Abstract
Argonaute (Ago) proteins bind to microRNA (miRNAs) and short interfering RNAs (siRNAs) and form the core components of effector complexes that mediate miRNA and siRNA function. Currently, there is a paucity of reliable antibodies against mammalian Ago proteins, thus precluding studies of endogenous Ago proteins from tissues. Here we report the development of 2A8, a novel anti-Ago monoclonal antibody that recognizes human and mouse Ago proteins and efficiently immunoprecipitates miRNAs. We report the characterization of 2A8 and its use to clone miRNAs from human brain and from preparations of human polymorphonuclear leukocytes (neutrophils), which revealed a prevalent miRNA with unusual features.
Keywords: microRNA, miRNA, Argonaute, Ago, RNAi, antibody
INTRODUCTION
MicroRNAs (miRNAs) are one of the most prevalent small (∼22 nucleotide [nt]) regulatory RNA classes in animals (for reviews, see Nelson et al. 2003; Cao et al. 2006). MiRNAs exert their influence through complementary base-pairing with specific “target” mRNAs, leading in turn to degradation or translational repression of the targeted mRNA. MiRNAs have been shown to play important roles in cell fate determination, development, apoptosis, and other fundamental biological processes (for reviews, see Nelson et al. 2003; Ambros 2004; Kim 2005; Wienholds and Plasterk 2005; Hwang and Mendell 2006).
A number of different proteins function in miRNA processing. Animal miRNAs are first processed in the nucleus from large precursor primary RNAs (pri-miRNAs), into smaller stem–loop structures called pre-miRNAs (Lee et al. 2003; Denli et al. 2004; Han et al. 2004). After export to the cytoplasm via Exportin 5 (Yi et al. 2003), pre-miRNAs are endonucleolytically processed by the Dicer protein, and the mature miRNA is loaded to Argonaute (Ago) proteins (Carmell and Hannon 2004; Tijsterman and Plasterk 2004; Hammond 2005). Ago proteins are a family of ∼95 kDa proteins that are divided in two subclades, Ago and Piwi, and contain two characteristic domains known as PAZ and PIWI (Carmell et al. 2002). These domains of Ago proteins interact with different parts of the mature miRNA; the PAZ domain interacts with the 3′-end of the miRNA, whereas the PIWI domain interacts with the 5′ phosphate and 5′-proximal region of the miRNA and guides target mRNA recognition (Hammond 2005). In mammals, there are four Ago proteins (Ago1–4) (Carmell et al. 2002; Sasaki et al. 2003). The genes for Ago1, Ago3, and Ago4 reside in tandem on human chromosome 1 (1p34) and mouse chromosome 4 (Sasaki et al. 2003). Ago2 is exceptional for the gene's chromosomal location (8q24 in humans) and also because Ago2 protein is the only mammalian Ago protein capable of endonucleolytic cleavage of target mRNAs catalyzed by its active PIWI nuclease domain (Hammond 2005). Piwi proteins bind to piwi-associated RNAs (piRNAs), which are slightly longer than miRNAs (26–29 nt), and are expressed almost exclusively in the germline (for review, see O'Donnell and Boeke 2007).
Currently, there is a paucity of antibodies against mammalian Ago proteins, limiting biochemical studies of endogenous Ago proteins and precluding isolation of Ago-associated miRNAs from tissues. To surmount these obstacles, we have developed a monoclonal antibody against mammalian Ago proteins. The antibody, which is termed 2A8, recognizes all four human and mouse Ago proteins, and coimmunoprecipitates miRNAs.
RESULTS AND DISCUSSION
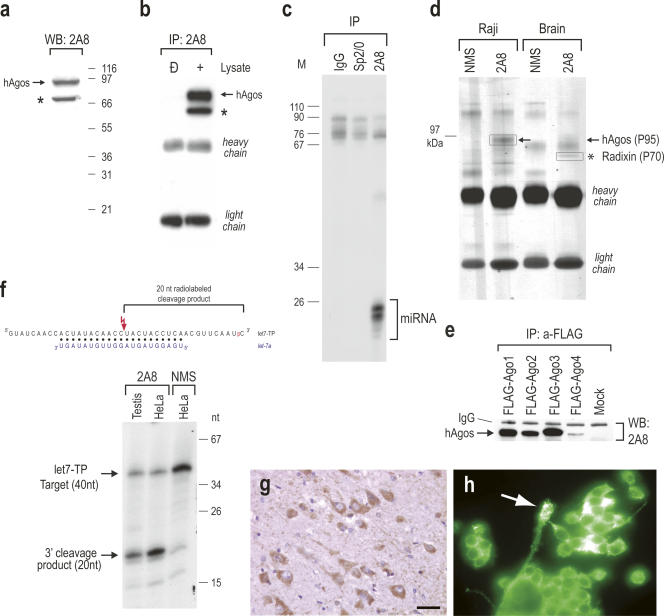
2A8 was raised in mice immunized with recombinant human Ago2 protein and its properties are shown in Figure 1. By Western blot and immunoprecipitation experiments from human cells, 2A8 recognized a band at ∼95 kDa, corresponding to Ago proteins and a cross-reacting band at ∼70 kDa (Fig. 1a,b). 2A8 immunoprecipitated efficiently miRNAs from human cells (Fig. 1c). To identify the antigens recognized by 2A8, we performed immunoprecipitations with 2A8 from human Raji cells or from human brain. We analyzed the immunoprecipitates on 4%–12% NuPAGE and we visualized the proteins by Coomassie Brilliant Blue staining. Nonimmune mouse serum served as a negative control. As shown in Figure 1d, there were two prominent bands in 2A8 immunoprecipitates, one at ∼95 kDa (P95) and another one at ∼70 kDa (P70), confirming the results of the immunoprecipitation–Western blot experiment shown in Figure 1b. We then identified the proteins contained in these bands by trypsin digestion followed by mass spectrometry. Numerous peptides corresponding to human Ago proteins 1, 2, and 3 were obtained from the P95 band, while peptides corresponding to radixin were obtained from the P70 band (see Table 1). To verify these findings, we examined whether 2A8 was able to recognize individual Ago proteins. We transfected 293T cells with FLAG-Ago1, FLAG-Ago2, FLAG-Ago3, FLAG-Ago4, or with empty vector and performed immunoprecipitations with anti-FLAG antibody and analyzed the immunoprecipitates by Western blotting with 2A8. As shown in Figure 1e, 2A8 recognized all FLAG-tagged Ago proteins. The intensity of the band corresponding to FLAG-Ago4 was less than the other Agos but that was secondary to low levels of expression of the transfected FLAG-Ago4, as determined by Western blot using anti-FLAG antibody (P. Nelson and Z. Mourelatos, unpubl.). We have also confirmed that the ∼70 kDa band corresponded to radixin by Western blots of 2A8 immunoprecipitates with anti-radixin antibody (P. Nelson and Z. Mourelatos, unpubl.). However, anti-radixin antibodies did not co-immunoprecipitate Ago proteins (P. Nelson and Z. Mourelatos, unpubl.). Radixin is an actin-binding protein with no known role in RNA regulation (Bretscher 1999; Hoeflich and Ikura 2004). Hence, there is an apparent cross-reaction between 2A8 and radixin.
FIGURE 1.
Properties of 2A8, a novel anti-Ago monoclonal antibody. (a) 2A8 recognizes Ago proteins on Western blot. Cell lysate from human HeLa cells was resolved on a 4%–12% NuPAGE gel, blotted and probed with 2A8. Molecular mass markers (in kilodaltons) are shown on the right. hAgos are shown with arrow. Asterisk indicates cross-reaction with radixin. (b) 2A8 immunoprecipitates Ago proteins. 2A8 immunoprecipitates from human 293 cells were resolved on a 4%–12% NuPAGE gel, blotted, and probed with 2A8 ascites. hAgos are shown with arrow. Asterisk indicates cross-reaction with radixin. (c) 2A8 coimmunoprecipitates miRNAs. Immunoprecipitations (IP) from HeLa cells were performed with 2A8 or nonimmune mouse serum (IgG) or Sp2/0 (both negative controls). RNA was isolated from the immunoprecipitates, 3′-end-labeled with [5′-32P]-pCp and T4 RNA ligase, resolved by electrophoresis on a 20% denaturing polyacrylamide gel, and visualized by autoradiography. Nucleotide sizes of the radiolabeled marker (M) are shown on the left. (d) Identification of immunoprecipitated protein with mass spectrometry. Immunoprecipitations were performed from human Raji cells or from human brain with 2A8 or nonimmune mouse serum (NMS). Immunoprecipitates were resolved on a 4%–12% NuPAGE gel, stained with Coomasie blue, and the P95 and P70 bands (boxed) were excised from the gel, subjected to trypsin digestion and mass spectrometry. (e) 2A8 recognizes all human Ago proteins. 293 cells were transfected with FLAG-tagged Ago1–4 proteins or with empty vector (Mock), followed by immunoprecipitations with anti-FLAG antibody. Immunoprecipitates were probed with 2A8 on Western blot. (f) 2A8 immunoprecipitates are active in miRNA-directed target RNA cleavage. Schematic of the RNA target (let7-TP) and the base-pairing with let-7a (blue); the [5′-32P] of pCp is shown in red. Cleavage site is indicated with red lightning bolt. Immunoprecipitations were performed with 2A8 or nonimmune mouse serum (NMS) and the beads were incubated with 3′-end-radiolabeled let7-TP RNA target. The products of the reactions were analyzed on 20% denaturing polyacrylamide gel. Nucleotide sizes of the radiolabeled marker (M) are shown on the right. (g,h) Immunocytochemistry with 2A8 was performed in formalin-fixed and paraffin-embedded (FFPE) human brain tissue (g) or MN-1, mouse motor neuron-like cells (h), and revealed predominantly cytoplasmic staining. In human FFPE brain, neurons were prominently stained (g). Arrow in h indicates a growth cone-like process of MN-1 cells, which is also labeled. The bar in (g) is either 50 μm (g) or 30 μm (h).
TABLE 1.
Peptides sequences obtained from mass spectrometry of proteins from 2A8 immunoprecipitations
We then tested whether 2A8 immunoprecipitates were able to cleave a radiolabeled RNA target complementary to the endogenous let-7 miRNA (let7-TP target, shown in Fig. 1f). As shown in Figure 1f, 2A8 immunoprecipitates derived from HeLa cells or mouse testis efficiently cleaved the let7-TP target, demonstrating that the immunoprecipitated Ago2 protein was catalytically active. 2A8 recognizes mouse Ago protein on Western blots but it does not recognize mammalian Piwi proteins as it does not coimmunoprecipitate piwi-associated RNAs (piRNAs) from mouse testis (Z. Mourelatos, unpubl.).
We found that 2A8 can be used for immunocytochemistry in human brain and in cultured mouse cells (Fig. 1g,h). However, these data must be interpreted with appropriate caution, because 2A8 also recognizes radixin.
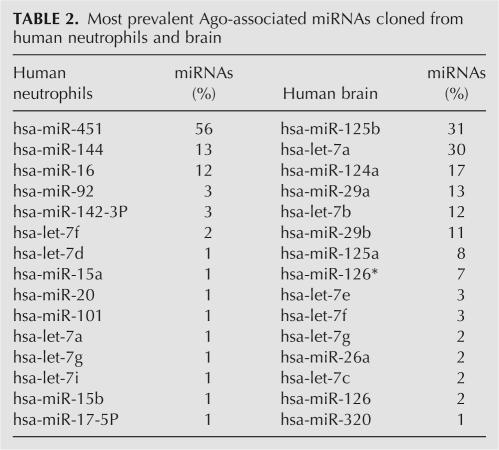
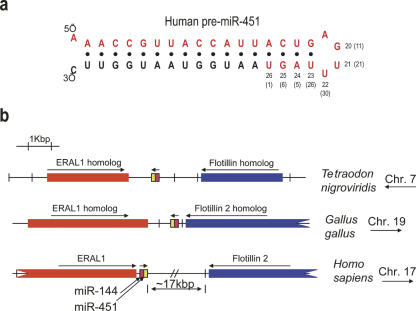
We exploited the ability of mAb 2A8 to efficiently coimmunoprecipitate miRNAs in order to isolate and clone the miRNAs bound to Ago proteins from human neutrophils. The neutrophils were isolated from the blood of an adult using a standard ficoll-based technique (Swiggard et al. 2004). The cloned miRNAs are shown in Table 2. We found that one particular miRNA was by far the most frequently cloned miRNA and corresponded to the recently discovered miR-451 (Fig. 2; Table 2). miR-451 was not among the earliest group of miRNAs isolated, and it has characteristics that are unusual. Berezikov et al. (2005) showed that miR-451 (which they had designated “Cand919”) was not expressed in mouse ES cells but was expressed in adult mouse brain and also in increasing amounts through embryonic stages (8.5 dpc < 12.5 dpc < 16.5 dpc). miR-451 has also been described by others (Altuvia et al. 2005; Sewer et al. 2005), but never in the context of human blood cells. We find that there is considerable heterogeneity of the 3′-end of miR-451 with many miR-451 molecules being longer than most miRNAs (Fig. 2a). Interestingly, mature miR-451 includes nucleotides that are on both sides of the pre-miRNA hairpin loop (Fig. 2a). Human miR-451 is a single-copy gene and the sequence of the mature miR-451 is conserved (absolutely conserved from humans to zebrafish) and located very close to the miR-144 gene on human chromosome 17 (Fig. 2b). Such genomic arrangement is conserved in other species (Fig. 2b). Interestingly, the second most commonly cloned Ago-associated miRNA from neutrophils was miR-144 (Table 2), suggesting that miR-451 and miR-144 may be coexpressed.
TABLE 2.
Most prevalent Ago-associated miRNAs cloned from human neutrophils and brain
FIGURE 2.
Cloning of miR-451 from immunopurified Agos from human neutrophils. (a) Sequence and secondary structure of human pre-miR-451. Mature miR-451 is shown in red. Nucleotide positions of the 3′-end of miR-451 are indicated. Percentages of clones that ended in specified nucleotide are shown in parentheses. Cloned miR-451 from human blood varied in length from 20 to 26 nt; 1% of clones were 26 nt long. (b) Genomic position of miR-451 gene in different species.
A recent paper (Zhan et al. 2007) demonstrated that miR-451 may play an important role in erythrocyte differentiation. A separate group also reported recently that miR-451 expression is very abundant in erythrocytes (Rathjen et al. 2006). It is possible that a fraction of miR-451 that we cloned from purified human neutrophils may represent contamination from erythrocytes. However, it is also possible that miR-451 is a miRNA that participates in cell differentiation for nonerythroid, myelogenous lineages; further studies are required to address this issue.
To determine whether 2A8 may be used to clone immunopurified miRNAs from solid human tissues, we performed immunoprecipitations of Ago proteins from human brain with 2A8, followed by isolation and deep sequencing of the coprecipitating miRNAs. The miRNAs expressed in brain were different from those in neutrophils and included miRNAs that are known to be expressed abundantly in brain, such as miR-124 and miR-125 (Table 2).
In summary, 2A8 will likely constitute an important new reagent in the technical toolkit for miRNA research.
MATERIALS AND METHODS
Antibody generation
mAb 2A8 against human Ago2 (hAgo2) protein was prepared by immunizing mice with recombinant human Ago2 protein. Briefly, a cDNA encoding amino acids 47–879 of hAgo2 was cloned between BamHI and HindIII sites of pET30a vector (EMD Biosciences/Novagen), in frame with upstream His and S-tags. The His-Ago2 construct was transformed in BL21DE3 Escherichia coli cells and inclusion bodies were isolated; recombinant His-hAgo2 was purified by electroelution from 10% preparative SDS-PAGE gels. The electroeluted protein was dialyzed with PBS, concentrated, and used to immunize BALB/C mice. Injections, hybridoma production, screening, and ascites production were done as described previously (Choi and Dreyfuss 1984). For hybridoma screening, a recombinant fusion protein between GST and aminoacids 518–897 of hAgo2 (GST–hAgo2-Cter) was used. The construct expressing GST–hAgo2-Cter was made by inserting the cDNA of hAgo2 coding for amino acids 518–897 in the BamHI-EcoRI sites of vector pGEX-6p-2 (GE Healthcare/Amersham). GST–hAgo2-Cter was purified from the soluble fraction using glutathione sepharose as per the manufacturer's instructions. A construct expressing GST–hAgo2-Nter was made by inserting the cDNA of hAgo2 coding for amino acids 47–518 in the BamHI-XhoI sites of pGEX-6p-2. GST–hAgo2-Nter protein was purified from the soluble fraction. Following the animal with the strongest Western blot signal, coimmunoprecipitation (Co-IP) experiments were performed as described previously (Mourelatos et al. 2002).
Cell culture and transfection
Human HeLa or 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 2 mM L-glutamine and 10% fetal bovine serum. Human Raji cells were grown in RPMI 1640 medium with 2 mM L-glutamine adjusted to contain 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, and 10% fetal bovine serum. 293T cells were transfected at 80% confluency in 100-mm dishes with 8 μg of FLAG-Ago1 or FLAG-Ago2 or FLAG-Ago3 or FLAG-Ago4 (Meister et al. 2004), or with empty vector using LF2000 (Invitrogen). Immunoprecipitations were carried out with M2 agarose (anti-FLAG coupled resin; SIGMA).
Western blots, immunoprecipitations, and cleavage assays
Western blots were performed as previously described using the ECL chemiluminescence kit (Mourelatos et al. 2002). 2A8 ascites was used at 1:300–1:500 dilution. Immunoprecipitations were performed essentially as described by Kiriakidou et al. (2005). For immunoprecipitation of endogenous Ago proteins, 2 μL of 2A8 ascites was used with 20 μL of protein-G agarose (Invitrogen) and lysate from cells or tissues. Cells or tissues were lysed in 20 mM Tris-HCL, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X100, 0.1 U/mL RNasin, and complete EDTA-free protease inhibitors. For cleavage experiments, the beads were washed extensively with the same buffer except that the NaCl concentration was raised to 600 mM and the final washes were with 20 mM HEPES-KOH pH 7.5, 100 mM KCl, and 0.5 mM DTT. Preparation of radiolabeled target complementary to let-7 and cleavage assays were performed as previously described (Kiriakidou et al. 2005). RNA from cleavage assays was resolved using 20% UREA-PAGE and visualized by autoradiography.
Proteomics
Immunoprecipitated proteins were resolved on 4%–12% NuPAGE gel (Invitrogen) and visualized by Coomassie Brilliant Blue staining. Bands were excised, digested with trypsin, and peptides were analyzed and identified by Microflow RP-HPLC/MS/MS.
Cloning of Ago-associated miRNAs from human neutrophils and human brain
Neutrophils were isolated using a standard technique that has been described previously (Swiggard et al. 2004), using Ficoll-Hypaque density centrifugation. Cloning Ago-associated miRNAs was performed as previously described (Maniataki et al. 2005). Briefly, miRNAs were coimmunoprecipitated using 2A8 from purified human neutrophils, isolated, and 3′-end-labeled with [32P]-pCp and T4 RNA ligase. The labeled miRNAs were then gel-purified and, after two sequential rounds of adapter ligation (3′ adapter: 5′-pUCCagatctatagtgtcacctaaatcgtatggaidT-3′; 5′ adapter: 5′-tccgaaattaaccctcactaaagGGA-3′; capitalized nucleotides are RNA), the ligated product was gel-purified, reverse-transcribed, amplified by PCR, and cloned. Plasmids isolated from 194 different colonies were used for sequencing. We also analyzed miRNAs from 2A8 immunoprecipitations using human brain (three pooled brains from Alzheimer's disease patients were used). Deep sequencing of the amplified PCR products was performed at 454 Life Sciences, Inc. From this sample, 12,576 different reads contained known miRNAs, and the top 15 of these are presented in Table 2.
Immunocytochemistry
Immunocytochemical staining was performed using conventional methods. Briefly, human brain tissue was fixed for 24 h and paraffin-embedded conventionally, and then sectioned at 5 μM on Probe-On slides. 2A8 was used at 1:500 dilution with an HRP-conjugated anti-mouse IgG secondary antibody (Jackson Immunoresearch). For immunofluorescence, MN-1 cells were fixed in 4% paraformaldehyde for 10 min; Alexa-488-labeled secondary antibody was used (Invitrogen).
ACKNOWLEDGMENTS
We thank Dr. T. Tuschl for the FLAG-Ago contructs and Dr. C.-X. Yuan of the Proteomics Core Facility of the Center for Cancer Pharmacology of the University of Pennsylvania for mass spectrometry. Studies with human tissues were IRB approved. Supported by NIH grants K08-NS050110 to P.T.N.; K08-AI63030 to M.K.; GM0720777, NS053839, and P30-HD026979 to Z.M.; and by the Philadelphia Foundation to Z.M. M.K. is a Pfizer Scholar.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.646007.
REFERENCES
- Altuvia, Y., Landgraf, P., Lithwick, G., Elefant, N., Pfeffer, S., Aravin, A., Brownstein, M.J., Tuschl, T., Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., Cuppen, E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr. Opin. Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- Cao, X., Yeo, G., Muotri, A.R., Kuwabara, T., Gage, F.H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Hannon, G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Choi, Y.D., Dreyfuss, G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F., Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich, K.P., Ikura, M. Radixin: Cytoskeletal adopter and signaling protein. Int. J. Biochem. Cell Biol. 2004;36:2131–2136. doi: 10.1016/j.biocel.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Hwang, H.W., Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N. Small RNAs: Classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P., Lamprinaki, S., Sharma, A., Mourelatos, Z. Detection of microRNAs and assays to monitor microRNA activities in vivo and in vitro. Methods Mol. Biol. 2005;309:295–310. doi: 10.1385/1-59259-935-4:295. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Maniataki, E., De Planell Saguer, M.D., Mourelatos, Z. Immunoprecipitation of microRNPs and directional cloning of microRNAs. Methods Mol. Biol. 2005;309:283–294. doi: 10.1385/1-59259-935-4:283. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E., Mourelatos, Z. The microRNA world: Small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- O'Donnell, K.A., Boeke, J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen, T., Nicol, C., McConkey, G., Dalmay, T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Shiohama, A., Minoshima, S., Shimizu, N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- Sewer, A., Paul, N., Landgraf, P., Aravin, A., Pfeffer, S., Brownstein, M.J., Tuschl, T., van Nimwegen, E., Zavolan, M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiggard, W.J., O'Doherty, U., McGain, D., Jeyakumar, D., Malim, M.H. Long HIV type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Res. Hum. Retroviruses. 2004;20:285–295. doi: 10.1089/088922204322996527. [DOI] [PubMed] [Google Scholar]
- Tijsterman, M., Plasterk, R.H. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, M., Miller, C.P., Papayannopoulou, T., Stamatoyannopoulos, G., Song, C.Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp. Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]