Abstract
Purpose
The genetic control of photoreceptor cell fate in the vertebrate retina is poorly understood. Published studies suggest that the genetic program underlying photoreceptor production involves neuroD, a proneural basic helix-loop-helix (bHLH) gene. The present study investigates whether neuroD is necessary for photoreceptor cell development, by using loss-of-function analyses.
Method
Engrailed-mediated active repression, antisense oligonucleotides, and small interfering RNA (siRNA) were used to attenuate neuroD expression and function in embryonic chick retina. The development of the retina was subsequently analyzed to determine whether these experimental manipulations would yield photoreceptor deficits in otherwise normal retina.
Results
Chick embryos infected with retroviruses expressing an active repression construct, En-NeuroDΔC, exhibited severe photoreceptor deficits. The outer nuclear layer (ONL) of the retina was no longer a contiguous structure, but became fragmented with regions that contained fewer or no photoreceptor cells. Photoreceptor deficiency was evident even before the retina became laminated, suggesting that active repression of NeuroD may have affected photoreceptor genesis. No deficiency was observed in other types of retinal cells. Culturing retinal cells in the presence of siRNA against neuroD resulted in a more than 50% reduction in the number of photoreceptor cells and an increase in the number of chx10+ cells. Subjecting the developing retina to antisense oligonucleotides against neuroD yielded fewer photoreceptor cells both in vivo and in vitro. Consistent with these observations, anti-NeuroD antibody specifically labeled the nuclei of the ONL.
Conclusions
The data suggest a specific and an essential role of neuroD in photoreceptor formation in the chick retina.
Photoreceptor cells of the vertebrate retina are a group of light-sensitive receptor neurons. During retinal neurogenesis photoreceptor cells arise from a pool of multipotent progenitors. How the photoreceptor’s fate is specified has been a long-standing question. Early studies suggested that lineage is not the primary deterministic factor in the vertebrate retina. Rather, the microenvironment signals multipotent progenitors to develop into the various retinal cell types, including photoreceptor cells.1–4 In addition, a concept has been put forth that the specification of photoreceptor cell fate is a postmitotic event4,5 and a photoreceptor precursor can be respecified to become a bipolar cell by ciliary neurotrophic factor (CNTF).6 During the past 10 years or so, a large body of information has been accumulated regarding extrinsic factors that may contribute to photoreceptor specification. These include cell–cell interactions,4 retinoic acid,7,8 growth factors,9,10 growth factor receptors,11,12 ligands of steroid–thyroid receptors,13 taurine,14 S-laminin,15 activin,16 and Hedgehog.17,18
Despite significant progress, however, our understanding of the control of the photoreceptor’s fate is still limited. It is now generally accepted that both intrinsic and extrinsic factors participate in the process (for recent reviews see Refs. 5,19), but the intrinsic factors underlying the photoreceptor’s fate remain elusive. Recent studies indicate that both Nrl, a gene encoding a leucine zipper transcription factor,20 and thyroid hormone receptor β221 play a role in subtype specification of photoreceptor cells.
Studies from our laboratory suggest that determination of the photoreceptor cell’s fate, at least in the chick retina, may involve neuroD, a basic helix-loop-helix (bHLH) gene homologous to the Drosophila proneural gene atonal.22,23 Retrovirus-driven neuroD expression in the developing chick retina results in an overproduction of photoreceptor cells.24 Furthermore, ectopic expression of neuroD in cultured RPE cells triggers RPE transdifferentiation into photoreceptor cells that express cone or rod opsin.24–26 These gain-of-function studies indicate that neuroD is sufficient to promote photoreceptor fate in the developing retina and in vitro in RPE cell culture.
To examine whether neuroD is necessary for photoreceptor production during chick retinal development, we performed loss-of-function studies using Drosophila Engrailed (En)–mediated active repression, antisense oligonucleotides, and small interfering RNA (siRNA). We report that all three methods of attenuating the expression and function of neuroD resulted in photoreceptor deficiency, without a concomitant reduction in other retinal neurons. The photoreceptor deficits were noticeable at early developmental stages, suggesting that photoreceptor genesis may be affected by decreased neuroD expression and function.
Methods
Chick Embryos
Fertilized, pathogen-free chicken (White Leghorn) eggs were purchased from Spafas (Preston, CT), incubated in a Petersime incubator (Gettysburg, OH), and used throughout the study. All usage of animals adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and was approved by the institutional review board at the University of Alabama at Birmingham.
Generation of Specific Antibody against Recombinant NeuroD
Because the bHLH domain of NeuroD shares sequence homology with several other bHLH proteins,27,28 we used the C-terminal, nonconserved region, excluding the bHLH domain, to produce specific antibodies against NeuroD. A XhoI-BglII fragment of 398 base pairs covering the C-terminal 102 amino acids of neuroD cDNA and 89 base pairs of noncoding sequence was fused, in frame, with the T7 phage gene 10 in a vector (pGEMEX2; Promega, Madison, WI). Recombinant fusion protein was partially purified, first by its insolubility and subsequently by SDS-PAGE. The corresponding protein band was cut out and used to immunize the rabbits. Before affinity purification with the NeuroD-gene 10 fusion protein, the antiserum was preabsorbed with total Escherichia coli lysate followed by reaction with partially purified gene 10 protein. Detailed procedures for purification of the fusion protein and affinity purification of antibodies were as previously described.29
Immunohistochemistry and In Situ Hybridization
Chick retinas were fixed at different developmental stages with ice-cold 4% paraformaldehyde. Cryosections of 8 to 10 μm were collected. Affinity-purified anti-NeuroD antibody was used at a dilution of 1:40. The following monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank (Iowa University, Iowa City, IA): anti-visinin (7G4; 1:500), anti-vimentin (H5; 1:500), antibody against a synaptic vesicle-specific membrane protein (mAb 48; 1:100), anti-LIM (4F2; 1:50), and anti-AP2α (3B5; 1:50). Additional antibodies included: anti-viral protein, p27 (1:500; Spafas), monoclonal antibody against Brn3a (1:200; Chemicon, Temecula, CA), polyclonal antibody against SNAP-25 (1:200; Affinity BioReagents, Deerfield, IL). Standard procedures for immunohistochemistry were followed. In situ hybridization was performed before immunohistochemistry, when both were performed on the same retinal sections. In situ hybridization with digoxigenin-labeled antisense RNA probes against visinin and chx10 was as previously described.24
Western Blot
Nuclei from E9 retina and from E9 heart were isolated by sucrose gradient centrifugation, and total nucleic extracts were prepared as described.30 Total nucleic protein was loaded at 1 μg per lane for SDS-PAGE. The standard Western blot procedure was performed, using affinity-purified anti-NeuroD antibody and the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences, Arlington Heights, IL).
Construction of Active Repression Constructs
Construction of RCAS expressing the repressor domain of Drosophila Engrailed has been previously described.31 The coding region of neuroD was PCR amplified from a cDNA clone with EcoRI and SalI introduced at the 5′ and 3′ ends, respectively. The DNA was fused in frame with the 3′ end of the repressor domain of En, producing En-NeuroD, in the shuttle vector Cla12Nco, and the sequence of the insert was verified. Deleting the XhoI and HindIII fragments from En-NeuroD resulted in En-NeuroDΔC. To generate En-cNSCL1 and En-cNSCL2, the neuroD sequence in Cla12Nco-En-NeuroD was replaced with the coding sequence of either cNSCL1 or cNSCL2. A ClaI fragment containing En-NeuroDΔC, En-cNSCL1, or En-cNSCL2 was digested from the shuttle vector and inserted into the retrovirus RCAS. Recombinant proviral DNA was transfected into chick embryonic fibroblast cells and retroviruses were harvested as previously described.24 Concentrated retroviruses expressing the various constructs were microinjected into the subretinal space at E2, as previously described.24
Oligonucleotides
Four phosphothiolated antisense oligonucleotides (5′-GTACGACTTGGTCATGGC-3′, 5′-TTCCGCAGCGCGTCCTCTT-3′, 5′-CGCCTTGGTCATCTTCTT-3′, and 5′-GATCTTGGAGAGTTTCT-3′) and their corresponding sense oligonucleotides were synthesized and HPLC-purified commercially by DNAgency (Malvern, PA). Solutions of 1 μL of the oligonucleotides at a concentration of 25 nM each in Medium 199 were microinjected into the subretinal space of embryonic day (E)4.5 embryos and repeated on E5.5. The injections were performed using a micromanipulated micropipette attached to a pneumatic pump with a positive-pressure delivery system. The entire head was fixed 24 hours after the second injection. Specimens were oriented and sectioned in such a way that anatomically equivalent regions of both retinas would be easily tracked for comparisons of gene expression in a pair-wise manner.
For experiments using retina explants, only two oligonucleotides corresponding to the sequence in the N-terminal region were used. Retinas were dissected at E4.5 and cultured in the presence of either antisense oligonucleotides (3 μM final concentration) or sense oligonucleotides at the same concentration. Hanks’ balanced salt solution (HBSS) was used as an additional control. Culture medium (Medium 199+ 10% fetal calf serum) with nucleotide additives was changed daily. After 3 days (equivalent to E7.5), retinal cells were dissociated, seeded for 4 hours onto polyornithine–treated dishes, fixed, and analyzed with immunocytochemistry and in situ hybridization. Positive cells were counted from three dishes, and the means and SDs were calculated on computer program (OriginPro 7; Microcal, Northampton, MA). The entire experiment, from retinal explant culture to cell counting was repeated at least three times.
siRNA
Annealed, dTdT-overhanging siRNA molecules targeting the neuroD sequences AAGGAGGAGGACCTGGAGGCT and AAGCCGCCGCACGCCTACGGC were synthesized by Xeragon (Germantown, MD). Control siRNA were fluorescein-labeled, 21 bp of random sequences provided by Xeragon. Cells of E5.5 retinas were dissociated and seeded in a polyornithine-treated 24-well plate for 3 hours in DMEM-12+10% fetal calf serum. siRNA was transfected into the retinal cells with transfection reagent (Oligofectamine), according to the procedure provided by Invitrogen (Carlsbad, CA). Briefly, 2.5 μL of 20 μg/mL siRNA and 3 μL of the transfection reagent were diluted to 50 μL and added to each well containing 300 μL of DMEM-F12. After 3 hours of incubation, the medium was replaced with that containing 10% serum, and the cells were cultured for 4 days with a change of medium after 2 days. Cells were fixed with 4% paraformaldehyde and subjected to immunostaining or in situ hybridization analyses. Data analysis was similar to that used in the experiments with antisense oligonucleotides.
Results
Photoreceptor Cell–Specific Expression of neuroD in the Developing Chick Retina
The bHLH domain of 59 amino acid residues and its adjacent 33 residues of NeuroD is highly homologous to several members of the bHLH protein family.27,28 To facilitate the generation of antibody specific to neuroD, we used the C-terminal, nonconserved region of 102 amino acids. We tested the specificity of the affinity-purified antibody against NeuroD, using cultured chick RPE cells infected with retrovirus RCAS expressing either neuroD (Figs. 1A, 1B) or green fluorescent protein (GFP; Figs. 1C, 1D). The antibody stained the nuclei of cultured RPE cells infected with RCAS-neuroD (Fig. 1A), but not those infected with RCAS-GFP (Fig. 1C). We also performed Western blot analysis with nucleic proteins isolated from E9 retina and E9 heart. The antibody recognized a protein with an estimated molecular weight consistent with that calculated for NeuroD protein. This protein was present in the nucleic extract of E9 retina and absent in that of E9 heart (Fig. 1E).
Figure 1.
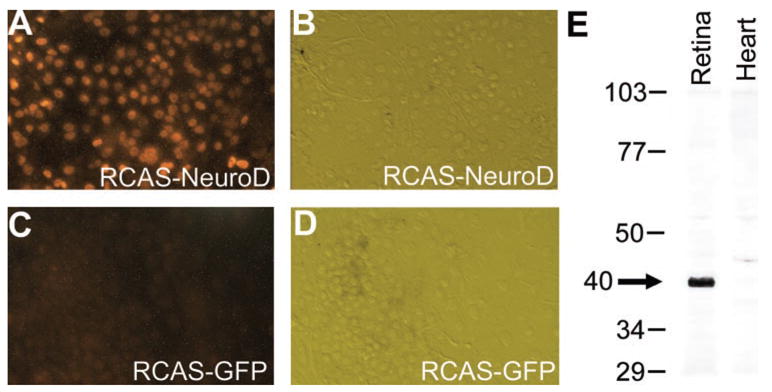
Immunostaining of cultured, retrovirus-infected RPE cells (A–D) and Western blot of nuclear extracts from the retina and the heart (E) with affinity-purified antibody against NeuroD. (A, C) Fluorescent images of the immunostaining. (C, D) Hoffman images of cells in (A, C). Numbers in (E) indicate molecular mass (in kilodaltons) standards. Arrow: major band present in E9 retina but absent from E9 heart.
The antibodies were then used to examine neuroD expression. In the chick retinas, photoreceptor cell birth peaks at E6 to E7, and newly born photoreceptor cells accumulate at the outer portion of the retinal neuroepithelium. Many infant photoreceptor cells express visinin, a gene encoding a calcium-binding protein present in cones. The chick retina becomes stratified at approximately at E9. At E6, NeuroD immunoreactive (NeuroD+) cells were located in the outer portion of the retinal neuroepithelium (Fig. 2A). This spatial location coincides with that of young photoreceptor cells that express visinin (data not shown).24 At E7, more NeuroD+ cells were detected (Fig. 2B). In a stratified retina, NeuroD protein was detected in the nuclei of cells in the outer nuclear layer (ONL), where photoreceptor cells are located (Fig. 2C). The inner nuclear layer (INL), where somas of horizontal neurons, bipolar neurons, Müller glia, and amacrine neurons reside, contained no immunoreactive nuclei. The nuclei in the ganglion cell layer (GCL) were also unstained. Thus, anti-NeuroD immunoreactivity was specifically detected in the nuclei of photoreceptor cells and their precursors.
Figure 2.
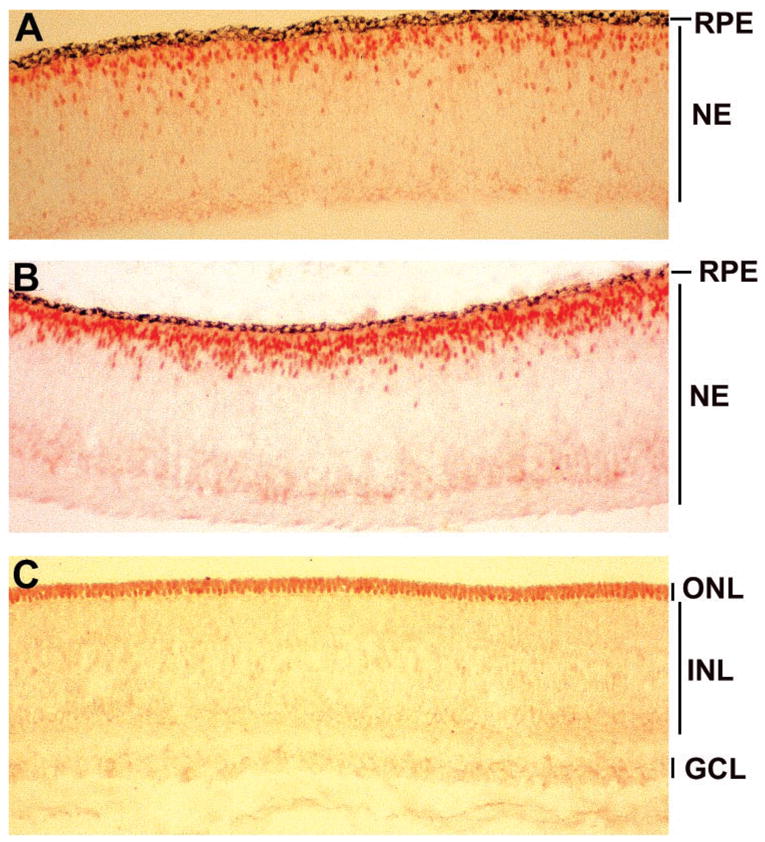
Expression of neuroD in embryonic chick retina. (A) Anti-NeuroD antibody labeled the nuclei of cells residing at the outer portion of the retinal neuroepithelium at E6. (B) At E7, more cells were labeled with the antibody, and the positive cells appeared to gather at the future position of the ONL. (C) At E9, only the nuclei in the ONL were stained with anti-NeuroD antibody. No nuclei in the INL or the GCL were stained. NE, retinal neuroepithelium; remaining abbreviations are defined in the article text. Magnification: × 50.
Photoreceptor Deficiency in Retinas Expressing En-NeuroDΔC
To study whether neuroD is necessary for photoreceptor formation in the chick retina, we performed loss-of-function experiments. Initially, we used a simple dominant negative construct that lacked 102 residues from the NeuroD C terminus. This C-terminal region is believed to contain the NeuroD activation domain.32 As predicted, this construct, NeuroDδC (Fig. 3A) was unable to induce RPE transdifferentiation toward photoreceptor cells.25 However, expression of the mutant construct driven by the retrovirus RCAS in the developing retina did not alter retinal neurogenesis in a detectable way (data not shown).
Figure 3.
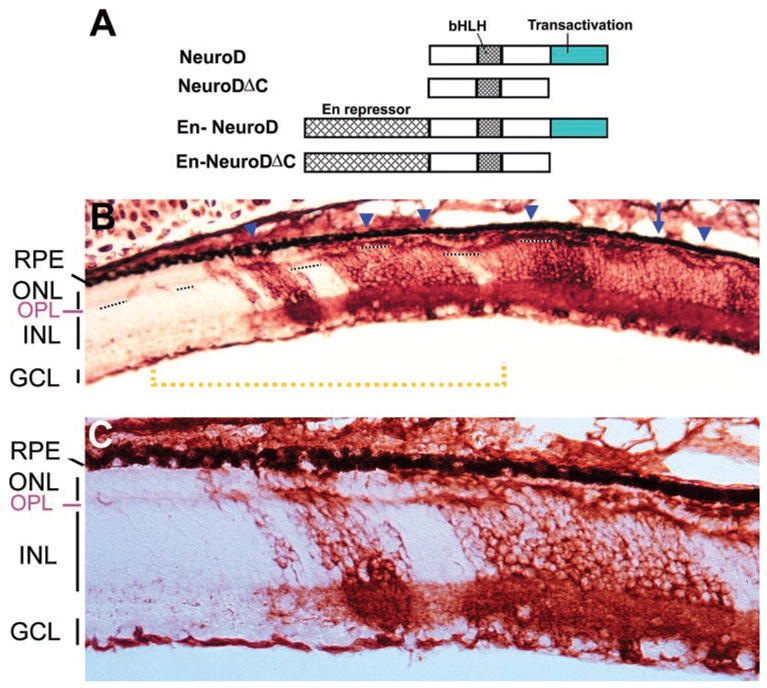
Reduction of the ONL in retina infected with RCAS-En-NeuroDΔC. (A) A schematic illustration of NeuroD active repression constructs. (B) An E17 retina partially infected with RCAS-En-NeuroDΔC. Short, dashed lines delineate the OPL, which, together with the RPE on the opposite side, demarcates the ONL. Arrow: region where the ONL was missing; arrowheads: regions where the ONL contained fewer cells. (C) A higher magnification of the region underlined by the yellow dashed line in (B) to show the reduction of the ONL thickness in regions infected with the virus. Magnification: (B) ×50; (C) ×100.
Subsequently, we exploited En-mediated active repression to repress NeuroD function. Drosophila Engrailed actively and specifically represses activated transcription.33,34 Its repressor domain can confer repression activity to heterologous DNA-binding domains when linked to them.33,34 We generated recombinant retrovirus RCAS-En-NeuroDΔC. RCAS-driven expression of En-NeuroDΔC in the developing chick retina resulted in gross alterations of the ONL (Figs. 3B, 3C). In a normal retina, the ONL is a contiguous structure with two rows of nuclei demarcated from the INL by a fibrous, outer plexiform layer (OPL). In retinas infected with RCAS-En-NeuroDΔC, the ONL contained many breaks where the layer was either nonexistent (Fig. 3B, arrow) or thinner, containing a single row of nuclei or only a few cells (Fig. 3B, arrowheads). The diminution of the ONL was observed only at places where the virus was detected (Figs. 3B, 3C, regions in red). The ONL of the uninfected regions remained normal.
The presence of breaks in the ONL indicated that photoreceptor deficits occurred in regions infected with RCAS-En-NeuroDΔC. We identified cone photoreceptor cells, which dominate the ONL of the chick retina, with in situ hybridization for visinin mRNA. The spatial distribution of visinin+ cells showed gross alterations, reminiscent of the diminution of the ONL. Some regions lacked visinin+ cells (Figs. 4A, 4D, arrows), and some regions contained a reduced number of visinin+ cells (Figs. 4A–C, arrowheads).
Figure 4.
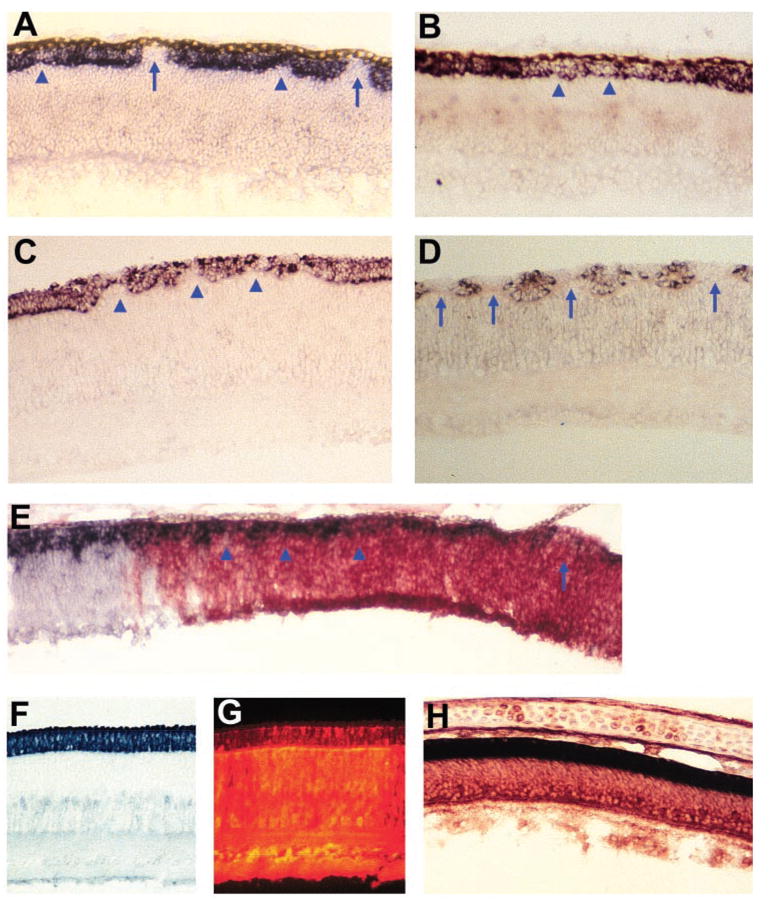
Photoreceptor deficits in retinas infected with RCAS-En-NeuroDΔC. (A, B) Cells expressing visinin in E9 retinas infected with RCAS-En-NeuroDΔC. (C, D) Cells expressing visinin in E12 retinas infected with RCAS-En-NeuroDΔC. (E) An E7 retina with its peripheral region infected with RCAS-En-NeuroDΔC. Photoreceptor cells are marked by visinin expression (in situ staining, dark stain). Viral infection is revealed by anti-p27 immunohistochemistry (red). (F) Visinin+ cells in an E12 retina infected with RCAS-En. (G) Anti-p27 immunofluorescent staining of the section in (F). Fluorescent signals in the ONL were quenched by the nonfluorescent, Visinin+ signals. (H) Double-labeling for visinin mRNA (dark stain) and p27 (red) of an E15 retina infected with RCAS-En-cNSCL1. Arrows: places where there was a total absence of visinin+ cells. Arrowheads: regions where fewer visinin+ cells were present. Magnification: ×50.
Retina of early developmental stages was used to examine the possibility that En-NeuroDΔC might affect the genesis of photoreceptor cells. If En-NeuroDΔC reduces photoreceptor genesis, then the number of visinin-expressing cells would be decreased in the neuroepithelial stage. We found that, in E7 retinal neuroepithelium, cells expressing visinin were absent or greatly reduced in their numbers when infected with RCAS-En-NeuroDΔC (Fig. 4E), whereas the adjacent, uninfected region contained much more visinin+ cells. To examine whether apoptosis plays a role in the decreased number of photoreceptor cells, TUNEL assays were performed, and no difference was observed in the number of TUNEL+ cells in E7 and E9 retinas infected with RCAS-En-NeuroDΔC and with RCAS GFP (data not shown).
To address the question of whether the photoreceptor deficiency was due to En-NeuroDΔC or to En or En fused with a bHLH gene, we generated recombinant RCAS expressing various constructs, including En alone, En-cNSCL1, or En-cNSCL2. Like neuroD, cNSCL1 and cNSCL2 are bHLH genes homologous to atonal and encoding class B bHLH proteins, which may form heterodimers with class A bHLH proteins and bind to the E box with a consensus sequence of CANNTG.35,36 All three bHLH genes are expressed in the retina with cell type specificities.37,38 Unlike neuroD, cNSCL1, and cNSCL2 are not expressed in photoreceptor cells. We found that retinas infected with RCAS-En, RCAS-En-cNSCL1, or RCAS-En-cNSCL2 appeared histologically normal. Their ONL was intact, had two rows of visinin+ cells, and was essentially indistinguishable from that of a normal retina (Figs. 4F–H; data not shown). Only rarely, a break in the ONL was observed. On average, one cross-section through or adjacent to the meridional plane contained 36- to 900-fold more breaks in the ONL of retinas infected with RCAS-En-NeuroDΔC than with En alone, En-cNSCL1, or En-cNSCL2 (Table 1). If normalized for 100% viral infection, the number of breaks in the ONL per cross section in En-NeuroDΔC retinas was 127- to 3018-fold more than that in the other constructs.
Table 1.
Number of breaks in the ONL of Chick Retinas Infected with RCAS Expressing Various En Constructs
| Construct | Breaks (n) | Sections (n) | Breaks per Section (n) | Embryos* (n) | Viral Infection (%) | Breaks per Section† (n) |
|---|---|---|---|---|---|---|
| En-NeuroDΔC | 996 | 55 | 18.11 | 9 | 20 | 90.55 |
| En | 1 | 44 | 0.02 | 6 | 60 | 0.03 |
| En-cNSCL1 | 11 | 22 | 0.50 | 3 | 70 | 0.71 |
| En-cNSCL2 | 2 | 48 | 0.04 | 6 | 80 | 0.05 |
Data were collected from E9–E18 chick embryos with the virus microinjected into the subretinal space at E2. Retinal cross sections were cut through or adjacent to the meridional plane.
Number of embryos, each with 5 to 8 retinal cross sections counted. Viral infection was the percentage of retinal cells staining positive for p27 and is shown as the average of n retinas.
Normalized for 100% viral infection.
Other Retinal Cells in Retinas Expressing En-NeuroDΔC
To determine whether En-NeuroDΔC specifically causes reduction in the number of photoreceptor cells, we examined other neurons of the retina infected with RCAS-En-NeuroDΔC. Despite the profound reduction of the ONL, the thickness of the INL and the GCL appeared unchanged in En-NeuroDΔC retinas (Figs. 3, 4, 5). Specific antibodies were used to identify the major types of retinal neurons and synaptic layers in the inner plexiform layer (IPL). Chick retinal horizontal cells were recognized with anti-LIM antibody (4F2). In uninfected regions, LIM+ cells were distributed in a single layer adjacent to the OPL. In regions infected with RCAS-En-NeuroDΔC, a single layer of LIM+ cells was present, even though they were localized in the breaks of the ONL (Figs. 5A, 5B). The density of AP2+ amacrine cells in the infected region was similar to that in the adjacent, uninfected regions (Figs. 5C, 5D). Likewise, ganglion cells, identified by the anti-Brn3a antibody, showed no reduction in their number (Figs. 5E, 5F). Bipolar cells constitute roughly half of the INL and express chx10. An equally thick band of chx10+ cells was present in the infected region as in the uninfected region (Fig. 5G). However, like horizontal cells, some of the chx10+ cells occupied the breaks of the ONL (Fig. 5G, arrows). Of note, the chx10+ cells at the breaks of the ONL remained contiguous with the chx10+ bipolar cells in the INL. Immunostaining for synaptic proteins with anti-SNAP-25 and mAb 48 showed no difference between infected and uninfected regions (Fig. 5H; data not shown).
Figure 5.
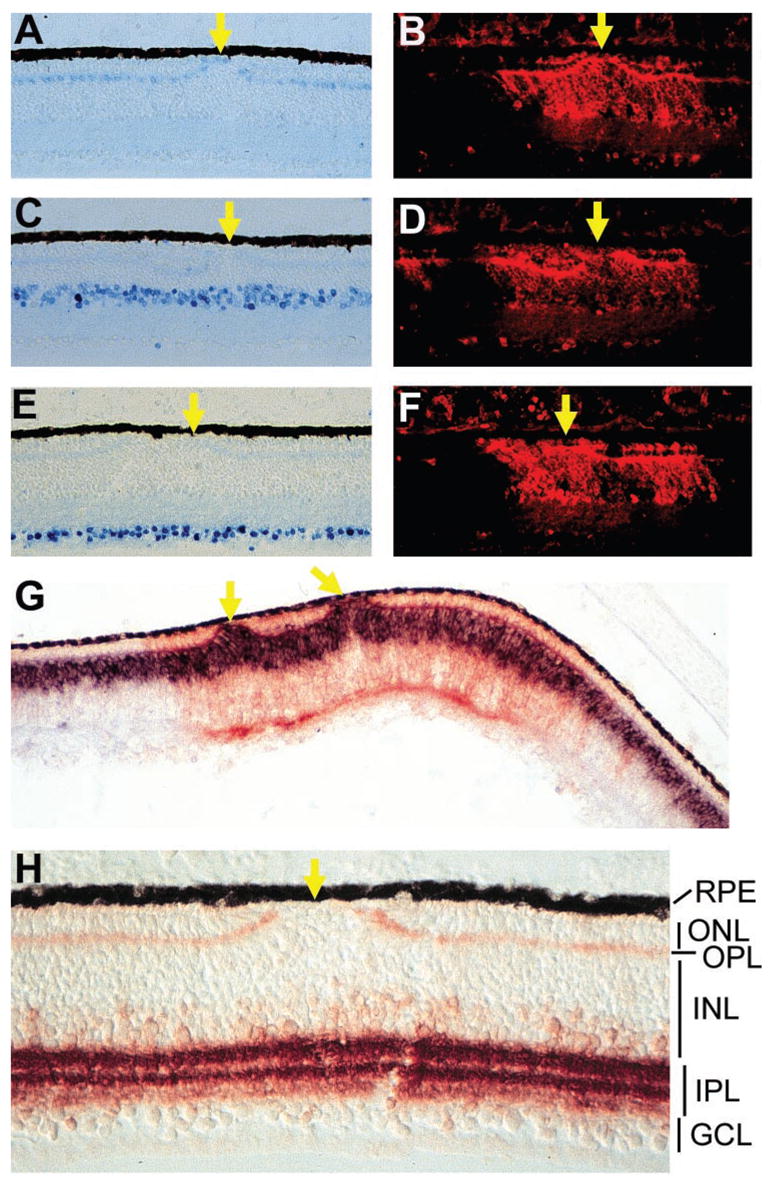
Nonphotoreceptor neurons in retinas infected with RCAS-En-NeuroDΔC. (A) Horizontal cells at E18 labeled with monoclonal antibody 4F2. (C) Amacrine cells at E18 identified with anti-Ap2 antibody. (E) Ganglion cells at E18 recognized by a monoclonal antibody against Brn3a. (B, D, F) Anti-p27 immunofluorescent staining of the sections in (A), (C), and (E), respectively, to mark regions infected with RCAS-En-NeuroDΔC. (G) Double labeling of an E12 retina for chx10 mRNA (blue) and p27 protein (red). (H) The IPL and the OPL at E18 immunostained with an antibody against SNAP-25. Arrows: places where fewer or no photoreceptor cells are present. Magnification: (A–G) ×50; (H) ×100.
Published studies reported an increase in Müller glial cells in explant culture of retinas from neuroD knockout mice.39 To examine whether the number of Müller glia was increased in En-NeuroDΔC retina, we performed double-labeling experiments for viral infection and Vimentin expression using retinas partially infected with RCAS-En-NeuroDΔC. Vimentin+ cells were present in regions infected with the RCAS-En-NeuroDΔC at densities similar to that of the adjacent, uninfected regions, despite the breaks in the ONL in the infected regions (Fig. 6).
Figure 6.
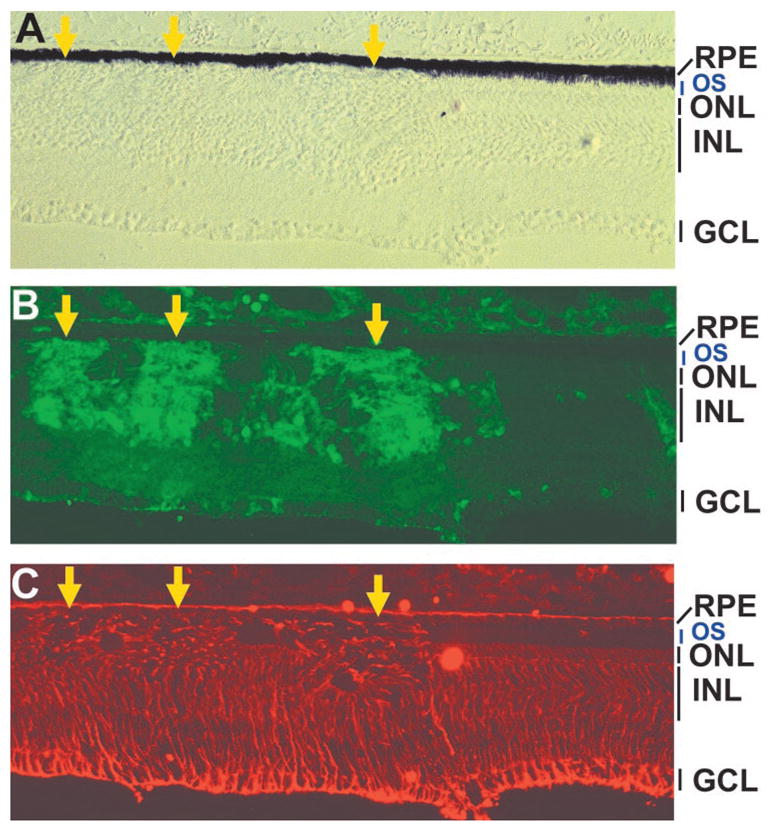
Müller glia (vimentin+) in E19 retina partially infected with RCAS-En-NeuroDΔC. (A) A DIC image of the retinal sections. (B, C) Double immunostaining for p27 protein (B) and vimentin (C). Arrows: virally infected regions with ONL deficits. OS: outer segments of photoreceptor cells.
Transient Presence of chx10+ Cells in the ONL
In a normal, stratified chick retina, chx10+ cells localize exclusively in the INL, and no such cells are found in the ONL. In retinas infected with RCAS-En-NeuroDΔC, however, chx10+ cells were detected in the ONL (Figs. 7A, 7B). Unlike those chx10+ cells that occupied the breaks of the ONL and were confined by the OPL (Fig. 5G), these chx10+ cells within the ONL were distributed in almost a single layer beyond the boundary defined by the OPL. The presence of chx10+ cells within the ONL occurred only in the infected regions, whereas in the adjacent, uninfected regions, chx10+ cells remained confined to the INL (Fig. 7A). The presence of chx10+ cells in the ONL appeared transient, and was observed only between E10 and E12.
Figure 7.
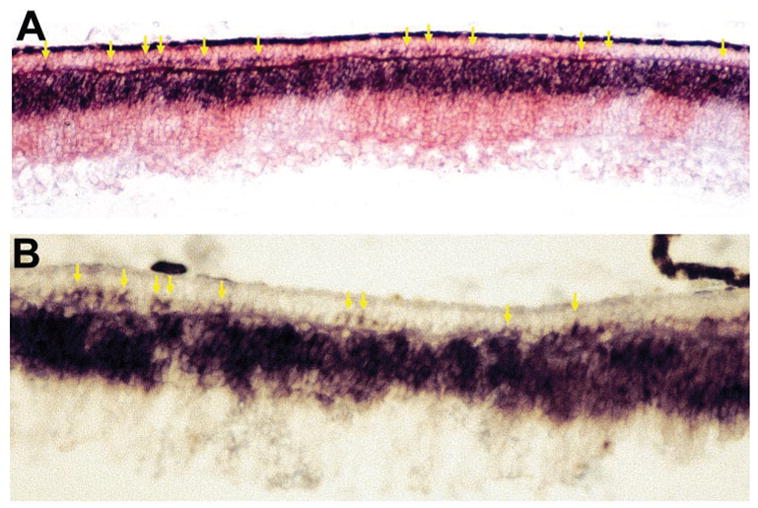
Chx10+ bipolar cells in the ONL in E10 retina infected with RCAS-En-NeuroDΔC. (A) Double labeling of an E12 retina for chx10 mRNA (blue) and p27 protein (red). (B) Chx10+ cells at a higher magnification. Arrows: ONL cells expressing chx10. Magnification: (A) ×50; (B) ×100.
Reduction in Photoreceptor Cells by Antisense Oligonucleotides
We also tested whether attenuating neuroD expression with antisense oligonucleotides specifically reduces the number of photoreceptor cells. We first microinjected antisense oligonucleotides into the subretinal space at E4.5 and E5.5, when neuroD expression begins to increase. Injections of corresponding sense oligonucleotides, as well as medium alone, were used as controls. Histologic analysis of gene expression was used, owing to the fact that only a small area of the retina was directly exposed to the injected solutions. Because retinal development proceeds from the fundus to the periphery, comparisons of gene expression were performed only with topo-graphically equivalent regions in the two retinas of a given animal, and at least four animals were examined in each group. Within 24 hours no gross abnormalities were observed in injected eyes, but by E8, when the retina begins to laminate, retinal folding and thinning were observed in some of the injected retinas, probably from repeated microinjections at the developmental stages. Therefore, the E6.5 retina was used in the analysis.
Microinjection of either buffer alone or the sense oligonucleotides had no detectable effect on the expression of neuroD and visinin (Figs. 8A, 8B, 8E, 8F). In contrast, a marked decrease in the neuroD mRNA was observed with injection of the antisense oligonucleotides (Figs. 8C, 8D). The antisense oligonucleotides also decreased the number of cells expressing visinin (Figs. 8G, 8H). Notably, the level of visinin mRNA appeared unaffected in those visinin+ cells of the injected retina (Fig. 8G), which might have committed earlier and thus avoided the treatment. The antisense oligonucleotides had no apparent effect on the number of Islet-1+ ganglion cells (Figs. 8I, 8J).
Figure 8.
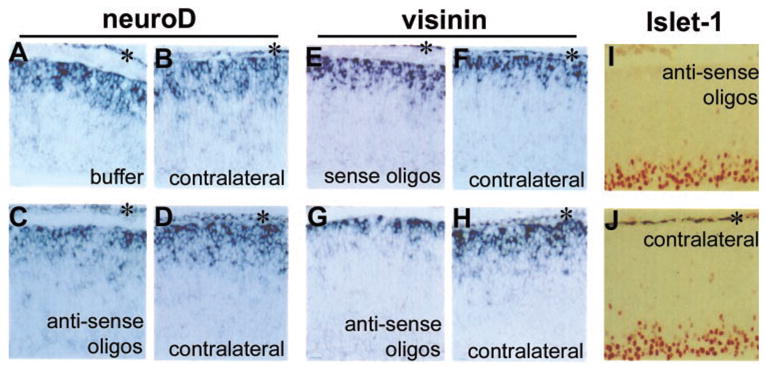
Effects of antisense oligonucleotides microinjected into the subretinal space. (A) In situ hybridization detection of neuroD mRNA in the retinal neuroepithelium with injection of medium alone and (B) in the contralateral eye. (C) neuroD mRNA in the retinal neuroepithelium injected with antisense oligonucleotides and (D) in the contralateral eye. (E) In situ hybridization detection of visinin mRNA in the retinal neuroepithelium with injection of sense oligonucleotides and (F) in the contralateral eye. (G) Visinin mRNA in the retinal neuroepithelium with injection of antisense oligonucleotides (H) in the contralateral eye. (I) Islet-1+ (immunostaining) cells in the retinal neuroepithelium injected with antisense oligonucleotides and (J) in the contralateral eye. (*) RPE. The gap between the RPE and retinal neuroepithelium in (A), (C), and (E) indicates retinal detachment from microinjection. Magnification: ×50.
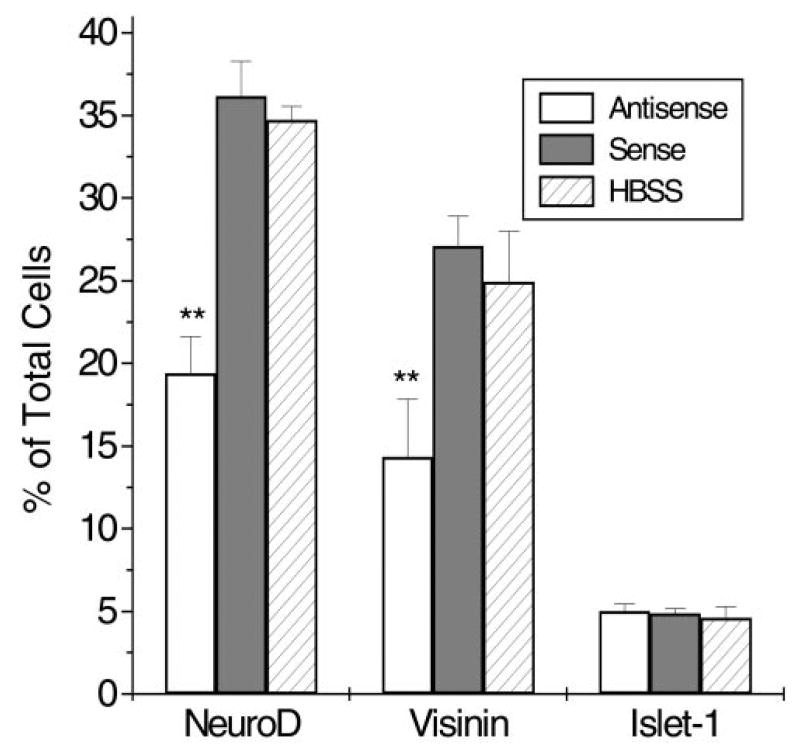
To analyze quantitatively the effect of the oligonucleotides on retinal cell production, we used retinal explants, the entirety of which is exposed to medium containing antisense oligonucleotides or control oligonucleotides. The number of photoreceptor cells was scored after dissociation of retinal cells. We found that the number of NeuroD+ cells, identified with the affinity-purified anti-NeuroD antibody, was reduced almost 50%, from 36.2% ± 2.1% of total cells in sense oligonucleotide control to 19.4% ± 2.2% (Fig. 9). A similar degree of reduction was observed with cells expressing visinin, from 27.1% ± 1.8% of total cells in the control to 14.3% ± 3.5%. The number of Islet-1+ ganglion cells remained unchanged (5.0% ± 0.4% and 4.9% ± 0.3%, respectively).
Figure 9.
Effects of antisense oligonucleotides added in the culture medium of retinal explants. Expression of neuroD and Islet-1 was revealed by immunostaining with specific antibodies and that of visinin by in situ hybridization. Data are the mean ± SD of the percentage of positive cells over total cells. **Statistically significant reduction (P = 0.01).
Effects of siRNA on Photoreceptor Formation
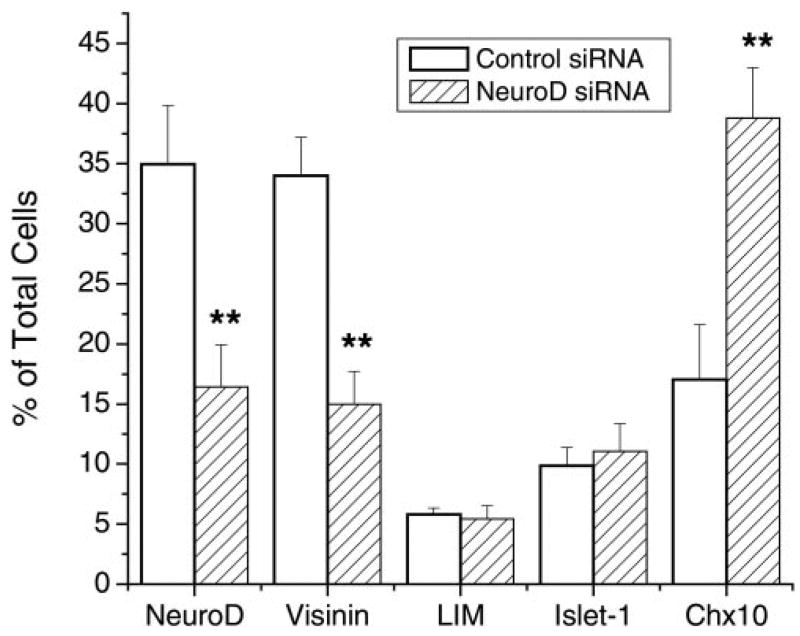
Recently reports have shown that siRNA can be a powerful tool in loss-of-function studies.40 We examined two siRNA molecules against neuroD on retinal neuron formation. E5.5 retinal cells were dissociated and transfected with siRNA. Examining cells for fluorescein (present in the control siRNA) indicated that nearly 100% of the cells were transfected (data not shown). Treatment of the retinal cells with siRNAs against neuroD resulted in a more than 50% reduction in the number of NeuroD+ cells and visinin+ cells, compared with control siRNA (Fig. 10). No reductions were observed in LIM+ horizontal cells or Islet-1+ ganglion cells. Notably, we detected a large increase in the number of chx10+ cells (Fig. 10).
Figure 10.
Effects of siRNA in the culture medium of dissociated retinal cells. Expression of neuroD, LIM, and Islet-1 was revealed by immunostaining with specific antibodies and that of visinin and chx10 by in situ hybridization. Data are the mean ± SD of the percentage of positive cells over total cells. **Statistically significant change (P = 0.01).
Discussion
Defining the retinal expression of neuroD is important in understanding its role in retinal development. We have shown that neuroD mRNA was detected in young photoreceptor cells and their precursors, and was undetectable in amacrine cells, as described by Morrow et al.39 and Inoue et al.,41 or in ganglion cells, as reported by Brown et al.42 Results from the current immunohistochemical staining showed that anti-NeuroD antibody specifically stained young photoreceptor cells and their precursors. These results are consistent with our previous in situ data, but show discrepancies with other reports. Species differences may contribute to these discrepancies. Furthermore, differences in detection methods, particularly differences in their specificity and sensitivity, may also play a role. We have noticed that when the hybridization stringency for visinin mRNA is lowered, some INL cells become positive, albeit faintly (Fig. 4B; data not shown). In addition the monoclonal antibody against visinin weakly stains a subpopulation of amacrine cells (Fig. 4F). It is not clear whether these cells are displaced photoreceptor cells that are transiently localized in the INL before migrating to their final destination of the ONL or certain amacrine cells express a low level of visinin or contain a visinin-like molecule. Hoover et al.43 found a few RXRγ+ cells at locations of amacrine cells in a developing retina, and they believed that these cells were photoreceptor cells and would eventually migrate into the ONL. If neuroD is weakly expressed in amacrine cells and if neuroD plays a role in amacrine cell specification as indicated by Morrow et al.,39 and Inoue et al.,41 active repression of NeuroD would be more easily achieved in amacrine cells than in photoreceptor cells, producing a detectable amacrine phenotype. However, our active repression studies showed a profound deficit of photoreceptor cells, with no deficiency of amacrine cells.
In our loss-of-function analyses, a simple dominant negative construct, NeuroDΔC, did not lead to noticeable alteration of retinal neurogenesis. This was probably due to inadequate inhibition of the wild-type NeuroD protein by the mutant one. En-mediated active repression of NeuroD, however, produced a profound retinal phenotype. This agrees with the observation by Badiani et al.44 that En-mediated active repression is much more potent than a conventional dominant negative construct. In retinas infected with RCAS-En-NeuroDΔC, there were severe photoreceptor deficits, and other retinal cell populations were not reduced. Photoreceptor deficiency was observed as early as the time of photoreceptor birth, suggesting that En-NeuroDΔC may have inhibited photoreceptor genesis. The specific reduction in the number of photoreceptor cells was also found with attenuating neuroD expression with either antisense oligonucleotides or siRNA. The results of all three lines of the loss-of-function investigation indicate that neuroD is necessary for photoreceptor cell generation in the chick retina and support the hypothesis that neuroD specifies the photoreceptor fate in the chick retina.
Although active repression of NeuroD led to a severe photoreceptor deficiency, only a mild reduction in ONL cells was noted in neuroD-null mice in one study,41 and no reduction was reported in another.39 Many factors may contribute to the conflicting observations. (1) Species differences may be a factor, considering that even the different genetic backgrounds of knockout mice may manifest various phenotypes, such as neonatal death of neuroD null mutants.45,46 It is possible that neuroD in mouse retina is involved in amacrine cell genesis,39,41 but not in photoreceptor genesis. (2) Differences in experimental approaches and conditions (e.g., in vivo versus cell or explant culture) may partially explain these discrepancies. For example, using in vivo lipofection of DNA, Kanekar et al.47 reported that neuroD decreases the number of photoreceptor cells and increases the number of bipolar and amacrine cells. NeuroD-promoting bipolar cells was also reported by Ahmad et al.,48 using retroviral-driven overexpression of neuroD in cultured retinal explants. Retroviral-driven overexpression of neuroD in postnatal mouse retina was found to promote amacrine cells and decrease bipolar cells by Morrow et al.,39 but was found to promote rod photoreceptor cells in explant culture.41 (3) It is conceivable that neuroD may be redundant in mouse photoreceptor formation. The bHLH gene hes5 is thought to be redundant in retinal gliogenesis.49 cAMP-response element modulator (CREM) has been proposed to compensate for cAMP response element binding (CREB) in CREB null mutants.50 In this case, a dominant negative construct of CREB produced the anticipated dwarf phenotype,51,52 even though CREB-null mice exhibit normal growth and development.50
On active repression of NeuroD protein, chx10+ cells were detected in the ONL, where photoreceptor cells reside. These chx10+ cells in the ONL might be authentic bipolar cells that have migrated past the OPL into the ONL. They might also be misfated or refated cells. It is possible that these cells originally were to become photoreceptor cells, but switched to chx10+ bipolar cells due to the repression of NeuroD function by Engrailed. This scenario is further favored by the increase in chx10+ cells in the neuroD siRNA experiment. Bipolar cell population was reported to be increased in mice without neuroD.39 Thus, it is possible that reducing neuroD activity promotes a switch in fate from a photoreceptor cell to a bipolar cell. It is also possible, however, that those chx10+ cells were merely progenitor cells unable to proceed to differentiation as photoreceptor cells, because of NeuroD repression.
En-mediated active repression has been used successfully to block the function of several transcription factors in various systems. 44,53–61 Published studies indicate that En-mediated active repression is specific. In this study, we show that three controls (En alone, En-cNSCL1, and En-cNSCL2), did not cause photoreceptor deficiency, whereas En-NeuroDΔC did. In a separate study, we generated En-cbx(HD) to block the function of a homeobox gene cbx.31 En-cbx(HD) impairs embryonic survival, but has no effect on ONL morphology or photoreceptor generation, providing further support to the specificity of En-NeuroDΔC causing photoreceptor deficiency. Additional evidence supporting the specificity of En-NeuroDΔC include that (1) retinal neurons other than photoreceptor cells were not reduced by En-NeuroDΔC; (2) both in situ hybridization and immunohistochemistry showed that neuroD expression was specific to photoreceptor cells and their precursors; and (3) the phenotypes observed with En-NeuroDΔC were exactly the reverse of those in gain-of-function studies, in which photoreceptor cells were overproduced in the retina infected with RCAS-neuroD.24 Together with the antisense oligonucleotides and siRNA data, our studies suggest that neuroD plays an important role in photoreceptor formation in the chick retina.
Acknowledgments
The authors thank Stephen Hughes for RCAS and Cla12Nco; and the following scientists for developing the monoclonal antibodies used in the study: Constance Cepko (7G4, anti-visinin), Thomas Jessell (4F2, anti-LIM), Jeremy Nathans (anti-Brn3a), Louise Reichardt (mAb 48, against a synaptic vesicle-specific membrane protein), Joshua Sanes (H5, anti-Vimentin), and Trevor Williams (3B5, anti-AP2α).
Supported by National Eye Institute Grant EY11640; the Research to Prevent Blindness Dolly Green Scholar Award; unrestricted grants to University of Alabama at Birmingham Department of Ophthalmology from Research to Prevent Blindness; and EyeSight Foundation of Alabama Grant 01-7.
Footnotes
Disclosure: R.-T. Yan, None; S.-Z. Wang, None
References
- 1.Turner DL, Cepko CL. A common progenitor for neurons and glial persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 2.Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 3.Raymond PA, Reifler MJ, Rivlin PK. Regeneration of goldfish retina: rod precursors are a likely source of regenerated cells. J Neurobiol. 1988;19:431–463. doi: 10.1002/neu.480190504. [DOI] [PubMed] [Google Scholar]
- 4.Adler R, Hatlee M. Plasticity and differentiation of embryonic retinal cells after terminal mitosis. Science. 1989;243:391–393. doi: 10.1126/science.2911751. [DOI] [PubMed] [Google Scholar]
- 5.Adler R. A model of retinal cell differentiation in the chick embryo. Prog Retin Eye Res. 2000;19:529–557. doi: 10.1016/s1350-9462(00)00008-2. [DOI] [PubMed] [Google Scholar]
- 6.Ezzeddine ZD, Yang X, DeChiara T, Yancopoulos G, Cepko CL. Postmitotic cells fated to become rod photoreceptors can be respecified by CNTF treatment of the retina. Development. 1997;124:1055–1067. doi: 10.1242/dev.124.5.1055. [DOI] [PubMed] [Google Scholar]
- 7.Kelley MW, Turner JK, Reh TA. Retinoic acid promotes differentiation of photoreceptors in vitro. Development. 1994;120:2091–2102. doi: 10.1242/dev.120.8.2091. [DOI] [PubMed] [Google Scholar]
- 8.Hyatt GA, Schmitt EA, Fadool JM, Dowling JE. Retinoic acid alters photoreceptor development in vivo. Proc Natl Acad Sci USA. 1996;93:13298–13303. doi: 10.1073/pnas.93.23.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks D, Courtois Y. Fibroblast growth factor stimulates photoreceptor differentiation in vitro. J Neurosci. 1992;12:2022–2033. doi: 10.1523/JNEUROSCI.12-06-02022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Sanchez C, Lopez-Carranza A, Alarcon C, de La Rosa EJ, de Pablo F. Autocrine/paracrine role of insulin-related growth factors in neurogenesis: local expression and effects on cell proliferation and differentiation in retina. Proc Natl Acad Sci USA. 1995;92:9834–9838. doi: 10.1073/pnas.92.21.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillien L. Changes in retinal cell fate induced by overexpression of EGF receptor. Nature. 1995;377:158–162. doi: 10.1038/377158a0. [DOI] [PubMed] [Google Scholar]
- 12.McFarlane S, Zuber ME, Holt CE. A role for the fibroblast growth factor receptor in cell fate decisions in the developing vertebrate retina. Development. 1998;125:3967–3975. doi: 10.1242/dev.125.20.3967. [DOI] [PubMed] [Google Scholar]
- 13.Kelley MW, Turner JK, Reh TA. Ligands of steroid/thyroid receptors induce cone photoreceptors in vertebrate retina. Development. 1995;121:3777–3785. doi: 10.1242/dev.121.11.3777. [DOI] [PubMed] [Google Scholar]
- 14.Altshuler D, Lo Turco JJ, Rush J, Cepko C. Taurine promotes the differentiation of a vertebrate retinal cell type in vitro. Development. 1993;119:1317–1328. doi: 10.1242/dev.119.4.1317. [DOI] [PubMed] [Google Scholar]
- 15.Hunter DD, Murphy MD, Olsson CV, Brunken WJ. S-laminin expression in adult and developing retinae: a potential cue for photoreceptor morphogenesis. Neuron. 1992;8:399–413. doi: 10.1016/0896-6273(92)90269-j. [DOI] [PubMed] [Google Scholar]
- 16.Davis AA, Matzuk MM, Reh TA. Activin A promotes progenitor differentiation into photoreceptors in rodent retina. Mol Cell Neurosci. 2000;15:11–21. doi: 10.1006/mcne.1999.0806. [DOI] [PubMed] [Google Scholar]
- 17.Levine EM, Roelink H, Turner J, Reh TA. Sonic hedgehog promotes rod photoreceptor differentiation in mammalian retinal cells in vitro. J Neurosci. 1997;17:6277–6288. doi: 10.1523/JNEUROSCI.17-16-06277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenkamp DL, Frey RA, Prabhudesai SN, Raymond PA. Function for Hedgehog genes in zebrafish retinal development. Dev Biol. 2000;220:238–252. doi: 10.1006/dbio.2000.9629. [DOI] [PubMed] [Google Scholar]
- 19.Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- 20.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 21.Ng L, Hurley JB, Dierks B, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 22.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 23.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 24.Yan R-T, Wang S-Z. neuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J Neurobiol. 1998;36:485–496. [PMC free article] [PubMed] [Google Scholar]
- 25.Yan R-T, Wang S-Z. Expression of an array of photoreceptor genes in chick embryonic RPE cell cultures under the induction of neuroD. Neurosci Letters. 2000;280:83–86. doi: 10.1016/s0304-3940(99)01003-4. [DOI] [PubMed] [Google Scholar]
- 26.Yan R-T, Wang S-Z. Differential induction of gene expression by basic fibroblast growth factor and neuroD in cultured retinal pigment epithelial cells. Vis Neurosci. 2000;17:157–164. doi: 10.1017/s0952523800171172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick MB, Tamimi RM, Snider L, Asakura A, Bergstrom D, Tapscott SJ. NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 29.Wang S-Z, Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg ME, Bender TP. Preparation and analysis of RNA. In: Ausubel FM, Brent B, Kingston RE, et al., editors. Current Protocols in Molecular Biology. Boston MA: Massachusetts General Hospital; 2000. pp. 4.0.1–4.10.10. [Google Scholar]
- 31.Li C-M, Yan R-T, Wang S-Z. Chick homeobox gene cbx and its role in retinal development. Mech Dev. 2002;116:85–94. doi: 10.1016/s0925-4773(02)00148-x. [DOI] [PubMed] [Google Scholar]
- 32.Sharma A, Moore M, Marcora E, et al. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol. 1999;19:704–713. doi: 10.1128/mcb.19.1.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:427–433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han K, Manley JL. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uittenbogaard M, Peavy DR, Chiaramello A. Expression of the bHLH gene NSCL-1 suggests a role in regulating cerebellar granule cell growth and differentiation. J Neurosci Res. 1999;57:770–781. [PubMed] [Google Scholar]
- 36.Ray SK, Nishitani J, Petry MW, Fessing MY, Leiter AB. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol Cell Biol. 2003;23:259–271. doi: 10.1128/MCB.23.1.259-271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C-M, Yan R-T, Wang S-Z. Misexpression of cNSCL1 disrupts retinal development. Mol Cell Neurosci. 1999;14:17–27. doi: 10.1006/mcne.1999.0765. [DOI] [PubMed] [Google Scholar]
- 38.Li C-M, Yan R-T, Wang S-Z. Misexpression of chick NSCL2 causes atrophy of Müller glia and photoreceptor cells. Invest Ophthalmol Vis Sci. 2001;42:3103–3109. [PMC free article] [PubMed] [Google Scholar]
- 39.Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 40.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 41.Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- 42.Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 43.Hoover F, Seleiro EA, Kielland A, Brickell PM, Glover JC. Retinoid X receptor gamma gene transcripts are expressed by a subset of early generated retinal cells and eventually restricted to photoreceptors. J Comp Neurol. 1998;391:204–213. [PubMed] [Google Scholar]
- 44.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 45.Naya FJ, Huang HP, Qiu Y, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M, Pleasure SJ, Collins AE, et al. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc Natl Acad Sci USA. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanekar S, Perron M, Dorsky R, et al. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad I, Acharya HR, Rogers JA, Shibata A, Smithgall TE, Dooley CM. The role of NeuroD as a differentiation factor in the mammalian retina. J Mol Neurosci. 1999;11:165–178. doi: 10.1385/JMN:11:2:165. [DOI] [PubMed] [Google Scholar]
- 49.Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- 50.Hummler E, Cole TJ, Blendy JA, et al. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Struthers RS, Vale WW, Arias C, Sawchenko PE, Montminy MR. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non-phosphorylatable CREB mutant. Nature. 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- 52.Barton K, Muthusamy N, Chanyangam M, Fischer C, Clendenin C, Leiden JM. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature. 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 53.Fan MJ, Sokol SY. A role for Siamois in Spermann organizer formation. Development. 1997;124:2581–2589. doi: 10.1242/dev.124.13.2581. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Wikramanayake AH, Klein WH. Requirement of SpOtx in cell fate decisions in the sea urchin embryo and possible role as a mediator of beta-catenin signaling. Dev Biol. 1999;212:425–439. doi: 10.1006/dbio.1999.9360. [DOI] [PubMed] [Google Scholar]
- 55.Wei Z, Angerer LM, Angerer RC. Spatially regulated SpEts4 transcription factor activity along the sea urchin embryo animal-vegetal axis. Development. 1999;126:1729–1737. doi: 10.1242/dev.126.8.1729. [DOI] [PubMed] [Google Scholar]
- 56.Zuber ME, Perron M, Philpott A, Bang A, Harris WA. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]
- 57.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 58.Becker JR, Dorman CM, McClafferty TM, Johnson SE. Characterization of a dominant inhibitory E47 protein that suppresses C2C12 myogenesis. Exp Cell Res. 2001;267:135–143. doi: 10.1006/excr.2001.5249. [DOI] [PubMed] [Google Scholar]
- 59.Yu X, St Amand TR, Wang S, et al. Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development. 2001;128:1005–1013. doi: 10.1242/dev.128.6.1005. [DOI] [PubMed] [Google Scholar]
- 60.Jamali M, Rogerson PJ, Wilton S, Skerjanc IS. Nkx2–5 activity is essential for cardiomyogenesis. J Biol Chem. 2001;276:42252–42258. doi: 10.1074/jbc.M107814200. [DOI] [PubMed] [Google Scholar]
- 61.Gammill LS, Sive H. otx2 expression in the ectoderm activates anterior neural determination and is required for Xenopus cement gland formation. Dev Biol. 2001;240:223–236. doi: 10.1006/dbio.2001.0470. [DOI] [PubMed] [Google Scholar]