swapping and to the acquired biological properties (19). It was reported that cross-linked dimer and trimer of RNase A also exhibit these properties (39–41). It is unknown, however, if the cross-linked dimer is 3D domain-swapped and, if so, if it has the same structure as the unlinked RNase A dimer reported here. Because the RNase A dimer and the dimeric BS-RNase have different orientations of the subunits, it is necessary to ask if these acquired biological functions are the result of the relative orientation of the subunits or if domain swapping is sufficient for these properties. Structural studies of these cross-linked oligomers may illuminate the mechanisms of these novel properties.
We conclude that there are at least two distinct structures for RNase domain-swapped dimers and that studies of dimers need to be interpreted in this context. Two mutant RNase A dimers that have been studied previously (19) may have the intersubunit orientation seen in the BS-RNase dimer rather than in the RNase A dimer of this paper. The first of these is the A19P mutant of RNase A. From their studies, Di Donato et al. (19) concluded that Pro-19 in BS-RNase favored domain swapping. However, they also found that A19P mutation on RNase A decreased the yield of dimer formation (19). Our structure offers a reasonable explanation for this apparent inconsistency. The introduction of Pro-19 into RNase A could introduce a turn and rigidity into the hinge loop. If this occurs, the A19P RNase A mutant may form a domain-swapped dimer with the “open interface” between the two α-helices, as in BS-RNase. The lower yield of dimer might then be due to either of the following factors: the small interaction surface, the lack of hydrogen bonds, and the absence of intersubunit disulfide bonds between these two helices of the A19P RNase A mutant, or the lack of the hydrogen bonds of the “open interface” of the RNase A dimer.
The second mutant RNase A that could conceivably form a dimer similar to BS-RNase is the K31C/S32C RNase A (19). This mutant formed 15% domain-swapped dimer spontaneously after long equilibration. This disulfide-linked dimer may have a similar structure to BS-RNase because the two intersubunit disulfide bonds are likely to restrict the orientations of two subunits, preventing a structure resembling the RNase A dimer. This is consistent with the idea that local high concentration of monomers is necessary for the formation of the domain-swapped dimer.
The fact that the RNase A and BS-RNase dimers have similar “closed interfaces” but different “open interfaces” suggests that domain swapping is sufficient for proteins to form oligomers, but the precise orientation of the subunits is influenced by the interactions of the “open interface.” Mutations can influence the nature of the “open interface” and hence the overall structure.
The Binding of Congo Red to Crystals of the RNase A Dimer.
Two models of Congo Red binding to amyloid proteins have been proposed. One model suggests that Congo Red intercalates between two antiparallel beta-strands of amyloid, with its long axis parallel to the direction of beta-strands, which was supported by the complex of insulin with Congo Red (24). The other model proposes that Congo Red stacks on the beta-sheets in amyloid, with its long axis perpendicular to the direction of beta-strands, supported by the study of Congo Red binding to poly-Lys and poly-Ser (25). Two such potential binding sites for Congo Red may be offered by the structure of the RNase A dimer: one is between the two beta-strands at the “open interface;” the other is on the six-stranded beta-sheet formed from three strands of each subunit (Fig. 2C). Indeed, we observe that Congo Red stains crystals of the RNase A dimer. But no Congo Red molecule can be found in the structure of the complex of Congo Red with the RNase A dimer. Considering that RNase A, with a pI of 9.3, is positively charged at pH 7.5 and Congo Red has one negative charge at each end of the molecule, we suggest that Congo Red is bound to crystals of the RNase A dimer through nonspecific ionic interactions. It may be that specific interactions are required to develop the characteristic green birefringence of amyloid, which we do not observe.
Implications for Amyloid Proteins from the RNase A Dimer.
The formation of amyloid is a current topic of intense scientific investigation, and the RNase A dimer offers a structurally characterized system that shares some properties of amyloids and prions. These amyloid-like properties include formation of aggregates and staining by Congo Red. A prion-like property is that the subtilisin-sensitive RNase A monomer (6) becomes insensitive when “aggregated” into the domain-swapped dimer (42).
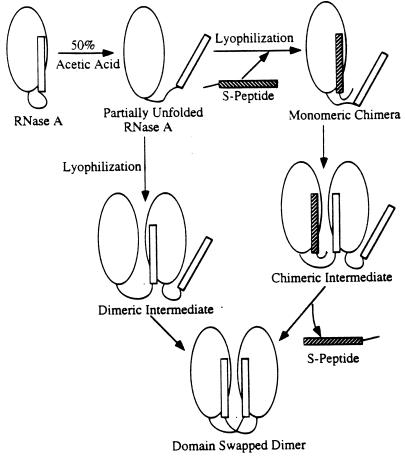
To explore the mechanism of formation of the RNase dimer, we examined the yield of dimer as a function of concentration of the S-peptide. This peptide consists of residues 1–20, which includes the swapped helical domain and part of the hinge loop (residues 16–22). The competition with the dimer formation by the S-peptide (Fig. 3) shows that when the molar ratio of the S-peptide to RNase A is lower than 1.0, the S-peptide slightly promotes the formation of the dimer. When the ratio is higher than 1.0, the S-peptide inhibits the formation of the dimer significantly. It is well established that after cleavage by subtilisin, the S-peptide remains associated with the S-protein, showing their strong binding affinity (6). Based on our results, we propose that during the formation of the dimer, the S-peptide binds to the RNase A monomer, forcing the N-terminal segment out of its binding site but remaining covalently attached to the rest of the RNase A monomer. We call this complex of intact RNase A with the S-peptide a monomeric chimera in Fig. 4. Because this segment cannot fold back into its original position, it has a greater chance to insert into another partially unfolded RNase A monomer to form an unstable complex called the chimeric intermediate in Fig. 4. When the two monomers bind, the N-terminal segment of the second monomer will replace the S-peptide to form the domain-swapped dimer, because the dimer is more stable than the chimeric intermediate. When the molar ratio of the S-peptide to RNase A is higher than 1.0, most RNase A monomers are occupied by the S-peptide so that the chance of forming the dimer decreases significantly.
Figure 4.
Schematic illustration of competition of the S-peptide (residues 1–20) with the formation of the domain-swapped RNase A dimer. At low concentrations of the S-peptide, the S-peptide binds to a partially unfolded RNase A monomer (Top Center) and displaces its N-terminal helix, creating a monomeric chimera (Top Right), which has the potential to bind to another partially unfolded monomer to form an unstable chimeric intermediate (Right Center). Thus, low concentrations of the S-peptide might favor formation of the domain-swapped dimer. At higher concentrations of the S-peptide, most RNase A monomers are occupied by the S-peptide, preventing the formation of the chimeric intermediate (Right Center) or the dimeric intermediate (Left Center), thus diminishing the yield of the domain-swapped dimer.
The effect of the S-peptide on the formation of the domain-swapped RNase dimer has a possible implication for the formation of protein aggregates by domain swapping, including possibly amyloid aggregates. Amyloid fibrils in Ig light chain amyloidosis are composed of intact Ig light chains, their variable domains, or both (13). Reduction of disulfide bonds in the Bence Jones protein DIA led to the formation of amyloid-like fibrils. A mechanism identical to domain swapping was proposed for the fibril formation (13). Based on our results and previous models, we propose a mechanism for amyloid formation. The process is initiated by the cleavage of a fragment or domain of a protein. Under reductive or destabilizing conditions, this fragment or domain can bind to an intact molecule of the same protein, replacing the domain and permitting the same domain to insert into another molecule, and so on, to form a fibril. At present, the mechanism of amyloid formation remains unclear, but perhaps further studies of 3D domain swapping in proteins that are well characterized structurally can illuminate this phenomenon.
Acknowledgments
We thank Drs. Daniel H. Anderson, Melanie J. Bennett, and Orly Dim for their thoughtful suggestions; we also thank the National Science Foundation and Department of Energy for support.
ABBREVIATIONS
- RNase A
bovine pancreatic ribonuclease A
- BS-RNase
bull seminal ribonuclease
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1A2W).
References
- 1.Sela M, White F H, Anfinsen C B. Science. 1957;125:691–692. doi: 10.1126/science.125.3250.691. [DOI] [PubMed] [Google Scholar]
- 2.Hirs C H W, Moore S, Stein W H. J Biol Chem. 1960;235:633–647. [PubMed] [Google Scholar]
- 3.Hirschman R, Nutt R F, Verber D F, Vitali R A, Vorga S L, Jacob T A, Holly F W, Denkewalter R G. J Am Chem Soc. 1969;91:507–508. doi: 10.1021/ja01030a055. [DOI] [PubMed] [Google Scholar]
- 4.Kartha G, Bello J, Harker D. Nature (London) 1967;213:862–865. doi: 10.1038/213862a0. [DOI] [PubMed] [Google Scholar]
- 5.Wyckoff H W, Hardman K D, Allewell N M, Inagam T, Johnson L N, Richards F M. J Biol Chem. 1967;242:3984–3988. [PubMed] [Google Scholar]
- 6.Richards F, Vithayathil P. J Biol Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 7.Crestfield, A. M., Stein, W. H. & Moore, S. (1962) Arch. Biochem. Biophys., Suppl. 1, 217–222. [PubMed]
- 8.Bennett M J, Choe S, Eisenberg D S. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlunegger M, Bennett M, Eisenberg D. In: Advances in Protein Chemistry. Richards F M, Eisenberg D S, Kim P S, editors. Vol. 50. New York: Academic; 1997. pp. 61–122. [DOI] [PubMed] [Google Scholar]
- 10.Pei X Y, Holliger P, Murzin A G, Williams R L. Proc Natl Acad Sci USA. 1997;94:9637–9642. doi: 10.1073/pnas.94.18.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett M J, Schlunegger M P, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parge H E, Arvai A S, Murtari D J, Reed S I, Tainer J A. Science. 1993;262:387–394. doi: 10.1126/science.8211159. [DOI] [PubMed] [Google Scholar]
- 13.Klafki H-W, Pick A I, Pardowitz I, Cole T, Awni L A, Barnikol H-U, Mayer F, Kratzin H D, Hilschmann N. Biol Chem Hoppe-Seyler. 1993;374:1117–1122. doi: 10.1515/bchm3.1993.374.7-12.1117. [DOI] [PubMed] [Google Scholar]
- 14.Mazzarella L, Capasso S, Demasi D, Di Lorenzo G, Mattia C A, Zagari A. Acta Crystallogr D. 1993;49:389–402. doi: 10.1107/S0907444993003403. [DOI] [PubMed] [Google Scholar]
- 15.Piccoli R, Di Donato A, D’Alessio G. Biochem J. 1988;253:329–336. doi: 10.1042/bj2530329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vescia S, Tramontano D. Mol Cell Biol. 1980;36:125–128. doi: 10.1007/BF02357027. [DOI] [PubMed] [Google Scholar]
- 17.Cafaro V, De Lorenzo C, Piccoli R, Bracale A, Mastronicola M R, Di Donato A, D’Alessio G. FEBS Lett. 1995;359:31–34. doi: 10.1016/0014-5793(94)01450-f. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Soucek J, Matousek J, Raines R T. J Biol Chem. 1995;270:31097–31102. doi: 10.1074/jbc.270.52.31097. [DOI] [PubMed] [Google Scholar]
- 19.Di Donato A, Cafaro V, Romeo I, D’Alessio G. Protein Sci. 1995;4:1470–1477. doi: 10.1002/pro.5560040804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazzarella L, Vitagliano L, Zagari A. Proc Natl Acad Sci USA. 1995;92:3799–3803. doi: 10.1073/pnas.92.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergdoll M, Remy M-H, Cagnon C, Masson J-M, Dumas P. Structure. 1997;5:391–401. doi: 10.1016/s0969-2126(97)00196-2. [DOI] [PubMed] [Google Scholar]
- 22.Wlodawer A, Svensson L, Sjolin L, Gilliland G. Biochemistry. 1988;27:2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- 23.Puchtler H, Sweat F, Levine M. J Histochem Cytochem. 1962;10:355–364. [Google Scholar]
- 24.Trunell W G, Finch J T. J Mol Biol. 1992;227:1205–1223. doi: 10.1016/0022-2836(92)90532-o. [DOI] [PubMed] [Google Scholar]
- 25.Klunk W E, Pettegrew J W, Abraham D J. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 28.Birdsall D L, McPherson A. J Biol Chem. 1992;267:22230–22236. [PubMed] [Google Scholar]
- 29.Brunger A T. A System for X-Ray Crystallography and NMR: X-PLOR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 30.Jones T A. J Appl Crystallogr. 1978;11:268–272. [Google Scholar]
- 31.McRee D E. Practical Protein Crystallography. San Diego: Academic; 1993. pp. 303–386. [Google Scholar]
- 32.Hodel A, Kim S H, Brunger A T. Acta Crystallogr A. 1992;48:851–858. [Google Scholar]
- 33.Evans S E. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R A, Mac Arthur M V, Moss D D, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Luthy R, Bowie J U, Eisenberg D. Nature (London) 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 36.Colovos C, Yeates T O. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 38.Satow Y, Cohen G H, Padlan E A, Davies D R. J Mol Biol. 1986;190:593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- 39.Piccoli R, D’Alessio G. J Biol Chem. 1984;259:693–695. [PubMed] [Google Scholar]
- 40.Bartholeyns J, Baudhuin P. Proc Natl Acad Sci USA. 1976;73:573–576. doi: 10.1073/pnas.73.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotte G, Testolin L, Costanzo C, Sorrentino S, Armato U, Libonati M. FEBS Lett. 1997;415:308–312. doi: 10.1016/s0014-5793(97)01147-2. [DOI] [PubMed] [Google Scholar]
- 42.Libonati M, Taniguchi T, Leone E. Biochim Biophys Acta. 1973;317:160–163. doi: 10.1016/0005-2795(73)90208-0. [DOI] [PubMed] [Google Scholar]