Abstract
Recombinant adeno-associated virus 2 (AAV) vectors transduction efficiency varies greatly in different cell types. We have described that a cellular protein, FKBP52, in its phosphorylated form interacts with the D-sequence in the viral inverted terminal repeat, inhibits viral second strand DNA synthesis, and limits transgene expression. Here we investigated the role of cellular heat-shock protein 90 (HSP90) in AAV transduction because FKBP52 forms a complex with HSP90, and because heat-shock treatment augments AAV transduction efficiency. Heat-shock treatment of HeLa cells resulted in tyrosine dephosphorylation of FKBP52, led to stabilization of the FKBP52-HSP90 complex, and resulted in ∼6-fold increase in AAV transduction. However, when HeLa cells were pre-treated with tyrphostin 23, a specific inhibitor of cellular epidermal growth factor receptor tyrosine kinase, which phosphorylates FKBP52 at tyrosine residues, heat-shock treatment resulted in a further 18-fold increase in AAV transduction. HSP90 was shown to be a part of the FKBP52-AAV D-sequence complex, but HSP90 by itself did not bind to the D-sequence. Geldanamycin treatment, which disrupts the HSP90-FKBP52 complex, resulted in >22-fold increase in AAV transduction in heat-shock-treated cells compared with heat shock alone. Deliberate overexpression of the human HSP90 gene resulted in a significant decrease in AAV-mediated transduction in tyrphostin 23-treated cells, whereas down-modulation of HSP90 levels led to a decrease in HSP90-FKBP52-AAV D-sequence complex formation, resulting in a significant increase in AAV transduction following pre-treatment with tyrphostin 23. These studies suggest that the observed increase in AAV transduction efficiency following heat-shock treatment is unlikely to be mediated by HSP90 alone and that increased levels of HSP90, in the absence of heat shock, facilitate binding of FKBP52 to the AAV D-sequence, thereby leading to inhibition of AAV-mediated transgene expression. These studies have implications in the optimal use of recombinant AAV vectors in human gene therapy.
Adeno-associated virus (AAV),1 type 2, a non-pathogenic human parvovirus, has gained attention as a vector for gene transfer and gene therapy (1-25). Although recombinant AAV vectors are currently in use in phase I/II clinical trials for gene therapy of cystic fibrosis and hemophilia B (1, 5, 8, 11, 22), and have been shown to transduce a wide variety of cells and tissues in vitro and in vivo (2-4, 6, 7, 9, 10, 12-21, 23-25), their transduction efficiency varies widely in different cell types. We and others have undertaken systematic studies to delineate various steps in the life cycle of AAV, which include viral binding and entry (26-28), intracellular trafficking (29-35), nuclear transport (29, 30, 34-37), and viral second strand DNA synthesis (38-47). The viral second strand DNA synthesis has been described by two independent laboratories to be the rate-limiting step, which leads to inefficient transduction by AAV vectors (38, 39). We have reported that a 52-kDa cellular protein, FKBP52, which binds the immunosuppressant drug FK506 (48, 49), interacts specifically with the D-sequence within the inverted terminal repeat of the AAV genome, is phosphorylated at tyrosine residues by the cellular epidermal growth factor receptor protein-tyrosine kinase (EGFR-PTK), and inhibits the viral second strand DNA synthesis leading to inefficient transgene expression (40-47). FKBP52 is a cellular chaperone protein (48-50) and has been documented to interact with a 90-kDa cellular heat-shock protein (HSP90) to form a complex, which has been shown to mediate import of a number of cellular and viral proteins to the nucleus (51-55). Although AAV transduction efficiency can be increased following heat-shock treatment (38, 56), it remains unclear what role, if any, HSP90 plays in modulating FKBP52 interaction with the AAV D-sequence, and whether FKBP52-HSP90 complex is involved in modulating AAV-mediated transgene expression.
In this report, we present evidence that HSP90 is a part of the FKBP52-AAV D-sequence complex, but HSP90 itself does not bind to the D-sequence directly, although it can be phosphorylated at both serine/threonine and tyrosine residues in vitro. We also demonstrate that the poor binding of unphosphorylated FKBP52 to the D-sequence can be significantly enhanced in the presence of unphosphorylated HSP90. Furthermore, heat-shock treatment of HeLa cells, which leads to overexpression of HSP90, also leads to tyrosine dephosphorylation of FKBP52, which in turn results in an increase in AAV transduction efficiency. Treatment with geldanamycin, a specific inhibitor of HSP90 which dissociates HSP90 from FKBP52, also increases AAV transduction efficiency in heat-shock-treated cells. These data led to the hypothesis that increased levels of HSP90 in heat-shock-treated cells facilitate the dissociation of FKBP52 from AAV D-sequence and thereby lead to a more efficient viral second strand DNA synthesis and, consequently, efficient transgene expression. This hypothesis was tested by deliberate overexpression of the human HSP90 gene in HeLa cells. However, this led to a significant decrease in AAV transduction efficiency. In contrast, stable transfection of the HSP90 gene in the antisense orientation reduced the levels of HSP90 in these cells, which led to a decrease in the HSP90-FKBP52-AAV D-sequence complex formation, and resulted in a significant increase in AAV-mediated transgene expression. These studies corroborate the notion that although HSP90 is a part of the complex between FKBP52 and the AAV D-sequence, the observed increase in AAV transduction efficiency following heat-shock treatment is unlikely to be mediated by increased levels of HSP90 alone. Our data also suggest that increased levels of HSP90, in the absence of heat-shock, facilitate binding of FKBP52 to the AAV D-sequence, and thus lead to inhibition of AAV-mediated transgene expression. Conversely, down-modulation of HSP90 levels decrease the extent of the complex formation between serine/threonine-phosphorylated forms of FKBP52 and the AAV D-sequence, resulting in augmentation in AAV-mediated transgene expression. We propose a model that helps explain the available data on the interaction between FKBP52 and HSP90 and its role in AAV-mediated transgene expression.
EXPERIMENTAL PROCEDURES
Cells, Virus, Plasmids, Antibodies, and Chemicals
The human cervical carcinoma cell line, HeLa, was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained as mono-layer cultures in Iscove's-modified Dulbecco's medium (IMDM) supplemented with 10% newborn calf serum and 1% (by volume) 100× stock solution of antibiotics (10,000 units of penicillin + 10,000 μg of streptomycin). Highly purified stocks of a recombinant AAV vector containing the β-galactosidase (lacZ) reporter gene driven by the cytomegalovirus (CMV) immediate-early promoter (vCMVp-lacZ) were generated as described previously (14, 15). Physical particle titers of recombinant vector stocks were determined by quantitative DNA slot blot analysis, as described previously (57). Plasmids pQE-30 and pCMV-IRES2-EGFP were purchased from Qiagen (Valencia, CA) and Clontech (Palo Alto, CA), respectively. The plasmid pH6HSP90 was generously provided by Dr. Takayuki K. Nemoto (Nagasaki University, Nagasaki, Japan). Antibodies specific for human FKBP52, HSP90 (goat polyclonal IgG), and normal goat IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies specific for FKBP52 and HSP90 (mouse monoclonal IgG1) were obtained from Calbiochem. All chemicals used were purchased from Calbiochem or Sigma.
Preparation of Whole Cell Extracts (WCE)
WCE from HeLa cells were prepared according to the method of Muller (58). In some experiments, WCE were prepared from HeLa cells following heat-shock treatment at 42.5 °C for 4 h. Total protein concentration was determined by the Bio-Rad protein assay kit (Bio-Rad), and the extracts were frozen in liquid N2 and stored at −70 °C.
Electrophoretic Mobility Shift Assays (EMSA)
EMSA were performed as described previously (45, 47). Briefly, DNA-binding reactions were performed in a volume of 20 μl with 2 μg of poly(dI-dC), 2 μg of bovine serum albumin, and 12% glycerol in Tris buffer (10 mm Tris-HCl, 0.5 mm dithiothreitol, 25 mm NaCl, 5 mm MgCl2, 0.05% Nonidet P-40, pH 8.0). Ten μg of proteins of each WCE were pre-incubated for 10 min at 25 °C followed by the addition of 10,000 cpm of 32P-labeled D-sequence synthetic oligonucleotide (5′-AGGAACCCCTAGTGATGGAG-3′) in the reaction mixture. The binding reaction was allowed to proceed for 30 min at 25 °C. In some experiments, specific FKBP52 and HSP90 antibodies (goat IgG) were used in supershift assays as described previously (43, 59, 60). Bound complexes were separated from the free probe on low ionic strength 4% polyacrylamide gels with Tris-glycine/EDTA buffer, pH 8.5, containing 50 mm Tris-HCl, 380 mm glycine, and 2 mm EDTA. Following electrophoresis, the gel was dried in vacuum and autoradiographed at −70 °C using Kodak X-Omat film. Densito-metric scanning of autoradiographs for the quantitation of FKBP52-D-sequence complex formation was evaluated using ImageJ analysis software (National Institutes of Health, Bethesda).
In Vitro Phosphorylation Assays
In vitro phosphorylation by casein kinase II (CKII, Calbiochem) was carried out as described previously by McElhinny et al. (61), Russo et al. (62), and Qing et al. (43). Briefly, the complete reaction mixture contained 1 μg of the purified FKBP52 or HSP90 protein (Stressgen, Victoria, British Columbia, Canada), 20 mm Tris-HCl, 50 mm KCl, 10 mm MgCl2, 50 mm Na3VO4, 10 μCi (0.37 mBq) of [γ-32P]ATP, and 500 units (500,000 units/ml) of purified CKII. In vitro phosphorylation by epidermal growth factor receptor tyrosine kinase (EGFR-PTK) was carried out as described previously by Weber et al. (63), Cybulsky et al. (64), and Qing et al. (43), with the following modifications. The complete reaction mixture contained 1 μg of the purified FKBP52 or HSP90 protein, 20 mm HEPES, 4 mm MgCl2, 10 mm MnCl2, 50 mm Na3VO4, 10 μCi (0.37 mBq) of [γ-32P]ATP, and 10 units (15,000 units/mg) of purified EGFR-PTK (Sigma) with the appropriate controls as indicated. The reaction mixtures were incubated at 30 °C for 1 h. The phosphorylated FKBP52 or HSP90 proteins were separated from free [γ-32P]ATP on 10% SDS-polyacrylamide gels followed by autoradiography with Kodak X-Omat film at −70 °C. In some experiments, in vitro phosphorylation assays by both CKII and EGFR-PTK were also carried out by adding 200 μm ATP instead of [γ-32P]ATP. The phosphorylated FKBP52 or HSP90 proteins were then used in EMSA with the radiolabeled AAV D-sequence probe as described above.
In Vitro Binding Assays
In vitro binding assays with purified HSP90 and FKBP52 were carried out as described previously (49). Briefly, 2 μg of FKBP52 and 3 μg of HSP90 were mixed in a buffer containing 25 mm HEPES, 15 mm Tris, 50 mm KCl, 30 mm NaCl, 2.5 mm MgCl2, 1 mm EDTA, 10 mm 2-mercaptoethanol, 0.01% Tween 20, and 10% glycerol, pH 7.6, and incubated at 30 °C for 1 h. The mixture was analyzed on 10% native polyacrylamide gels and used in EMSA with the radiolabeled AAV D-sequence probe as described above.
Preparation of Whole Cell Lysates (WCL) and Co-immunoprecipitation
WCL were prepared as described previously (45), with the following modifications. Briefly, HeLa cells were either mock-treated or treated with 10 μm geldanamycin or 1 μm radicicol and lysed in NETN buffer (0.5% Nonidet P-40 (Roche Applied Science), 1 mm EDTA, 50 mm Tris-HCl, pH 8.0, 120 mm NaCl) containing 1 mm dithiothreitol, 10 mm NaF, 2 mm Na3VO4, 0.5 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin. Equivalent amounts (∼500 μg) of WCLs were cleared of nonspecific binding by incubation with 0.25 μg of normal goat IgG together with 20 μl of protein G-agarose beads for 30 min at 4 °C in an orbital shaker. After pre-clearing, 2 μg of polyclonal anti-FKBP52 antibody (goat IgG) or 2 μg of normal goat IgG (as a negative control) were added and incubated at 4 °C for 1 h, followed by precipitation with protein G-agarose beads at 4 °C for 12 h in the shaker. Pellets were collected by centrifugation at 2,500 rpm for 5 min at 4 °C and washed four times with phosphate-buffered saline. After the final wash, the supernatant was aspirated and discarded, and the pellet was resuspended in 40 μl of 1× SDS sample buffer. Twenty μl of resuspended pellet solutions were used for Western blotting with monoclonal anti-FKBP52 and anti-HSP90 antibodies (mouse IgG1) as described below.
Western Blot Analyses
Western blotting was performed as described previously (65). For immunoprecipitation, resuspended pellet solutions were boiled for 2–3 min. Twenty μl of sample was separated by 10% SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Membranes were blocked at 4 °C for 12 h with 5% nonfat milk in 1× Tris-buffered saline (TBS, 20 mm Tris-HCl, pH 7.5, 150 mm NaCl). Membranes were treated with monoclonal anti-FKBP52 antibody (1:1,000 dilution) or monoclonal anti-HSP90 antibody followed by horseradish peroxidase-conjugated anti-mouse IgG1 (1:7,000 dilution). To determine the levels of human HSP90 in transfected HeLa cells, ∼1 × 106 cells were seeded in culture dishes for 24 h, and WCLs were prepared as described above. Equivalent amounts (∼10 μg) of WCLs were electrophoresed on SDS-polyacrylamide gels and transferred to Immobilon-P membranes. Membranes were treated with polyclonal anti-HSP90 antibody (1:500 dilution) followed by horseradish peroxidase-conjugated anti-goat IgG (1:7,000 dilution). Immunoreactive bands were visualized using chemiluminescence (ECL-Plus, Amer-sham Biosciences) and evaluated using ImageJ analysis software as described above.
Construction of Sense and Antisense Plasmid Vectors and Stable Transfection
The eukaryotic expression plasmids containing the cDNA sequence for human HSP90, in both sense and antisense orientations, under the control of the CMV immediate-early promoter were constructed by standard methods (26). Briefly, HSP90 cDNA was obtained from plasmid pH6HSP90 following digestion with BamHI and SmaI and subcloned into the pCMV-IRES2-EGFP vector at the SmaI site by the blunt-ended cloning method. Approximately 4 × 105 HeLa cells were seeded into each well in a 6-well plate in IMDM with 10% newborn calf serum for 24 h prior to transfection. Approximately 4 μg of each plasmid carrying either the sense or the antisense HSP90 sequences were added to cells using LipofectAMINE™ Reagent (Invitrogen) according to the manufacturer's instructions. Twenty-four hours post-transfection, cells were cultured in the selective medium containing 400 μg/ml geneticin (G418) (Invitrogen). Clones containing HSP90 sense and antisense vectors were obtained after 14 days of selection. Positive clones were identified by Western blotting using anti-HSP90 antibody as described above. Densitometric scanning was performed to determine the levels of HSP90 in control HeLa cells and in each individual clone.
Recombinant AAV Vector Transduction Assay
Approximately 1 × 105 HeLa cells were plated in each well in a 12-well plate and incubated at 37 °C for 12 h. Cells were washed once with IMDM and then infected at 37 °C for 2 h with 2 × 103 particles per cell of a recombinant AAV-lacZ vector as described previously (43). Cells were incubated in complete IMDM containing 10% newborn calf serum and 1% antibiotics for 48 h. The β-galactosidase activity was measured using the Galacto-Light Plus chemiluminescent reporter assay (Tropix, Inc., Bedford, MA) according to the manufacturer's instructions. Data were expressed as relative light units per μg of total protein and were within the linear range of the assay.
RESULTS
HSP90 Is a Part of the FKBP52-AAV D-sequence Complex
In our studies published previously (42-45), we documented that phosphorylated forms of cellular FKBP52 protein bind efficiently to the single-stranded D-sequence in the viral inverted terminal repeat. Because heat-shock treatment has been shown to augment AAV transduction efficiency (38, 56), and because FKBP52 is known to form a complex with a cellular heat-shock protein, HSP90 (48, 50), we addressed the following two questions. First, does the heat-shock treatment lead to dephosphorylation of FKBP52? Second, is HSP90 a part of the FKBP52-AAV D-sequence complex? EMSA were performed using WCE prepared from HeLa cells because these cells contain FKBP52 predominantly phosphorylated at tyrosine residues (42-45). These results are shown in Fig. 1. Consistent with previously published data (42-45), the AAV D-sequence probe (lane 1) formed a complex with the tyrosine-phosphorylated form of FKBP52 (closed arrow, lane 2). When WCE prepared from HeLa cells treated at 42.5 °C for 4 h (heat-shock) were used, the same probe formed a complex that migrated faster because of tyrosine dephosphorylation of FKBP52 (closed arrowhead, lane 3). We have previously identified this band to be the serine/threonine-phosphorylated form of FKBP52 (43). Because the serine/threonine-phosphorylated form of FKBP52 is less inhibitory than the tyrosine-phosphorylated form for AAV second strand synthesis (43), these data are consistent with the conclusion that heat-shock treatment augments AAV transduction efficiency by mediating tyrosine dephosphorylation of FKBP52. When anti-FKBP52 antibodies were included in these assays, a supershift was seen with WCE prepared from untreated HeLa cells (open arrow, lane 4), which is consistent with our previously published reports (42, 43). It is interesting to note, however, that when WCE prepared from HeLa cells following heat-shock treatment were used, the inclusion of anti-FKBP52 antibodies led to a supershifted band that migrated slower than that with untreated HeLa cells (open arrowhead, lane 5). Because heat-shock treatment leads to an increase in HSP90 levels (66, 67), we suspected that this supershift was due most likely to the interaction of HSP90 with the FKBP52-AAV D-sequence complex. This possibility was tested by using anti-HSP90 antibodies in these assays. As is evident, anti-HSP90 antibodies also caused a supershift with WCE prepared from untreated HeLa cells that was indistinguishable from that with anti-FKBP52 antibodies (lane 6). Similar results were obtained when WCE prepared from HeLa cells following heat-shock were used (lane 7). These results corroborate the notion that (i) heat shock leads to tyrosine dephosphorylation of FKBP52, (ii) HSP90 is a part of the FKBP52-AAV D-sequence complex, and (iii) both tyrosine- and serine/threonine-phosphorylated forms of FKBP52 can interact with the AAV D-sequence-HSP90 complex. However, it remained unclear whether HSP90 interacts with the AAV D-sequence directly or indirectly through FKBP52. This was experimentally tested as follows.
Fig. 1. Electrophoretic mobility shift assays for the tyrosine phosphorylation status of FKBP52 in HeLa cells following heat-shock treatment and the involvement of HSP90 in the complex formation with the AAV D-sequence.

The 20-nucleotide 32P-labeled AAV D-sequence probe (lane 1) was incubated with WCE prepared from HeLa cells without treatment (lane 2), or following heat-shock treatment (lane 3). Untreated HeLa cells contain FKBP52 phosphorylated predominantly at tyrosine residues (closed arrow). In heat-shock-treated cells, FKBP52 undergoes tyrosine dephosphorylation (closed arrow). The FKBP52-AAV D-sequence complex is supershifted with incubation with anti-FKBP52 antibody (lane 4, open arrow). Following heat-shock treatment, the supershifted band migrates slower (lane 5, open arrowhead). Similar supershifted bands are seen with anti-HSP90 antibodies (lanes 6 and 7).
HSP90 Can Be Phosphorylated at Both Serine/Threonine and Tyrosine Residues but Does Not Bind to AAV D-sequence Directly
Previous studies (49, 68) have shown that HSP90 can be phosphorylated at serine/threonine residues by casein kinase II (CKII) in vitro. Whether HSP90 can also be phosphorylated at tyrosine residues has not been reported. We have reported previously (43) that FKBP52 can be phosphorylated at both serine/threonine and tyrosine residues by CKII and EGFR protein-tyrosine kinase, respectively. By using the same in vitro phosphorylation assays, we first examined whether purified HSP90 could be phosphorylated by EGFR. Purified FKBP52 was used as a positive control. These results, shown in Fig. 2A, clearly indicate that purified FKBP52 (lanes 2 and 7) and HSP90 (lanes 3 and 8) can be readily phosphorylated in vitro by both CKII and EGFR at serine/threonine and tyrosine residues, respectively. It was of interest to next determine whether the purified HSP90, with and without prior phosphorylation at serine/threonine or tyrosine residues, could bind to the AAV D-sequence. Purified FKBP52 and HSP90 proteins, with and without prior in vitro phosphorylation by CKII or EGFR, were used in EMSA with the D-sequence probe. These results are shown in Fig. 2B. As can be seen, although the free probe (lane 1) did not interact with CKII (lane 2), and little binding of unphosphorylated FKBP52 to the D-sequence probe occurred (closed arrowhead, lane 3), FKBP52 phosphorylated at serine/threonine residues by CKII formed a complex with the probe (arrow, lane 4), which is consistent with our report published previously (43). It is evident, however, that neither unphosphorylated HSP90 (lane 5), nor serine/threonine-phosphorylated HSP90 (lane 6), nor tyrosine-phosphorylated HSP90 (lane 8) interacts with the AAV D-sequence probe. We conclude from these results that HSP90 is a part of the FKBP52-AAV D-sequence complex, but by itself does not bind to the AAV D-sequence.
Fig. 2. Electrophoretic mobility shift assays for interaction between AAV D-sequence and unphosphorylated or phosphorylated forms of purified FKBP52 and HSP90.
A, autoradiographic image of SDS-polyacrylamide gel for in vitro phosphorylation of purified FKBP52 and HSP90 by CKII and EGFR-PTK. CKII was incubated in the absence (lane 1) or presence of 1 μg each of FKBP52 (lane 2) or HSP90 (lane 3). FKBP52 and HSP90 were incubated in the absence of a protein kinase (lanes 4 and 5). Similarly, EGFR-PTK was incubated in the absence (lane 6) or presence of FKBP52 (lane 7) or HSP90 (lane 8) as described under ”Experimental Procedures.“ The arrows indicate the phosphorylated forms of FKBP52 and HSP90, and the closed and open arrowheads denote the autophosphorylated CKII and EGFR-PTK, respectively. B, the radiolabeled AAV D-sequence probe (lane 1) does not interact with casein kinase II (lane 2) and interacts poorly with unphosphorylated FKBP52 (closed arrowhead, lane 3), although prior in vitro phosphorylation of FKBP52 with CKII allows the complex formation (arrow, lane 4). HSP90, on the other hand, fails to interact with AAV D-sequence either in its unphosphorylated form (lane 5) or following phosphorylation at serine/threonine residues by CKII (lane 6), or at tyrosine residues by EGFR protein-tyrosine kinase (lane 8). EGFR alone does not interact with the D-sequence (lane 7).
Heat-shock Treatment Promotes the FKBP52-HSP90 Complex Formation, and HSP90 Can Be Dissociated from FKBP52 Complex by Treatment with Geldanamycin
Because all the data obtained thus far were from cell-free studies in vitro, we next examined the effect of heat-shock treatment on the FKBP52-HSP90 complex in intact cells. The following two types of experiments were carried out. In the first set, HeLa cells were either mock-treated or treated with geldanamycin, a benzoquinone ansamycin, which is known to dissociate the FKBP52-HSP90 complex (51, 69-71). In the second set, HeLa cells were subjected to heat-shock treatment at 42.5 °C for 4 h. WCL were prepared, and equivalent amounts of proteins were analyzed on Western blots using anti-HSP90 or anti-FKBP52 antibodies. Equivalent amounts of WCL were also immunoprecipitated first with anti-FKBP52 antibodies followed by Western blot analyses. These results are shown in Fig. 3A. As can be seen, although the total amount of HSP90 in untreated HeLa cells (lane 1) remained unchanged in geldanamycin-treated cells (lane 2), heat-shock treatment caused a slight increase in the HSP90 level (lane 3), as has been reported previously (66, 72). On the other hand, the total amount of FKBP52 did not change significantly in any of the conditions (Fig. 3B, lanes 1–3). When WCL were immunoprecipitated with anti-FKBP52 antibodies prior to Western blot analyses, the total amount of HSP90 in geldanamycin-treated HeLa cells (Fig. 3A, lane 6) was significantly reduced compared with that in untreated cells (lane 5), and the heat-shock treatment significantly increased the complex formation between HSP90 and FKBP52 (lane 7). The total amounts of FKBP52 were approximately the same (Fig. 3B, lanes 5–7). Immunoprecipitation with normal goat IgG (lane 4) was performed as a negative control. These results confirm that heat shock increases HSP90 expression, which in turn allows for a more efficient complex formation between FKBP52 and HSP90, and that this complex can be dissociated following treatment with geldanamycin.
Fig. 3. Autoradiographic image of SDS-polyacrylamide gels for co-immunoprecipitation of FKBP52-HSP90 complex in vivo.
A, Western blot (WB) analyses of whole cell lysates (WCL) prepared from untreated HeLa cells (lane 1) and following treatment with geldanamycin (lane 2) or heat shock (lane 3) were probed with anti-HSP90 antibody. The same WCL were also first immunoprecipitated (IP) with either goat IgG or anti-FKBP52 antibody followed by Western blot analyses (lanes 4–7). The immunoprecipitated HSP90 band is denoted by the left arrow. B, an identical blot to that in A was probed with anti-FKBP52 antibody. The right arrow indicates the immunoprecipitated FKBP52 band.
Dissociation of HSP90-FKBP52-AAV D-sequence Complex Leads to a Significant Increase in AAV-mediated Transgene Expression but Only in Tyrphostin 23- and Heat-shock-treated Cells
Based on our experiments, to be described later (see Fig. 7A), in which we observed that HSP90 stabilizes the FKBP52-AAV D-sequence complex and prevents FKBP52 from dissociating from the D-sequence, we reasoned that increased levels of HSP90 would inhibit AAV second strand synthesis and therefore decrease AAV-mediated transgene expression. We wished to determine whether dissociation of the HSP90-FKBP52 complex would lead to an increase in AAV transduction efficiency. HeLa cells were treated with geldanamycin, known to dissociate the FKBP52-HSP90 complex (51, 69-71) (Fig. 3), and transduced with the recombinant AAV-lacZ vector, and the transgene expression was determined 48 h post-transduction. Mock-transduced cells, and cells treated with tyrphostin 23, a specific inhibitor of EGFR-PTK (41), and heat shock, both known to augment AAV transduction efficiency (38, 41, 56), were used as appropriate controls. These results are shown in Fig. 4. Consistent with studies published previously (38, 41, 56), both tyrphostin 23 and heat-shock treatments led to ∼6-fold increase in AAV transduction efficiency. Surprisingly, geldanamycin treatment had little effect. Treatment with radicicol, another HSP90 inhibitor, also did not affect the transduction efficiency. Most interesting, however, in heat-shock-treated cells, both tyrphostin 23 and geldanamycin increased the transduction efficiency ∼18- and ∼22-fold, respectively. A significant increase in the transduction efficiency was also observed with radicicol in heat-shock-treated cells, and treatment with tyrphostin 23 + geldanamycin also led to an increase in the AAV transduction efficiency. The simplest explanation to account for these results is that tyrosine dephosphorylation of FKBP52, which is mediated by both tyrphostin 23 and heat-shock treatments, is a prerequisite for geldanamycin-mediated dissociation of the FKBP52-HSP90 complex, which in turn leads to efficient viral second strand DNA synthesis resulting in an increase in the AAV transduction efficiency. However, it is intriguing that the heat-shock treatment increases AAV transduction efficiency, presumably through increased levels of HSP90, which should stabilize the FKBP52-AAV D-sequence complex and thus inhibit the viral second strand synthesis and transgene expression. This apparent paradox was pursued experimentally as follows.
Fig. 7. Electrophoretic mobility shift assays for the role of HSP90 levels on the interaction between AAV D-sequence and unphosphorylated and tyrosine- and serine/threonine-phosphorylated forms of FKBP52.

A, although the free probe (lane 1) interacts poorly with FKBP52 (closed arrowhead, lane 2) and does not interact with HSP90 (lane 3), its interaction is significantly pronounced when FKBP52 is complexed with HSP90 (arrow, lane 4). These assays were performed as described under “Experimental Procedures.” B, EMSA were performed using WCE prepared from HeLa cells, or HeLa cells stably transfected with empty vector, or HSP90 sense or antisense HSP90 expression vectors. C, WCE prepared from cells treated with either 500 μm tyrphostin 23 or heat-shock were used. The probe formed a complex with tyrosine-dephosphorylated FKBP52 in each case (closed arrowhead, lanes 1–6). In the HSP90 sense clone, pretreatment with tyrphostin 23 or heat shock led to a slight increase in the extent of the complex formation between tyrosine-dephosphorylated FKBP52 and the AAV D-sequence (lanes 3 and 4), compared with control cells (lanes 1 and 2). In the HSP90 antisense clone, although heat-shock had no significant effect (lane 6), pretreatment with tyrphostin 23 significantly decreased the extent of the complex formation between serine/threonine-phosphorylated FKBP52 and the AAV D-sequence (lane 5).
Fig. 4. Comparative analyses of AAV-mediated transduction efficiency in HeLa cells following various treatments.
Equivalent numbers of cells were either mock-treated or treated with tyrphostin 23, heat shock, geldanamycin, or radicicol, and various combinations thereof, and either mock-infected (Mock) or infected (Control) with the recombinant AAV-lacZ vector under identical conditions. Transgene expression was determined 48 h post-infection. Transgene expression was evaluated 48 h post-infection. These data, expressed as relative light units (RLU) per μg of total protein, were within the linear range of the assay and represent average values from two separate experiments performed in triplicate. *, p < 0.05; **, p < 0.001 versus control; ‡, p < 0.001 versus tyrphostin 23.
AAV Transduction Efficiency Is Inhibited by Deliberate Overexpression of HSP90 and Is Significantly Augmented Following Down-modulation of HSP90 Levels
The heat-shock treatment undoubtedly has a pleiotropic effect on HeLa cells, and although increased levels of HSP90 is one of the consequences of this treatment, it remains unclear whether the increased HSP90 levels directly mediate the observed increase in AAV transduction efficiency. In order to directly examine the role of HSP90 on AAV-mediated transgene expression in intact cells, we constructed eukaryotic expression plasmids containing the human HSP90 gene, in both sense and antisense orientations, under the control of the CMV promoter. HeLa cells were stably transfected with these plasmids separately, and individual clones were isolated. WCLs prepared from these clones were analyzed to detect the HSP90 levels by Western blot analyses using anti-HSP90 antibodies. Three representative clones from each group were pretreated with tyrphostin 23 and transduced with the recombinant AAV-lacZ vector. Transgene expression was determined 48 h post-transduction. Mock-transfected HeLa cells, with or without pre-treatment with tyrphostin 23, were used as appropriate controls. These results are shown in Fig. 5A. As can be seen, whereas tyrphostin 23 treatment led to ∼6-fold increase in AAV transduction efficiency in control HeLa cells, in each of the clones overexpressing HSP90 (Fig. 5B, lanes 2–4), the transgene expression was significantly inhibited, even following pre-treatment with tyrphostin. In contrast, in clones in which the endogenous levels of HSP90 were down-regulated (Fig. 5B, lanes 5–7), an ∼10–15-fold increase in AAV transduction efficiency was observed. These data strongly suggest that there is an inverse correlation between HSP90 levels and the efficiency of AAV-mediated transgene expression and that the observed increase in AAV transduction efficiency following the heat-shock treatment is unlikely to be mediated by increased levels of HSP90 alone.
Fig. 5. Comparative analyses of AAV-mediated transduction efficiency in HeLa cell clones stably transfected with HSP90 expression plasmids in sense and antisense orientations.

A, HeLa cells were either mock-infected (Mock) or infected in the absence (Control) or presence of tyrphostin 23 as appropriate controls. Three individual HeLa cell sense and antisense HSP90 clones were infected with the recombinant AAV-lacZ vector under identical conditions, and transgene expression was evaluated 48 h post-infection as described in the legend to Fig. 4. *, p < 0.001 versus control; ‡, p < 0.001 versus tyrphostin 23. B, Western blot analysis to document the levels of expression of HSP90 in control HeLa cells (lane 1) and in each of the three sense (lanes 2–4, average increase ∼2.2-fold) and antisense (lanes 5–7, average decrease ∼2.4-fold) HSP90 clones.
Down-modulation of HSP90 Levels Decreases the HSP90-FKBP52-AAV D-sequence Complex Formation
Because we documented that the heat-shock treatment resulted in an increase in the FKBP52-HSP90 complex formation in intact cells (Fig. 3), and that down-modulation of HSP90 levels augmented AAV transduction efficiency (Fig. 5), it was next of interest to examine the role of HSP90 in the FKBP52-AAV D-sequence complex formation in cells that either overexpressed HSP90 or in which the levels of HSP90 were down-regulated. WCE prepared from HeLa cell clones stably transfected with a control plasmid vector and with sense- or antisense HSP90 expression plasmid vectors were used in supershift EMSA using anti-HSP90 antibodies. These results are shown in Fig. 6. As can be seen, although the supershifted band by anti-HSP90 antibody in HeLa HSP90-sense clone (lane 4) remained unchanged compared with that in HeLa empty vector clone (lane 3), down-modulation of HSP90 levels in HeLa HSP90 antisense clone resulted in a significant decrease in the HSP90-FKBP52-AAV D-sequence complex formation (open arrowhead, lane 5). On the other hand, the supershifted band caused by anti-FKBP52 antibody did not change significantly in any of the conditions (data not shown, but see Fig. 7). Although not quantitative, we conclude from these data that HSP90 levels have a significant effect on the HSP90-FKBP52-AAV D-sequence complex formation. Whether alterations in the levels of HSP90 also affected the extent of tyrosine-dephosphorylated FKBP52-AAV D-sequence complex formation, which would correlate with AAV transduction efficiency, was tested as follows.
Fig. 6. Electrophoretic mobility shift assays for HSP90-FKBP52-AAV D-sequence complex formation in sense-HSP90 and antisense-HSP90 HeLa cells clones.

The 20-nucleotide 32P-labeled AAV D-sequence probe (lane 1) was incubated with WCEs prepared from HeLa cells without treatment (lane 2), or HeLa cells stably transfected with the empty vector, or sense- or antisense-HSP90 expression vectors in the presence of anti-HSP90 antibody (lanes 3–5). HeLa cells contain FKBP52 phosphorylated predominantly at tyrosine residues (arrow). Down-modulation of HSP90 levels in HeLa HSP90 antisense clone leads to a significant decrease in the supershift band formation following incubation with anti-HSP90 antibody (lane 5, open arrowhead).
Down-modulation of HSP90 Levels Reduce the Extent of the Tyrosine-dephosphorylated FKBP52-AAV D-sequence Complex Formation
Although unphosphorylated FKBP52 binds to the AAV D-sequence poorly (43), and HSP90 levels have little effect on the extent of the tyrosine-phosphorylated FKBP52-AAV D-sequence complex formation (Fig. 6), we wished to determine whether HSP90 had any effect on unphosphorylated or tyrosine-dephosphorylated FKBP52-AAV D-sequence complex formation. The following two sets of experiments were performed. In the first set, purified FKBP52 and HSP90 proteins, in their unphosphorylated forms, were used in EMSA with the AAV D-sequence probe. Consistent with our previous studies (43), the results, shown in Fig. 7A, document that although FKBP52 bound poorly with the probe (lane 2, closed arrowhead), and HSP90 did not interact with the probe at all (lane 3), the FKBP52-HSP90 complex interacted efficiently with the AAV D-sequence probe (arrow, lane 4). Because HSP90 stabilizes the binding of unphosphorylated FKBP52 with the AAV D-sequence, increased levels of HSP90 would be expected to lead to inhibition of AAV-mediated transgene expression, as has been observed (Fig. 5). Because these data were obtained from cell-free studies in vitro, in the second set of experiments, we examined whether HSP90 levels also affect the FKBP52-AAV D-sequence complex in intact cells. EMSA were performed using WCE prepared from control HeLa cells or HSP90 sense-and antisense HeLa cell clones. These results are shown in Fig. 7B. The AAV D-sequence probe (lane 1) formed a complex with the tyrosine-phosphorylated form of FKBP52 (arrow, lanes 2–5), the relative levels of which were determined by densitometric scanning to be very similar (densities of 4,721, 4,261, 4,164, and 4,425, respectively). Thus, HSP90 levels have no significant effect on tyrosine-phosphorylated forms of FKBP52 complexed with the AAV D-sequence. However, as can be seen in Fig. 7C, when cells were treated either with tyrphostin 23 or heat shock, the probe formed a complex that migrated faster because of tyrosine dephosphorylation of FKBP52 (closed arrowhead, lanes 1–6), consistent with our previous studies (41-45). It is interesting that in HSP90 sense clone, pretreatment with tyrphostin 23 or heat shock led to an increase in the FKBP52-AAV D-sequence complex formation (lane 3 and 4; densities of 5,871 and 5,753, respectively) compared with control cells (lanes 1 and 2; densities of 4,172 and 4,309, respectively). Moreover, in HSP90 antisense clone, although heat-shock treatment did not significantly change the extent of the complex formation (lane 6, density of 4,581), pretreatment with tyrphostin 23 significantly decreased the complex formation (lane 5, density of 1,705). Therefore, we conclude that increased levels of HSP90 can facilitate the binding of unphosphorylated FKBP52 to the AAV D-sequence as well as stabilize the tyrosine-dephosphorylated FKBP52-AAV D-sequence complex formation resulting in inefficient AAV second strand DNA synthesis and, consequently, transgene expression in the absence of heat shock. Down-modulation of HSP90 levels, on the other hand, leads to a significant decrease in the FKBP52-AAV D-sequence complex formation leading to augmentation in AAV transduction efficiency.
DISCUSSION
A reasonable picture of various steps in the life cycle of AAV has emerged in the recent past. For example, a cell surface receptor, heparan sulfate proteoglycan for viral binding (28), and co-receptors, fibroblast growth factor receptor 1 and/or αVβ5 integrin for viral entry (26, 27), have been identified. Detailed studies on the mechanisms of intracellular viral trafficking and nuclear transport have also been published (29-37). We and others (38-47) have invested considerable time and efforts in delineating the requirements and optimal conditions for the viral second strand DNA synthesis because the AAV genome is single-stranded and, therefore, transcriptionally inactive. During the course of these studies, we identified that phosphorylated forms of FKBP52 form a complex with the AAV D-sequence and play a critical role in regulating AAV transduction efficiency (42-45).
FKBP52 has also been shown to be a chaperone protein and plays a role in protein folding and delivery (48, 50, 51). HSP90, a ubiquitous molecular chaperone, can bind to FKBP52 directly and form a heterocomplex with other proteins and receptors (48-51, 53–55, 70). The FKBP52-HSP90 complex mediates cytoplasmic transport of a number of cellular and viral proteins to the nucleus (50, 51, 53-55). The pursuit of the questions whether HSP90 also interacts with the FKBP52-AAV D-sequence, and whether such an interaction is involved in the regulation of AAV second strand DNA synthesis, has led to the present studies, in which we have documented the following. (i) HSP90 is a part of the FKBP52-AAV D-sequence complex, but it does not interact with the D-sequence directly. (ii) HSP90 facilitates FKBP52 binding to the D-sequence. (iii) HSP90 can be dissociated from the FKBP52-AAV D-sequence complex, but this dissociation alone is insufficient to exert any effect on AAV transduction efficiency. (iv) Heat-shock treatment, which results in dephosphorylation of FKBP52, acts synergistically with dissociation of the FKBP52-HSP90 complex to cause a maximal increase in AAV transduction efficiency. (v) Heat-shock treatment, which also increases HSP90 levels, is maximally effective when FKBP52 is present in its tyrosine-dephosphorylated form. (vi) Deliberate overexpression of the human HSP90 gene leads to stabilization of the HSP90-FKBP52-AAV D-sequence complex and results in a significant decrease in AAV transduction efficiency. (vii) Down-regulation of the endogenous levels of HSP90 reduces the extent of the FKBP52-AAV D-sequence complex formation, which results in a significant increase in AAV transduction efficiency. Thus, although increased levels of HSP90 following heat-shock treatment are unlikely to be a major player in the observed increase in AAV transduction efficiency, in the absence of heat shock, increased levels of HSP90 lead to stabilization of the FKBP52-AAV D-sequence complex formation, presumably both serine/threonine-phosphorylated as well as unphosphorylated FKBP52, resulting in a significant inhibition in AAV-mediated transduction. On the other hand, decreased levels of HSP90 disrupt the tyrosine-dephosphorylated FKBP52-AAV D-sequence complex formation, thereby leading to increased transgene expression.
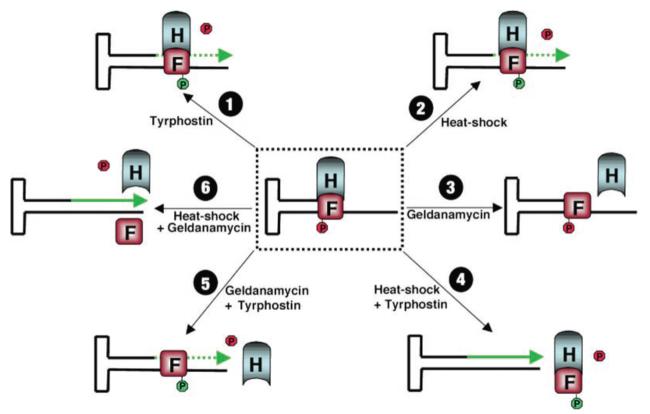
Based on all available data, we propose a model, shown in Fig. 8, that helps explain the complex interactions among FKBP52, HSP90, and the AAV D-sequence. In this model, the single-stranded D-sequence in the AAV inverted terminal repeat forms a complex with FKBP52 (Fig. 8, block F), which is phosphorylated predominantly at tyrosine residues (Fig. 8, red symbols) in these cells. HSP90 Fig. 8, block H) is also involved in the complex formation, which blocks the viral 3′-end and thus prevents the second strand DNA synthesis Fig. 8, middle dotted box). Following pre-treatment with tyrphostin (step 1), FKBP52 undergoes tyrosine dephosphorylation, is phosphorylated at serine/threonine residues (Fig. 8, green symbols), and the complex remains bound to the AAV D-sequence, but a low level viral second strand DNA synthesis leads to a modest increase in AAV transduction efficiency. The outcome is essentially the same following heat-shock treatment (step 2) as in (step 1). Treatment with geldanamycin (step 3) has little effect on AAV transduction efficiency because the tyrosine-phosphorylated form of FKBP52 still remains bound to the AAV D-sequence, even though the FKBP52-HSP90 complex is dissociated. Following heat-shock and tyrphostin treatments (step 4), the FKBP52-HSP90 complex is dissociated from the AAV D-sequence, thus allowing efficient viral second strand DNA synthesis and, therefore, maximal increase in AAV transduction efficiency. Following treatments with geldanamycin and tyrphostin (step 5), the FKBP52-HSP90 complex is dissociated; FKBP52 undergoes tyrosine dephosphorylation, but the serine/threonine-phosphorylated form of FKBP52 remains bound to the AAV D-sequence and allows low levels of viral second strand DNA synthesis and transgene expression. Finally, in cells pre-treated with heat shock and geldanamycin (step 6), the FKBP52-HSP90 complex is dissociated from the AAV D-sequence, which in turn allows efficient viral second strand DNA synthesis and maximal transgene expression.
Fig. 8. A model for the role of cellular FKBP52 (block F) and HSP90 (block H) proteins in AAV second strand DNA synthesis and transgene expression.
See text for details.
In our recent studies with FKBP52-knockout mice, we have observed that AAV fails to traffic efficiently in primary hematopoietic cells from these mice (86), whereas viral trafficking to the nucleus is significantly increased in cells from transgenic mice carrying the T-cell protein tyrosine phosphatase gene, which we have shown previously (43, 44) to catalyze tyrosine dephosphorylation of FKBP52. These data suggest that in addition to its crucial role in the viral second strand DNA synthesis, FKBP52 is also involved in intracellular trafficking and/or nuclear transport of AAV, especially because FKBP52 binds to HSP90, and because HSP90 also localizes at the nuclear pore complexes and facilitates the nuclear export of ribosomal proteins (73). Thus, the HSP90-FKBP52 complex might also be involved in intracellular trafficking of AAV. In this context, it is noteworthy that cellular stress, which is known to augment AAV transduction efficiency (38, 56), also regulates nucleocytoplasmic distribution of T-cell protein tyrosine phosphatase, which down-regulates signaling from EGFR (72), a known regulator of AAV transgene expression (40-45). Furthermore, the FKBP52-HSP90 complex is involved in interaction with dynein, a nucleotide-sensitive microtubule-associated protein (51), which has been shown previously to be involved in intracellular trafficking of adenovirus type 2, JC virus, simian virus 40, and retrovirus (74-77), and dynamin, another nucleotide-sensitive microtubule-associated protein that has been shown to be involved in intracellular trafficking of AAV (30).
The recently described role of HSP90 as a capacitor for morphological evolution (78-80) may be of particular relevance to the cryptic life cycle of AAV. For example, during a natural course of infection, the wild-type AAV has been shown to establish a latent infection (81-84), and we hypothesize that the proviral integration and/or the maintenance of viral latency might be mediated by HSP90. Under conditions of cellular stress, known to lead to the rescue and replication of the proviral AAV genome (13, 46, 56, 81), HSP90, tied up in its protective role (85), may no longer be able to maintain the latent phenotype. Further studies are warranted to explore this possibility.
Our ongoing studies on intracellular co-localization of FKBP52, HSP90, and AAV, using immunofluorescence and confocal microscopy, protein-protein interactions, and the development of HSP90-knockout mice, should allow us to gain a better understanding of the role of FKBP52 and HSP90 in various steps in the life cycle AAV, which may have implications in the use of recombinant AAV vectors in human gene therapy.
Acknowledgments
We thank Dr. Takayuki K. Nemoto for generously providing the pH6HSP90 plasmid, and Drs. Jacqueline A. Hobbs and Kirsten A. Weigel-Kelley for a critical review of this manuscript.
Footnotes
The abbreviations used are: AAV, adeno-associated virus type 2; EMSA, electrophoretic mobility shift assays; HSP90, heat-shock protein 90; FKBP52, FK506-binding protein; WCE, whole cell extracts; CKII, casein kinase II; EGFR epidermal growth factor receptor; PTK, protein-tyrosine kinase; WCLs, whole cell lysates; IMDM, Iscove's-modified Dulbecco's medium; CMV, cytomegalovirus.
This work was supported in part by United States Public Health Service Grants R01 EB-002073 and HL-65570 from the National Institutes of Health (to A. S.).
REFERENCES
- 1.Aitken ML, Moss RB, Waltz DA, Dovey ME, Tonelli MR, Mcnamara SC, Gibson RL, Ramsey BW, Carter BJ, Reynolds TC. Hum. Gene Ther. 2001;12:1907–1916. doi: 10.1089/104303401753153956. [DOI] [PubMed] [Google Scholar]
- 2.Carter BJ, Flotte TR. Curr. Top. Microbiol. Immunol. 1996;218:119–144. doi: 10.1007/978-3-642-80207-2_8. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee S, Lu D, Podsakoff G, Wong KK., Jr. Ann. N. Y. Acad. Sci. 1995;770:79–90. doi: 10.1111/j.1749-6632.1995.tb31045.x. [DOI] [PubMed] [Google Scholar]
- 4.Flotte TR, Afione SA, Conrad C, McGrath SA, Solow R, Oka H, Zeitlin PL, Guggino B, Carter BJ. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flotte TR, Carter BJ, Conrad CK, Guggino WB, Reynolds TC, Rosenstein B, Taylor G, Walden S, Wetzel R. Hum. Gene Ther. 1996;7:1145–1159. doi: 10.1089/hum.1996.7.9-1145. [DOI] [PubMed] [Google Scholar]
- 6.Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL, During MJ. Nat. Genet. 1994;8:148–153. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 7.Kaplitt MG, Xiao X, Samulski RJ, Li J, Ojamaa K, Klein IL, Makimura H, Kaplitt MJ, Strumpf RK, Diethrich EB. Ann. Thorac. Surg. 1996;62:1669–1676. doi: 10.1016/s0003-4975(96)00946-0. [DOI] [PubMed] [Google Scholar]
- 8.Kay MA, Manno CS, Ragni MV, Larson PJ, Couto LB, McClelland A, Glader B, Chew AJ, Tai SJ, Herzog RW, Arruda V, Johnson F, Scallan C, Skarsgard E, Flake AW, High KA. Nat. Genet. 2000;28:257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 9.Kessler PD, Podsakoff GM, Chen X, McQuiston SA, Colosi PC, Matelis LA, Kurtzman GJ, Byrne BJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koeberl DD, Alexander IE, Halbert CL, Russell DW, Miller AD. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1426–1431. doi: 10.1073/pnas.94.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manno CS, Shew AJ, Hutchison S, Larson PJ, Herzog RW, Arruda VR, Tai SJ, Ragni MV, Thompson A, Ozelo M, Couto LB, Leonard DG, Johnson FA, McClelland A, Scallan C, Skarsgard E, Flake AW, Kay MA, High KA, Glader B. Blood. 2003;101:2963–2972. doi: 10.1182/blood-2002-10-3296. [DOI] [PubMed] [Google Scholar]
- 12.McCown TJ, Xiao X, Li J, Breese GR, Samulski RJ. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- 13.Muzyczka N. Curr. Top. Microbiol. Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 14.Nathwani AC, Hanawa H, Vandergriff J, Kelly P, Vanin EF, Nienhuis AW. Gene Ther. 2000;7:183–195. doi: 10.1038/sj.gt.3301068. [DOI] [PubMed] [Google Scholar]
- 15.Ping P, Yang Q, Hammond HK. Microcirculation. 1996;3:225–228. doi: 10.3109/10739689609148292. [DOI] [PubMed] [Google Scholar]
- 16.Ponnazhagan S, Mukherjee P, Wang X-S, Qing KY, Kube DM, Mah C, Kurpad C, Yoder MC, Srour EF, Srivastava A. J. Virol. 1997;71:8262–8267. doi: 10.1128/jvi.71.11.8262-8267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponnazhagan S, Mukherjee P, Yoder MC, Wang X-S, Zhou SZ, Kaplan J, Wadsworth S, Srivastava A. Gene (Amst. 1997;190:203–210. doi: 10.1016/s0378-1119(96)00576-8. [DOI] [PubMed] [Google Scholar]
- 18.Ponnazhagan S, Yoder MC, Srivastava A. J. Virol. 1997;71:3098–3104. doi: 10.1128/jvi.71.4.3098-3104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder RO, Miao CH, Patijn GA, Spratt SK, Danos O, Nagy D, Gown AM, Winther B, Meuse L, Cohen LK, Thompson AR, Kay MA. Nat. Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava A, Wang X-S, Ponnazhagan S, Zhou SZ, Yoder MC. Curr. Top. Microbiol. Immunol. 1996;218:93–117. doi: 10.1007/978-3-642-80207-2_7. [DOI] [PubMed] [Google Scholar]
- 21.Tan MQ, Qing KY, Zhou SZ, Yoder MC, Srivastava A. Mol. Ther. 2001;3:940–946. doi: 10.1006/mthe.2001.0346. [DOI] [PubMed] [Google Scholar]
- 22.Wagner JA, Nepomuceno IB, Messner AH, Moran ML, Batson EP, Dimiceli S, Brown BW, Desch JK, Norbash AM, Conrad CK, Guggino WB, Flotte TR, Wine JJ, Carter BJ, Reynolds TC, Moss RB, Gardner P. Hum. Gene Ther. 2002;13:1349–1359. doi: 10.1089/104303402760128577. [DOI] [PubMed] [Google Scholar]
- 23.Walsh CE, Liu JM, Miller JL, Nienhuis AW, Samulski RJ. Proc. Soc. Exp. Biol. Med. 1993;204:289–300. doi: 10.3181/00379727-204-43665. [DOI] [PubMed] [Google Scholar]
- 24.Xiao X, Li J, Samulski RJ. J. Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou SZ, Li Q, Stamatoyannopoulos G, Srivastava A. Gene Ther. 1996;3:223–229. [PubMed] [Google Scholar]
- 26.Qing KY, Mah C, Hansen J, Zhou SZ, Dwarki VJ, Srivastava A. Nat. Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 27.Summerford C, Bartlett JS, Samulski RJ. Nat. Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 28.Summerford C, Samulski RJ. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett JS, Wilcher R, Samulski RJ. J. Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. J. Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. J. Clin. Investig. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen J, Qing KY, Kwon H-J, Mah C, Srivastava A. J. Virol. 2000;74:992–997. doi: 10.1128/jvi.74.2.992-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen J, Qing KY, Srivastava A. J. Virol. 2001;75:4080–4090. doi: 10.1128/JVI.75.9.4080-4090.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanlioglu S, Benson PK, Yang J, Atkinson EM, Reynolds T, Engelhardt JF. J. Virol. 2000;74:9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seisenberger G, Ried MU, Endress T, Buning H, Hallek M, Brauchle C. Science. 2001;294:1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 36.Hansen J, Qing KY, Srivastava A. Mol. Ther. 2001;4:289–296. doi: 10.1006/mthe.2001.0457. [DOI] [PubMed] [Google Scholar]
- 37.Xiao W, Warrington KH, Jr., Hearing P, Hughes J, Muzyczka N. J. Virol. 2002;76:11505–11517. doi: 10.1128/JVI.76.22.11505-11517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrari FK, Samulski T, Shenk T, Samulski RJ. J. Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher KJ, Gao G-P, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. J. Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mah C, Qing KY, Hansen J, Khuntrirat B, Yoder MC, Srivastava A. Gene Ther. Mol. Biol. 1999;3:57–65. [Google Scholar]
- 41.Mah C, Qing KY, Khuntrirat B, Ponnazhagan S, Wang X-S, Kube DM, Yoder MC, Srivastava A. J. Virol. 1998;72:9835–9841. doi: 10.1128/jvi.72.12.9835-9843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qing KY, Khuntrirat B, Mah C, Kube DM, Wang X-S, Ponnazhagan S, Zhou SZ, Dwarki VJ, Yoder MC, Srivastava A. J. Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qing KY, Hansen J, Weigel-Kelley KA, Tan MQ, Zhou SZ, Srivastava A. J. Virol. 2001;75:8968–8976. doi: 10.1128/JVI.75.19.8968-8976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qing KY, Li WM, Zhong L, Tan MQ, Hansen J, Weigel-Kelley KA, Chen LY, Yoder MC, Srivastava A. J. Virol. 2003;77:2741–2746. doi: 10.1128/JVI.77.4.2741-2746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qing KY, Wang X-S, Kube DM, Ponnazhagan S, Bajpai A, Srivastava A. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10879–10884. doi: 10.1073/pnas.94.20.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X-S, Ponnazhagan S, Srivastava A. J. Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X-S, Qing KY, Ponnazhagan S, Srivastava A. J. Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czar MJ, Lyons RH, Welsh MJ, Renoir J-M, Pratt WB. Mol. Endocrinol. 1995;9:1549–1560. doi: 10.1210/mend.9.11.8584032. [DOI] [PubMed] [Google Scholar]
- 49.Miyata Y, Chambraud B, Radanyi C, Leclerc J, Lebeau M-C, Renoir K-M, Shirai R, Catelli M-G, Yahara I, Baulieu E-E. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14500–14505. doi: 10.1073/pnas.94.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Czar MJ, Owens-Grillo JK, Dittmar KD, Hutchison KA, Zacharek AM, Leach KL, Deibel MR, Pratt WB. J. Biol. Chem. 1994;269:11155–11161. [PubMed] [Google Scholar]
- 51.Galigniana MD, Radanyi C, Renoir JM, Housley PR, Pratt WB. J. Biol. Chem. 2001;276:14884–14889. doi: 10.1074/jbc.M010809200. [DOI] [PubMed] [Google Scholar]
- 52.Hung JJ, Chung CS, Chang W. J. Virol. 2002;76:1379–1390. doi: 10.1128/JVI.76.3.1379-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratt WB. Proc. Soc. Exp. Biol. Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 54.Pratt WB, Toft DO. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 55.Pratt WB, Silverstein AM, Galigniana MD. Cell. Signal. 1999;11:839–851. doi: 10.1016/s0898-6568(99)00064-9. [DOI] [PubMed] [Google Scholar]
- 56.Yakinoglu AO, Heilbronn R, Burkle A, Schlehofer JR, zur Hausen H. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 57.Kube DM, Ponnazhagan S, Srivastava A. J. Virol. 1997;71:7361–7371. doi: 10.1128/jvi.71.10.7361-7371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller MT. J. Virol. 1987;61:858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johansson E, Hjortsberg K, Thelander L. J. Biol. Chem. 1998;273:29812–29816. doi: 10.1074/jbc.273.45.29816. [DOI] [PubMed] [Google Scholar]
- 60.Knössl M, Löwer R, Löwer J. J. Virol. 1999;73:1254–1261. doi: 10.1128/jvi.73.2.1254-1261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McElhinny JA, Trushin SA, Bren GD, Chester N, Paya CV. Mol. Cell. Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russo GL, Vandenberg MT, Yu IJ, Bae YS, Franza BR, Jr., Marshak DR. J. Biol. Chem. 1992;267:20317–20325. [PubMed] [Google Scholar]
- 63.Weber W, Bertics PJ, Gill GN. J. Biol. Chem. 1984;259:14631–14636. [PubMed] [Google Scholar]
- 64.Cybulsky AV, Goodyear PR, McTavish AJ. Am. J. Physiol. 1994;267:F428–F436. doi: 10.1152/ajprenal.1994.267.3.F428. [DOI] [PubMed] [Google Scholar]
- 65.Zhong L, Su JY. Anesthesiology. 2002;96:148–154. doi: 10.1097/00000542-200201000-00028. [DOI] [PubMed] [Google Scholar]
- 66.Lindquist S, Craig EA. Annu. Rev. Genet. 1988;27:631–667. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 67.Parsell DA, Lindquist S. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 68.Zhao YG, Gilmore R, Leone G, Coffey MC, Weber B, Lee PWK. J. Biol. Chem. 2001;276:32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 69.Grenert JP, Sullivan WP, Fadden P, Haystead TAJ, Clark J, Sausville E, Neckers LM, Toft DO. J. Biol. Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 70.Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pear LH. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 71.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri ES, Litwack G, Toft D. J. Biol. Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 72.Lam MH, Michell BJ, Fodero-Tavoletti MT, Kemp BE, Tonks NK, Tiganis T. J. Biol. Chem. 2001;276:37700–37707. doi: 10.1074/jbc.M105128200. [DOI] [PubMed] [Google Scholar]
- 73.Schlatter H, Langer T, Rosmus S, Onneken ML, Fasold H. Biochem. J. 2002;362:675–684. doi: 10.1042/0264-6021:3620675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ashok A, Atwood WJ. J. Virol. 2003;77:1347–1356. doi: 10.1128/JVI.77.2.1347-1356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giannakakou P, Nakano M, Nicolaou KC, O'Brate A, Yu J, Blagosklonny MV, Greber UF, Fojo T. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10855–10860. doi: 10.1073/pnas.132275599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petit C, Giron ML, Tobaly-Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H, Saib A. J. Cell Sci. 2003;116:3433–3442. doi: 10.1242/jcs.00613. [DOI] [PubMed] [Google Scholar]
- 77.Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP, Greber UF. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Queitsch C, Sangster TA, Lindquist S. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 79.Rutherford SL, Lindquist S. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 80.Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Nat. Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 81.Berns KI, Giraud C. Curr. Top. Microbiol. Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 82.Kotin RM, Menninger JC, Ward DC, Berns KI. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 83.Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter LA, Laughlin CA, McLaughlin SK, Muzyczka N, Rocchi M, Berns KI. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samulski RJ, Zhu X, Xiao X, Brook JD, Houseman DE, Epstein N, Hunter LA. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pennisi E. Science. 1998;282:1796. doi: 10.1126/science.282.5395.1796a. [DOI] [PubMed] [Google Scholar]
- 86.Yang Z, Zhong L, Li W, Qing KY, Li Y, Chen L, Tan MQ, Yoder MC, Shou W, Srivastava A. Mol. Ther. 2003;7:S14. [Google Scholar]