Abstract
Caenorhabditis elegans embryos establish cortical domains of PAR proteins of reproducible size before asymmetric cell division. The ways in which the size of these domains is set remain unknown. Here we identify the GTPase-activating proteins (GAPs) RGA-3 and RGA-4, which regulate the activity of the small GTPase RHO-1. rga-3/4(RNAi) embryos have a hypercontractile cortex, and the initial relative size of their anterior and posterior PAR domains is altered. Thus, RHO-1 activity appears to control the level of cortical contractility and concomitantly the size of cortical domains. These data support the idea that in C. elegans embryos the initial size of the PAR domains is set by regulating the contractile activity of the acto-myosin cytoskeleton through the activity of RHO-1. RGA-3/4 have functions different from CYK-4, the other known GAP required for the first cell division, showing that different GAPs cooperate to control the activity of the acto-myosin cytoskeleton in the first cell division of C. elegans embryos.
Keywords: cell polarity, cortex, NMY-2, contractility
Cell polarization allows a spatial differentiation of cellular functions. For instance, cells polarize to migrate toward a signal, to secrete molecules in a biased direction, and to divide asymmetrically to generate different daughter cells. Polarity establishment depends on the segregation of the cell cortex into different cortical domains, which then dictate asymmetric functions within the cytoplasm. The formation of cortical domains involves two interlinked problems: How does a cell initially set the size of its domains, and how is the size of the domains maintained during asymmetric functioning? Recent work in a number of organisms has identified many of the components of these cortical domains (1–4). However, it remains unclear how these components determine the size of the domains. How can the collective activity of molecules determine the size of a domain that might be several orders of magnitude larger than the molecules themselves?
The formation of polarity in Caenorhabditis elegans embryos is an excellent system to study the establishment of cortical domains. The polarization of the cortex is marked by the formation of PAR protein domains: An anterior domain comprising PAR-3, PAR-6, and atypical PKC (PKC-3), together with the Rho GTPase CDC-42; and a posterior domain comprising PAR-1 and PAR-2. After meiosis, the members of the anterior PAR complex are localized all over the cortex. When symmetry is broken by the centrosome, the anterior complex retracts toward the anterior of the embryo. Concomitantly, the posterior PAR complex fills in the cortical area left by the retraction of the anterior PAR complex, thereby establishing the polarity of a one-cell embryo (5, 6). During the subsequent maintenance phase, the two PAR domains have consistent sizes {a variation of ≈2% between embryos [unpublished data and supporting information (SI) Text]} until cytokinesis. These cortical domains control the asymmetric localization of cell fate determinants and dictate an unequal cell division.
Establishment of PAR polarity requires the acto-myosin cytoskeleton. If the acto-myosin network is globally disrupted, symmetry is not broken and the anterior PAR complex does not segregate into an anterior domain (7–14). How does the acto-myosin cytoskeleton regulate the size of the PAR domains? An important clue to this problem comes from the analysis of the contractility of the cortex and of the acto-myosin distribution in the cortex. Shortly after meiosis, the cortex is uniformly contractile, and the acto-myosin contractile elements are evenly distributed. Contractile polarity is formed through the segregation of the acto-myosin network toward the anterior pole, resulting in a contractile anterior domain and a posterior noncontractile (“smooth”) domain. The boundary between these two domains is transiently marked by an ingression called pseudocleavage furrow (PCF) located in the middle of the embryo. Thus, the contractile and noncontractile domains occupy opposite halves of the embryo.
The anterior PAR complex localizes to the contractile domain, whereas posterior PAR proteins are confined to the noncontractile domain. The precise coincidence of the PAR domains and the contractile domains suggests that the formation of these two domains is linked. Although positive feedback from the PAR domains has been proposed to maintain contractility in the anterior domain (15), its mechanism remains so far unclear. Thus, it has been proposed that the size of the contractile domains could determine the size of the PAR domains.
How does the embryo set the size of the contractile domains? A popular model is based on the mobility of the contractile elements in the cortex and proposes that, in C. elegans embryos, the acto-myosin network is initially uniformly distributed over the cortex and therefore contracts isotropically. The contractile network generates tension throughout the embryo cortex. Local inhibition of acto-myosin contractility in the cortical region overlying the centrosomes provides the symmetry-breaking event, inducing a local relaxation of cortical tension. Because the acto-myosin network is now anisotropic, it contracts toward the anterior. This cortical relaxation model (5, 16) proposes that the degree of contractility of the acto-myosin cytoskeleton determines the respective sizes of anterior and posterior domains. If contractility is high, more cortical tension would be generated and thus symmetry breaking would result in a large relaxed domain. In contrast, low contractility would prevent the relaxed domain from spreading.
Although this model is attractive, there are few data so far to show that the degree of contractility of the cortex determines the size of cortical or PAR domains. To experimentally test the cortical relaxation model, one would need to modulate the activity of the acto-myosin cortex and see how this affects the domain size. In all systems studied, the acto-myosin cytoskeleton is regulated by the activity of the small GTPase Rho (17). Rho GTPases cycle between a GTP-bound (active) form and a GDP-bound (inactive) form. GTPase-activating proteins (GAP) enhance intrinsic GTP hydrolysis, which inactivates the GTPase. Guanine nucleotide exchange factors (GEF) catalyze the exchange of the GDP for a new GTP molecule, thus reactivating the GTPase. In C. elegans RNAi of rho-1 or its GEF ect-2 abolishes cortical contractility and disrupts the organization of the acto-myosin cytoskeleton (10–12). As a consequence, anterior and posterior PAR domains can overlap. Although these data underscore that the acto-myosin meshwork is required for the formation of cortical polarity, there is still no evidence that the degree of contractility determines the size of the domains, as proposed in the cortical relaxation model.
Here we report the identification of two GAPs for RHO-1, RGA-3 and RGA-4, which control the contractile activity of the cortex and the size of the anterior and posterior PAR domains during the first cell cycle of C. elegans embryos. rga-3/4(RNAi) embryos display increased contractility and a corresponding increase in the size of the noncontractile domain, lending experimental support to the cortical relaxation model. Indeed, RGA-3/4 appears to specify domain size, at least in part, through a role in organization of the myosin II meshwork.
Results
In wild-type embryos, the entire cortex contracts after the end of meiosis. These contractions can be seen by Nomarski microscopy as small cortical invaginations that persist from 5 seconds to several minutes (5). After polarity initiation, the posterior cortex “smoothes” as contraction ceases in this region. The PCF marks the boundary between the smooth noncontractile posterior and the contractile anterior cortex, and thus its position provides a good marker for the anterior and posterior domain size. We reasoned that genes required for positioning the PCF would be good candidates for regulators of domain size.
In a genome-wide RNAi phenotype database (18) we identified two genes that had a dramatic effect on cortical activity after RNAi: rga-3 and rga-4. The PCF formed further to the anterior than in wild type, giving rise to a larger posterior and a smaller anterior cortical domain. Furthermore, the anterior domain underwent violent contractions that often resulted in formation of cytoplasts (Fig. 1A). Examination of the database suggested that this phenotype was unique to the activity of these two genes. rga-3 and rga-4 are 79% homologous at the protein level. The RNAi reagent used in these experiments is predicted to target both genes. In an attempt to isolate the function of RGA-3 from RGA-4, we designed two RNAs targeting nonhomologous regions in the 3′ UTR of rga-3 and rga-4. RNAi of either 3′ UTR resulted in a weaker phenotype than RNAi of a region in the ORF, which is predicted to target both genes (SI Text). Therefore, both genes likely contribute to PCF positioning in embryos, and thus we refer to the RNAi phenotype as rga-3/4(RNAi).
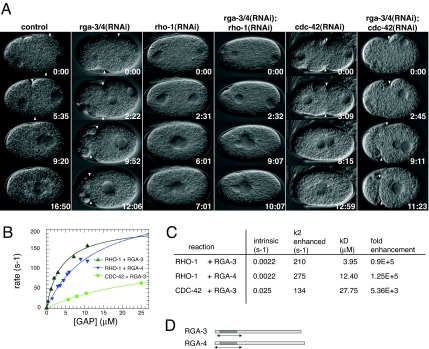
Fig. 1.
RGA-3 and RGA-4 are GAPs for RHO-1. (A) Time-lapse differential interference contrast images of control and RNAi embryos during polarity establishment. In this and subsequent figures the embryos are ≈50 μm long. Embryo posterior is to the right. In rga-3/4(RNAi) embryos, the cortex is hypercontractile and the boundary between the contractile anterior cortex and the noncontractile posterior cortex (PCF, white arrowhead) is shifted toward the anterior. The relaxation of the PCF is delayed in rga-3/4(RNAi) embryos. In rho-1(RNAi) and rga-3/4(RNAi);rho-1(RNAi) embryos cortical contractility is abolished, whereas in rga-3/4(RNAi);cdc-42(RNAi) embryos the contractility resembles rga-3/4(RNAi) embryos. Times (minutes:seconds) are standardized to similar position of the pronuclei at the beginning of pronuclear migration. (B) Observed hydrolysis rates for 100 nM RHO-1 or CDC-42 in the presence of the indicated amounts of RGA-3 (green) or RGA-4 (blue). Solid lines represent hyperbolic fits to the data points, from which the values in C were extracted. (C) Comparison between intrinsic RHO-1/CDC-42 GTP hydrolysis rates and maximally attainable ones in the presence of RGA-3 or RGA-4 GAP domains. The binding constant (affinity) between the GTPase and the GAP is represented by kD. Fold enhancement is the ratio between the intrinsic and the maximally attainable, stimulated rate. (D) Predicted GAP domains (dark shading) and the soluble GAP domains used in this analysis (arrows).
RGA-3 and RGA-4 Are GAPs for the Small GTPase RHO-1.
Both genes encode proteins that contain a predicted GAP domain, and they are called RGA-3 and RGA-4 for RhoGAPs 3 and 4. Phylogenetic analysis revealed that both genes are most likely sequence orphans. The Drosophila melanogaster RhoGAP54D (DmCG4677) is the closest homolog to both proteins (B. Habermann, personal communication), but the function of this putative GAP is not known.
In C. elegans, RHO-1 and CDC-42 appear to be the only Rho GTPases that play essential roles in the one-cell embryo. To determine whether RGA-3 and RGA-4 are active GAPs and to test their specificity toward RHO-1 and CDC-42, we identified soluble fragments containing the GAP domains and expressed and purified them. We recorded the intrinsic hydrolysis rate of GTP by RHO-1 and CDC-42 with a fluorimetric assay and confirmed the results by an HPLC assay (SI Text and SI Fig. 7). We found that the intrinsic activity of CDC-42 (0.025 s−1) is ≈10 times larger than that of RHO-1 (0.0022 s−1) (Fig. 1C), similar to other systems (19). Those hydrolysis rates mean that both GTPases are capable of inactivating themselves, but very slowly. To assess the specificity of the GAPs, we reacted them with the GAP domain of RGA-3 and RGA-4 and monitored the change in the signal of a fluorescent GTP analogue, mantGTP, as a measure of GTP hydrolysis. In the presence of RGA-3 or RGA-4, the catalytic activity of RHO-1 could be enhanced as much as 100,000-fold, whereas RGA-3 enhanced the catalytic activity of CDC-42 only 5,000-fold (Fig. 1 B and C), namely, 20-fold less. Moreover, RGA-3 has a higher affinity (lower kD) for RHO-1 than for CDC-42. Thus, our kinetics experiments suggest that RGA-3 and RGA-4 are more likely to be regulators for RHO-1 than for CDC-42.
To confirm these kinetics results, we performed double RNAi experiments between RGA-3/4 and RHO-1 or CDC-42 (Fig. 1A). rga-3/4(RNAi);rho-1(RNAi) abolished contractility (n = 12), suggesting that the rga-3/4(RNAi) phenotype requires RHO-1. In rga-3/4(RNAi);cdc-42(RNAi) embryos cortical contractility resembled rga-3/4(RNAi) embryos (n = 11), showing that RGA-3/4 regulate contractility independent of CDC-42. Defects in PAR protein localization observed in cdc-42(RNAi) embryos (12, 20–22), such as meiotic PAR-2 cycle failure (12), are not seen in rga- 3/4(RNAi) embryos (data not shown). Taken together, the in vitro and in vivo experiments strongly suggest that RGA-3 and RGA-4 are acting as GAPs for RHO-1 in the embryo.
RGA-3/4 Are Required to Organize Myosin in the Cortex.
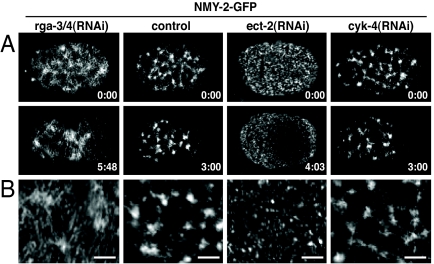
In rga-3/4(RNAi) embryos, contractile activity of the cortex is increased (Fig. 1A). Because RHO-1 is known to regulate the dynamics of acto-myosin contractile elements (10–12), we next asked whether the increased contractile activity in rga-3/4(RNAi) embryos was reflected in changes in the organization of the acto-myosin cortex. For this purpose we looked at the distribution of nonmuscle myosin II (NMY-2) using a GFP-NMY-2 fusion protein in rga-3/4(RNAi) embryos and compared it to control and ect-2(RNAi) embryos (Fig. 2). This allowed us to study the effects of increasing or reducing the activity of RHO-1 on NMY-2 organization. In control embryos, NMY-2 forms patches interconnected by small filaments, which assemble into a contractile network (Fig. 2 and SI Movie 1). These patches are constantly dissolving and reforming during the polarization of the embryo (15). As previously shown, in the absence of RHO-1 activity [rho-1(RNAi) or ect-2(RNAi)], NMY-2-GFP is organized into small foci, and no patches or filaments are seen (Fig. 2 and SI Movie 2) (10–12). Here we show that, in an rga-3/4(RNAi) background, which would correspond to a gain-of-function RHO-1 activity, the meshwork appears denser and more filamentous than in control embryos (n = 9). However, as in control embryos, the myosin network moves toward the anterior in rga-3/4(RNAi) embryos (Fig. 2 and SI Movie 3). The most likely explanation for these data is that the GAP and the GEF, by controlling the level of RHO-1 activity, determine how cortical myosin II is organized.
Fig. 2.
RHO-1 activity is required to organize myosin in the cortex. (A) Time-lapse images of cortical confocal views of NMY-2-GFP of rga-3/4(RNAi), control, ect-2(RNAi), and cyk-4(RNAi) embryos during polarity establishment. Times (minutes:seconds) are relative to pronuclear appearance. Note that in ect-2(RNAi) embryos, the acto-myosin contractile meshwork does not form, but the myosin foci respond to the polarization signal from the centrosomes and move away from the polarization site. In cyk-4(RNAi) embryos NMY-2 organization appears normal. (B) Magnified section of the cortex showing NMY-2-GFP. (Scale bars: 4.5 μm.)
RGA-3/4 Are Required to Position the Boundary Between Anterior and Posterior PAR Cortical Domains.
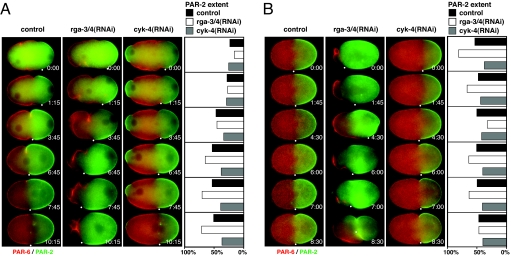
Previous work has suggested that, during polarity establishment, the segregation of the acto-myosin cytoskeleton to the anterior coordinates the segregation of the PAR proteins (3). The boundary of the PAR-2 domain coincides with the position of the PCF, and the formation/expansion of the PAR-2 domain appears to correlate with the movement of the PCF toward the anterior. To investigate whether the PCF mispositioning and high contractile activity in rga-3/4(RNAi) embryos affect the PAR domain formation, we analyzed the localization of the PAR domains using a strain in which PAR-2 was fused to GFP and PAR-6 was fused to mCherry (Fig. 3 and SI Movies 4 and 5). In GFP-PAR-2;mCherry-PAR-6 control embryos, the PCF and the PAR-2 domain expand to ≈55% of the embryo length (n = 7). In rga-3/4(RNAi) embryos after 28 h of RNAi, symmetry breaking proceeded as normal, with PAR-2 localizing at the posterior and PAR-6 retracting to the anterior (Fig. 3A). However, the PAR-2 domain continued to expand toward the anterior until it reached ≈75% of embryo length (n = 5) (Fig. 3A). Likewise, the PCF moved further to the anterior until it also reached ≈75% (n = 5). Importantly, although the size of the anterior and the posterior domains was changed, the PAR-2 and PAR-6 domains did not appear to overlap, showing that the boundary between anterior and posterior was formed (Fig. 3). However, after PCF regression, the position of the boundary between the PAR proteins in rga- 3/4(RNAi) embryos was unstable (n = 32) (Fig. 3B). The PAR-2–PAR-6 boundary moved posteriorly during mitosis, such that its position was similar to wild-type embryos by the time cytokinesis was complete, suggesting that RGA-3/4 independent mechanisms position the boundary during cell division. The cleavage furrow often formed in the anterior and subsequently moved around the embryo (SI Movie 5), suggesting that RGA-3/4 also play a role in stabilizing the position of the cytokinesis furrow.
Fig. 3.
RGA-3/4 control the position of the boundary between anterior and posterior PAR domains. Time-lapse images of control, rga-3/4(RNAi), and cyk-4(RNAi) embryos (after 28–31 h of RNAi incubation) expressing GFP-PAR-2 (green) and mCherry-PAR-6 (red) either during polarity establishment (A) or during the period from anaphase through cytokinesis (B) (SI Movies 4, 5, and 10). One boundary of each GFP-PAR-2 domain (white dot) was tracked manually over time (minutes:seconds). The distance between this boundary and the posterior pole was measured and is expressed as a fraction of total embryo length in percent. Zero percent corresponds to the posterior pole. The distance is shown for control (dark gray bars), rga-3/4(RNAi) (white bars), and cyk-4(RNAi) (light gray bars) embryos. During PAR domain establishment (A) the boundary between the PAR domains becomes positioned more anterior in rga-3/4(RNAi) embryos than in control and cyk-4(RNAi) embryos. Subsequently, this position is unstable in rga-3/4(RNAi) embryos (B). In cyk-4(RNAi) embryos a meiotic PAR-2 domain is formed (A), which often disappears before cytokinesis (B).
Although we have not been able to directly monitor RHO-1 activity, these data suggest that increasing contractility through increasing RHO-1 activity by rga-3/4(RNAi) does not disturb symmetry breaking or the boundary formation between the PAR domains. Rather, the RGA-3/4-regulated RHO-1 activity is required to set the relative size of the anterior and posterior PAR domains.
RGA-3 Is a Cortical Protein That Segregates to the Anterior During Polarization.
We generated an YFP-RGA-3 fusion protein to analyze the localization of RGA-3/4. YFP-RGA-3 forms a dynamic network throughout the entire cortex consisting of clustered foci interconnected by filaments. With onset of polarity this network segregates anteriorly (n = 17/17) (Fig. 4 and SI Movie 6). YFP-RGA-3 also localizes to the centrosomes (n = 7/7; data not shown). This localization pattern was confirmed by staining wild-type and rga-3(RNAi) embryos for RGA-3 (n = 35) (Fig. 4C).
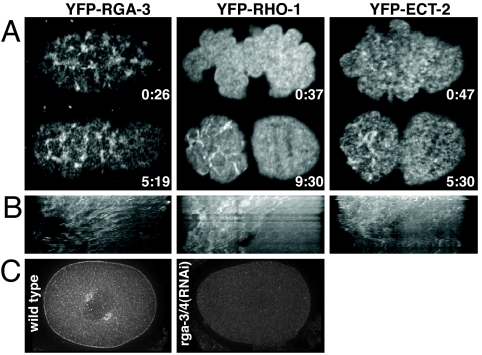
Fig. 4.
RGA-3 localizes to the cortex and the centrosomes. Shown are time-lapse images (A) and kymographs (B) of the cortical segregation of YFP-RGA-3, YFP-RHO-1, and YFP-ECT-2 to the anterior (SI Movies 6–8). Times (minutes:seconds) are relative to pronuclear appearance. (C) A wild-type embryo and a rga-3/4(RNAi) embryo stained with anti-RGA-3 antibody. RGA-3 localizes to the cortex as well as to centrosomes.
We produced kymographs to compare the dynamics of YFP-RGA-3 with the dynamics of YFP-RHO-1 and YFP-ECT-2 during polarity establishment (Fig. 4B). ECT-2 and RHO-1 have been reported to segregate to the anterior with polarity onset (11). Our data show that, as the cortex polarizes, YFP-RGA-3, YFP-ECT-2, and YFP-RHO-1 segregate to the anterior [Fig. 4B, YFP-RGA-3 (n = 17), YFP-RHO-1 (n = 3), YFP-ECT-2 (n = 20), and SI Movies 6–8]. It has been proposed that anterior segregation of RHO-1 and ECT-2 acts to keep the contractile activity of the anterior cortex high during polarity establishment (11). We show that, during polarity establishment, both positive and negative regulators of RHO-1 segregate to the anterior. Thus, RHO-1 activity is not regulated simply through spatial restriction of regulatory molecules; rather, the activity of the regulators must be controlled.
The Balance of RhoGAP RGA-3/4 and RhoGEF ECT-2 Activities Regulates the Size of Cortical Domains.
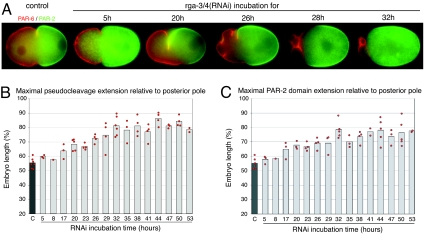
The analysis of rga-3/4(RNAi) embryos suggests that RHO-1 activity controls the posterior domain size. To test this idea more directly, we studied the relationship between the strength of the phenotype after rga-3/4(RNAi) and the size of the posterior domain by varying the time between injection of dsRNA into hermaphrodites and filming of newly fertilized embryos (23, 24). We varied the RNAi incubation time between 5 and 53 h and measured the maximal extension of the PCF and of the PAR-2 domain from the posterior pole. Fig. 5A shows representative embryos at selected time points, and Fig. 5 B and C shows the data collected during the course of the experiment. The severity of the phenotype depended on how long the dsRNA was incubated. Up to 17 h, the only noticeable defect was a shift of both the PCF and the PAR-2 domain extent to ≈64% of embryo length (Fig. 5 B and C). With longer RNAi incubation, the position of the PCF as well as the PAR-2 domain extent were shifted further to the anterior and the cortical ruffling increased (Fig. 5A). The maximum extension of the PCF from the posterior pole reached a plateau after 32 h of RNAi, at 81 ± 7% of embryo length (n = 6) (Fig. 5B). Similarly, the size of the PAR-2 domain also varied with RNAi time, reaching a maximum after 32 h at 78 ± 6% (n = 5) (Fig. 5C).
Fig. 5.
Time course of RGA-3/4 depletion by RNAi. (A) PAR-2-GFP (green) and mCherry-PAR-6 (red) localization in control and in representative rga-3/4(RNAi) embryos at selected time points. Time is in hours (h) and represents the time between dsRNA injection and analysis. The severity of the phenotype depends on the length of RNAi incubation time: PAR-2 domain size increases, the PAR-6 domain shrinks, and the PCF becomes more asymmetric, such as one side of the furrow is more anterior and deeper than the other side. We further observed an increase in membrane blebbing and the formation of cytoplasts, which pinched off anteriorly but were often resorbed later. (B and C) Time course of rga-3/4(RNAi). The maximal extension of the PCF (B) and of the PAR-2 domain (C) from the posterior pole increased with longer rga-3/4(RNAi) incubation. Each bar represents the average value for each RNAi time point.
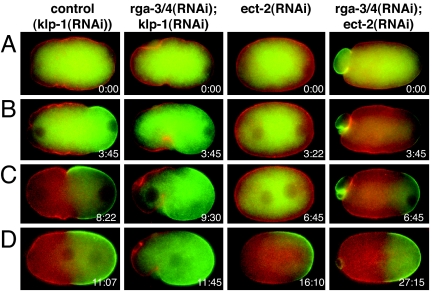
Taken together, our experiments show that the size of both contractile domains and PAR domains is controlled by RHO-1 activity. A likely interpretation of our experiments is that fine-tuning of RHO-1 activity by the activities of the GAPs (RGA-3/4) and GEF (ECT-2) is responsible for the extent of the contractile activity of the cortex and, hence, for the position of the boundary between the contractile and noncontractile domains. To test this idea, we depleted both RGA-3/4 and ECT-2 and analyzed the cortical contractility and PAR-2 and PAR-6 localization after 29–32 h of RNAi (Fig. 6 and SI Fig. 8). In this experiment we used kinesin-like protein 1 (klp-1) RNAi as a control RNAi (SI Text). We confirmed the previously published phenotype of ect-2(RNAi): Cortical contractility is abolished (n = 10) and cytokinesis fails (n = 7/10); PAR-6 is localized throughout the cortex, and PAR-2 localization to the cortex is delayed (13). Under these conditions the late PAR-2 initially overlaps with PAR-6, but PAR-6 slowly disappears from the PAR-2 region (SI Fig. 9). In embryos depleted of both RGA-3/4 and ECT-2, we observed reduced contractility (Fig. 6, SI Fig. 8, and SI Movie 9) compared with rga-3(RNAi) embryos and increased contractility compared with ect-2(RNAi) embryos (n = 8). The position of the contractile and noncontractile boundary is formed at 52 ± 8% (n = 6) in the double RNAi [compared with 81 ± 7% for rga-3/4(RNAi), n = 6]. Similarly, we observed a reduction of the maximal extent of PAR-2 (46 ± 8%, n = 6) compared with rga-3/4(RNAi) embryos (78 ± 6%, n = 5) and control embryos (55 ± 7%, n = 6). Thus, we conclude that ECT-2 and RGA-3/4 oppose each other to determine the position of the boundary between the PAR domains, presumably by modulating the activity of RHO-1.
Fig. 6.
Codepletion of ECT-2 and RGA-3/4 by RNAi. Shown are time-lapse images of control [klp-1(RNAi)], rga-3/4(RNAi);klp-1(RNAi), ect-2(RNAi), and rga-3/4(RNAi);ect-2(RNAi) embryos expressing GFP-PAR-2 (green) and mCherry-PAR-6 (red). In rga-3/4(RNAi);ect-2(RNAi) embryos (SI Movie 9) contractility is reduced compared with rga-3(RNAi) and increased compared with ect-2(RNAi) embryos. Polar body extrusion failed in rga-3/4(RNAi);ect-2(RNAi) embryos like in ect-2(RNAi) embryos under weaker RNAi conditions (data not shown). (A) Onset of posterior smoothing. (B) Beginning of PAR-2 domain formation. (C) Time point of maximal PCF ingression. (D) Time point of maximal PAR-2 extension. Times are in minutes:seconds.
The RhoGAPs CYK-4 and RGA-3/4 Regulate Different Aspects of Acto-Myosin Contractility in One-Cell Embryos.
Interestingly, another RhoGAP, CYK-4, has been proposed to regulate RHO-1 in C. elegans first cell division. Paternal-derived CYK-4 is required during symmetry breaking (10). The primary role of maternal-derived CYK-4 is in cytokinesis and polar body extrusion (25). We compared the role of maternal CYK-4 with RGA-3/4 in cortical contractility and PAR localization (Fig. 3). In cyk-4(RNAi) embryos, polar body extrusion and cytokinesis failed, as previously reported. In addition, we observed some meiotic PAR-2 left on the cortex (Fig. 3). However, symmetry breaking occurred on time, cortical contractility was unaffected (SI Movie 10), and the organization of NMY-2 appeared normal (Fig. 2). The maximal extent of the PCF and of the PAR-2 domain from the posterior pole were both slightly smaller in cyk-4(RNAi) embryos (PCF extent 45 ± 2%, PAR-2 extent 44 ± 3%, n = 7) than in control embryos (PCF extent 55 ± 3%, PAR-2 extent 55 ± 7%, n = 7) and rga-3/4(RNAi) embryos. The reason for this slightly smaller PAR-2 domain remains unclear but is probably related to residual PAR-2 left over from the meiotic failure. The different phenotypes of cyk-4(RNAi) and rga-3/4(RNAi) suggest that two different GAPs control different functions of RHO-1 in C. elegans embryos. Maternal CYK-4 is apparently involved in specifically controlling contractile events during cytokinesis, whereas RGA-3/4 is required for controlling global contractility during the whole first division.
Discussion
The current model for polarity establishment in C. elegans embryos proposes that the activity of the acto-myosin cortex regulates the distribution of the PAR proteins (1). In this model the cortex is an isotropically contractile meshwork before symmetry breaking. The anterior PAR proteins are thought to be associated with the contractile acto-myosin meshwork in part through CDC-42. During polarity establishment, disassembly of the meshwork in the region of the centrosome, which is thought to be triggered in part by the RhoGAP CYK-4 (11), causes the meshwork together with the PAR proteins to contract away from this region, forming an anterior cortical domain. An important prediction of this model is that the control of the extent of contractility would regulate the size of the contractile domain and thereby the PAR domain size. Our data provide support for this model by showing that increased contractility, through up-regulation of RHO-1 activity, results in a smaller anterior and concomitantly in a larger posterior PAR domain. Importantly, the exclusion of the positive regulator ECT-2 and the negative regulators RGA-3/4 from the posterior suggests that regulation of RHO-1 activity is required throughout the segregation of the acto-myosin meshwork to the anterior. Indeed, our data show that the balance of RGA-3/4 and ECT-2 activity can control the relative sizes of the cortical domains.
One interesting and unsolved problem is why the contractile network stops moving to the anterior after symmetry breaking. It seems likely that contractility is reduced by down-regulation of RHO-1 activity, which could be mediated by increasing RGA-3/4 activities, decreasing the ECT-2 activity, or both. The regulators of RHO-1 could be under cell cycle control, or formation of the contractile domain could modulate the activity of the regulators themselves. Interestingly, the first cell division of C. elegans embryos employs two different GAPs for RHO-1: CYK-4, which regulates symmetry breaking and cytokinesis, and RGA-3/4, which regulate the global contractile activity of the cortex. It seems that C. elegans has developed different GAPs to regulate RHO-1 at different times during the first division of the embryo. The interplay between the activities of these different GAPs will be essential for determining the organization of the acto-myosin cytoskeleton during the first asymmetric cell division of C. elegans embryos.
Materials and Methods
Worm Strains.
The following strains were used: N2 (wild type), TH110 (mCherry-PAR-6), JJ1473 (NMY-2-GFP), TH133 (YFP-RGA-3), TH87 (YFP-RHO-1), TH86 (YFP-ECT-2), TH129 (GFP-PAR-2), and TH120 (GFP-PAR-2;mCherry-PAR-6). Coding sequences for rga-3, rho-1, ect-2, par-2, and par-6 were identified by using WormBase, amplified by PCR (primer sequence in SI Text) using genomic (N2) DNA or cDNA, and inserted in pTH-YFP(N) or pAZ-mCherry (modified versions of the pAZ132 plasmid; gift from A. Pozniakovsky, Max Planck Institute of Molecular Cell Biology and Genetics) (26). Expression is driven by the pie-1 promoter. Transgenic worms were created by high-pressure biolistic bombardment. Strain TH120 was obtained by crossing strain TH129 with TH110.
RNA-Mediated Interference.
Primers used to amplify regions from N2 genomic DNA for production of dsRNA are listed in SI Text or described in ref. 12. Worms were incubated for 5–50 h after injection or feeding at 25°C. rga-3/4(RNAi) time course was performed by injection combined with feeding (27).
Time-Lapse Microscopy.
Worms were shifted to 25°C before recording. Worms were dissected, and embryos were mounted in 0.1 M NaCl and 4% sucrose with or without 2% agarose. Recordings were acquired at 10- to 15-second intervals (2 × 2 binning) with an Orca ER 12-bit digital camera (Hamamatsu, Bridgewater, NJ) mounted on a spinning disk confocal microscope (Axioplan, ×63 1.4 N.A. PlanApochromat objective and Yokogawa disk head; Zeiss). Illumination was via a 488-nm argon ion laser (Melles Griot, Carlsbad, CA). GFP-PAR-2;mCherry-PAR-6 time-lapse recordings were done on a wide-field microscope (Zeiss Axioplan II, 63 × 1.4 N.A. PlanApochromat objective, and a Hamamatsu Orca ER 12-bit digital camera). Image processing was done with MetaView (Universal Imaging, Sunnyvale, CA).
Immunofluorescence.
The RGA-3 antibody was raised against an RGA-3 fragment ≈200 aa long (primer sequence in SI Text), which shares 60% identity with RGA-4. The antibody was raised, purified (28), and visualized with FITC-conjugated secondary antibody (Jackson Immunochemicals, Suffolk, U.K.) (29). Imaging was performed on a DeltaVision system (Applied Biosystems, Issaquah, WA), and stacks were deconvolved (28).
Measurements of PCF Position and PAR-2 Domain Extent.
The extent of PCF and of the GFP-PAR-2 domain from the posterior pole was manually measured as a distance along the embryo anterior–posterior axis by using Metamorph and expressed as percentage of embryo length. In some rga-3/4(RNAi) embryos, PAR-2 retracted toward the posterior pole before PCF relaxed. We therefore measured the maximal extent of the PAR-2 domain and of the pseudocleavage furrow at different time points.
Kymograph Analysis.
Kymographs were generated from a straight line along the anterior–posterior axis with Metamorph from cortical YFP-RGA-3, YFP-RHO-1, and YFP-ECT-2 time-lapse recordings (11–14 minutes total).
Protein Expression and Purification.
RHO-1 and CDC-42 ORFs were cloned in pET-28, expressed, and purified on Ni2+ resin (Qiagen, Valencia, CA). The GAP domains were fused to GST and purified on GSH-Agarose. The GST tag was subsequently cleaved. Nucleotide exchange was performed under excess of GTP (or mantGTP; JenaBioscience, Jena, Germany) and EDTA. The protein–nucleotide complex was isolated on a NAP-5 gel filtration column. Nucleotide content was estimated by HPLC, and protein content was estimated by a Bradford reaction.
GTP Hydrolysis Assays.
For intrinsic GTP hydrolysis rate determination, the tryptophan fluorescence was measured. Long time-base spectra were acquired on a FluoroMax3 spectrophotometer (Horiba Jobin Yvon, Longjumeau, France). The HPLC assay was performed by mixing GTP-loaded GTPase with MgCl2 buffer; GDP was separated from GTP on a C18 column (4.5 μM Luna; Phenomenex) in a mobile phase consisting of 10 mM tetrabutylammonium bromide in 100 mM KPi (pH 6.0) and 10% CH3CN. GDP peak area was integrated and plotted as a function of the time point the sample was taken. For GAP-enhanced hydrolysis, the GTPase loaded with mantGTP was premixed with increasing amounts of GAP domains in presence of EDTA. The complex was rapidly mixed on a SX-18M system (Applied Photophysics, Surrey, U.K.) with a solution containing MgCl2; the fluorescence change of the mant label was monitored (excitation at 368 ± 7 nm, emitted light collected through a GG400 cutoff filter). Exponential fits to the average of three to six traces gave the observed hydrolysis rate for that particular reaction. In all assays, the buffer used was 40 mM Na Hepes (pH 7.4), 40 mM NaCl, and 5 mM DTT.
Supplementary Material
Acknowledgments
We thank C. Cowan, N. Goehring, and S. Grill for comments on the manuscript; K. Oegema (Ludwig Institute for Cancer Research, La Jolla, CA) for the RGA-3 antibody; J. Kong (Wellcome Trust Sanger Institute, Cambridge, U.K.) and A. Pozniakovsky for reagents; J. Howard and B. Janetzky for access to laboratory equipment; K. Alexandrov for advice; and S. Ernst and A. Zinke for worm bombardment and strain maintenance. C.H. was supported by an Ernst Schering postdoctoral fellowship.
Abbreviations
- PCF
pseudocleavage furrow
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor.
Note Added in Proof.
Similar results were obtained by Schmutz et al. (30).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706941104/DC1.
References
- 1.Cowan CR, Hyman AA. Development (Cambridge, UK) 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- 2.Knoblich JA. Nat Rev Mol Cell Biol. 2001;2:11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- 3.Munro EM. Curr Opin Cell Biol. 2006;18:86–94. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki A, Ohno S. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 5.Cowan CR, Hyman AA. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- 6.Schneider SQ, Bowerman B. Annu Rev Genet. 2003;37:221–249. doi: 10.1146/annurev.genet.37.110801.142443. [DOI] [PubMed] [Google Scholar]
- 7.Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Development (Cambridge, UK) 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo S, Kemphues KJ. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- 9.Hill DP, Strome S. Dev Biol. 1988;125:75–84. doi: 10.1016/0012-1606(88)90060-7. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins N, Saam JR, Mango SE. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- 11.Motegi F, Sugimoto A. Nat Cell Biol. 2006;8:978–985. doi: 10.1038/ncb1459. [DOI] [PubMed] [Google Scholar]
- 12.Schonegg S, Hyman AA. Development (Cambridge, UK) 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- 13.Severson AF, Bowerman B. J Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelton CA, Carter JC, Ellis GC, Bowerman B. J Cell Biol. 1999;146:439–451. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro E, Nance J, Priess JR. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Hird SN, White JG. J Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etienne-Manneville S, Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 18.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 19.Richnau N, Aspenstrom P. J Biol Chem. 2001;276:35060–35070. doi: 10.1074/jbc.M103540200. [DOI] [PubMed] [Google Scholar]
- 20.Aceto D, Beers M, Kemphues KJ. Dev Biol. 2006;299:386–397. doi: 10.1016/j.ydbio.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotta M, Abraham MC, Ahringer J. Curr Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 22.Kay AJ, Hunter CP. Curr Biol. 2001;11:474–481. doi: 10.1016/s0960-9822(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 23.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 24.Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 25.Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally K, Audhya A, Oegema K, McNally FJ. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmons L, Fire A. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 28.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. J Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmutz C, Stevens J, Spang A. Development (Cambridge, UK) 2007;134:3495–3505. doi: 10.1242/dev.000802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.