Abstract
We present a new approach to molecular classification based on mRNA comparisons. Our method, referred to as the top-scoring pair(s) (TSP) classifier, is motivated by current technical and practical limitations in using gene expression microarray data for class prediction, for example to detect disease, identify tumors or predict treatment response. Accurate statistical inference from such data is difficult due to the small number of observations, typically tens, relative to the large number of genes, typically thousands. Moreover, conventional methods from machine learning lead to decisions which are usually very difficult to interpret in simple or biologically meaningful terms. In contrast, the TSP classifier provides decision rules which i) involve very few genes and only relative expression values (e.g., comparing the mRNA counts within a single pair of genes); ii) are both accurate and transparent; and iii) provide specific hypotheses for follow-up studies. In particular, the TSP classifier achieves prediction rates with standard cancer data that are as high as those of previous studies which use considerably more genes and complex procedures. Finally, the TSP classifier is parameter-free, thus avoiding the type of over-fitting and inflated estimates of performance that result when all aspects of learning a predictor are not properly cross-validated.
Keywords: microarray data, class prediction, mRNA comparisons
1 Introduction
Using DNA microarray technology, it is now possible to measure the expression levels of thousands of genes simultaneously (Duggan et al. (1999); Lipshutz et al. (1999); Lockhart and Winzeler (2000)). However, the number of profile measurements per experimental study remains quite small, usually fewer than one hundred (Hughes et al. (2000); Pomeroy et al. (2002)). When measured against the complexity of the systems being studied, the amount of data available for modeling and inference is therefore severely limited. Discovery of the underlying structure within these data, in particular correlation patterns or even higher-dimensional interactions, is exceedingly difficult in this small-sample regime.
The small-sample dilemma in the statistical analysis of microarray data is well-documented in the literature (Dudoit and Fridlyand (2003); Sebastiani et al. (2003); Simon et al. (2003); West et al. (2001)). In view of well-known tradeoffs in computational learning theory between sample size and model complexity (Hastie et al. (2001)), and between “bias” and “variance” (Geman et al. (1992)), some simplifying assumptions (such as the reduction of the dimensionality of the data and/or the family of classifiers) appear necessary. Indeed, there is already evidence (Dudoit and Fridlyand (2003)) that relatively simple classification methods as in Tibshirani et al. (2002) are competitive with more complex ones. Our results support and extend this finding.
Another limitation of current methods for classifying gene expression profiles is the “black box” dilemma. Standard methods in statistical learning and pattern recognition are routinely applied to microarray data, examples being neural networks (Bicciato et al. (2003); Bloom et al. (2004); Khan et al. (2001)), decision trees (Boulestiex et al. (2003); Dettling and Buhlmann (2003); Zhang et al. (2003)) and support vector machines (Peng et al. (2003); Yeang et al. (2001)). Typically, this results in predictions based on nonlinear functions of many expression values, and consequently highly complex decision boundaries between the classes of interest. Such boundaries are then difficult to summarize in simple terms or to characterize in a manner which is biologically meaningful.
We address both problems – small samples and lack of interpretability – by basing our predictions entirely on pairwise comparisons. More specifically, we attempt to differentiate between two classes by finding pairs of genes whose expression levels typically invert from one class to the other. Our approach is a particular instance of a larger class of rank-based methods, all of which are characterized by immediately replacing expression levels by their corresponding ranks (i.e., most heavily expressed, second most heavily expressed, etc.) determined across all genes assayed using a single DNA microarray; this step is equivalent to recording the gene having a larger expression intensity for each pair of genes on a DNA microarray. Rank-based methods are therefore robust to quantization effects and are invariant to pre-processing designed to overcome chip-to-chip variation, such as normalization methods (Yang et al. (2001)), under the very mild (and nearly universally satisfied) assumption that the normalization procedure is monotonic in the expression values, and hence preserves ordering.
There is no question that information is lost using a rank-based procedure. However, the results reported here, obtained using several different data sets and based entirely on internal comparisons within the profile, demonstrate that the amount of information residing in the ordering of gene expression levels is more than sufficient to reliably perform classification and other tasks. Indeed, in some cases, accurate prediction can be achieved by comparing the expression levels of a single pair of genes.
In the following sections, we focus on a particularly simple example of a comparison-based approach to classifying gene expression profiles – the “top-scoring pair(s)” or TSP classifier. A family of gene pairs is isolated and then a profile is classified based on a decision rule which only involves comparing mRNA abundance in each pair and then aggregating the results. For the TSP classifier, the participating pairs are those which achieve the largest “score” relative to a simple measure of discrimination. Scores are estimated from the training data and there are no parameters to tune since the number of top-scoring pairs is completely determined by the data. In some cases, there is a single pair of genes achieving the top score and classification then amounts to choosing the class under which the observed ordering within this pair is most likely.
We demonstrate the efficacy of this method on several gene expression data sets involving breast, prostate and leukemia cancers. We also illustrate how the TSP method can generate a specific hypothesis for follow-up studies. Our classification rates are comparable to the best results reported in the literature; moreover, in some cases those results are not properly validated (e.g., by fully cross-validating parameter choices) and may be biased (Simon et al. (2003); Dudoit and Fridlyand (2003)). In addition, the TSP classifier usually employs considerably fewer genes and is easier to interpret.
Our approach to selecting informative pairs of genes is but one example of attempting to exploit information residing in gene-gene interactions. Despite the evident importance of capturing such joint statistics, the literature is sparse, due perhaps to the sample size limitations alluded to earlier. The co-expression of genes in the cell cycle is considered in Li (2002) but there is no connection with classification. The idea of evaluating genes in pairs in order to select discriminating genes for classifying microarray data first appears in Bø and Jonassen (2002). A subset of k gene pairs is selected based on a recursive procedure and a scoring criterion which involves diagonal linear discriminant analysis and a two-sample t-test. Using the resulting 2k genes in conjunction with standard classifiers, the authors demonstrate an improvement over feature selection based on individual genes. In particular, there is nothing rank-based and the pairing of genes is ignored during classification. They also report good prediction rates for two cancer data sets using a few tens of genes, although the parameter choices (such as k) are not part of the cross-validation. Our results corroborate their findings. Further, we show that the information in gene pair interactions can be directly exploited using simple comparisons for both scoring and decision-making.
In the following section we describe our rank-based approach to pair selection and classification in more detail. The experiments with three cancer data sets are presented in §3, including comparisons with other results in terms of accuracy and efficiency as well as a brief description of the biological relevance of the top-scoring pairs. We conclude in §4 with some remarks about ratios of concentrations, estimating prediction rates and sample size.
2 Comparison-Based Classification
Consider G genes whose expression levels X = {X1, X2, …, XG} are measured using DNA microarrays and regarded as random variables. Each profile X has a true class label in {1, 2, …, C}. For simplicity, we assume C = 2, although the results extend to higher numbers of classes. We focus on detecting “marker gene pairs” (i, j) for which there is a significant difference in the probability of Xi < Xj from class 1 to class 2. Profile classification is then based on this collection of distinguished pairs. Here, the quantities of interest are pij(c) = P (Xi < Xj|c), c = 1, 2, i.e., the probabilities of observing Xi < Xj in each class. These probabilities are estimated by the relative frequencies of occurrences of Xi < Xj within profiles and over experiments. Consequently, for our analysis it is sufficient to know the ranks of the expression values within profiles on each microarray. This approach differs from nonparametric methods for detecting differentially regulated genes (see, e.g., Sebastiani et al. (2003)) in which ranking is done across experiments for each fixed gene.
Let Δij = |pij(1) − pij(2)| denote the “score” of (i, j). An example of computing a score is provided in Table 1. We seek gene pairs with “large” scores.
Table 1.
An example of scoring a gene pair from the prostate study. Expression levels for 12,600 probes are obtained for 52 profiles associated with class 1 (prostate tumors) and 50 associated with class 2 (normal tissue). (See the Appendix for details on this study, which involves detecting prostate cancer.) For a particular pair (i,j) of genes we have identified, the 102 profiles are labeled according to the above 2 × 2 contingency table. These data lead to the probability estimates pij(1) = 50/52 and pij(2) = 3/50, which results in the score . Since pij(1) > pij(2), the classifier based on this gene pair votes for class 1 for a profile with Xi < Xj and for class 2 otherwise.
| Xi < Xj | Xi > Xj | ||
|---|---|---|---|
| class 1 | 50 | 2 | 52 |
| class 2 | 3 | 47 | 50 |
2.1 Gene Pair Selection
Detection of marker gene pairs is a problem in feature selection, and plays the same role in our analysis as finding individual marker genes does in more standard methods (Dudoit et al. (2002); Sebastiani et al. (2003); Tibshirani et al. (2002); Stolovitzky (2003)). One option for pair selection might be to first select differentially-regulated or “marker genes” and only then proceed from individual genes to gene pairs by restricting the search for marker pairs to pairs of these marker genes. But two major drawbacks would ensue: 1) such post-filtering results would no longer be invariant to normalization; and 2) by construction, only differentially expressed genes could appear in the selected comparisons, thereby possibly losing discriminating pairs in which at most one gene is itself differentially expressed. We therefore adopt a more straightforward method based on direct search: We estimate Δij for every distinct pair (i, j) and apply a selection rule based on the magnitude of Δij. An example of such a decision rule, and the one we use throughout this paper, is to rank the scores Δij from largest-to-smallest and select all pairs achieving the top score.
2.2 Classification
Pair selection results in a family P of distinguished pairs. Again, the gene pairs in P are precisely those whose score is maximal; hence the number of pairs in P is not a parameter of the system but rather data-driven. For medium sample sizes, there are usually only a few pairs which achieve the top score; for example, in two of the three experiments presented here there is only one such pair, and there are three pairs in the other experiment. For very small sample sizes there may be many pairs; an example is given at the end of §3.
Any standard classification algorithm may then be implemented using P as input. We are interested in algorithms for which classification decisions have a simple interpretation. Voting is an example of such a decision algorithm, where individual votes are driven by maximum likelihood. In this method, given a new expression profile X, an individual pair (i, j) in P votes for the class for which the observed ordering between Xi and Xj is more likely; see the example in Table 1. That is, if we observe Xi < Xj, then pair (i, j) votes for class 1 if pij(1) ≥ pij(2) and votes for class 2 otherwise. The class with the most votes is chosen. We refer to the resulting classifier as the top scoring pair(s) classifier, henceforth denoted TSP.
It is noteworthy that for classification based on a single gene pair, the sum of misclassification probabilities over the two classes can be expressed as 1 − Δij, which provides a natural justification for score maximization.
The procedure of tallying individual votes, while attractive from the point of view of simplicity (Dudoit et al. (2002)), also can be derived as a maximum likelihood rule under the simplifying assumptions that (i) individual comparisons are conditionally independent given the class, and (ii) for some p we have either pij(c) = p or pij(c) = 1 − p for all (i, j) ∈ P and both classes c = 1, 2.
2.3 Error Estimation
In estimating the (generalization) error rate of a classifier, gene pair selection was performed within the cross-validation loop. With n samples and (leave-one-out) cross validation (CV), this means choosing n separate subsets P, one for each profile “held out” during training, then classifying that profile. (Other methods for estimating the error rate could be considered; see §4.) In particular, both the actual top-score, as well as the set of pairs which achieve it, may vary with the sample left out. The estimated prediction rate is then 1 − e/n where e ∈ {1, …, n} is the number of errors observed in the cross-validation.
For our procedure there are no parameters to select inside the CV loop. For other procedures that do require parameters, e.g., k-nearest neighbors, random forests and support vector machines, the estimated prediction rates may be severely biased if performance is sensitive to these parameters and they are not properly cross-validated (using an inner CV loop to choose parameter values) (Dudoit and Fridlyand (2003); Simon et al. (2003); West et al. (2001)). The TSP classifier avoids this source of bias.
3 Experiments
The TSP classifier was evaluated on three class prediction problems: Predicting the status of lymph nodes in patients with breast tumors (Breast study); Classifying profiles into leukemia subtypes (Leukemia study); Distinguishing prostate tumors from normal profiles (Prostate study). Details involving these data (references, chips, samples sizes, web addresses, etc.) can be found in the Appendix.
3.1 Top-Scoring Pairs
There are three top-scoring pairs for the Leukemia data and only one for the Breast and Prostate data; the actual top scores, and corresponding gene pairs, are identified in Table 2, together with their individual t-statistics. Some of these genes would not be regarded as “differentially regulated” on the basis of their individual t-statistics. Notice that the same gene may appear in more than one pair.
Table 2.
The top scoring pair(s) for each study, together with the top score and individual t-statistics.
| Problem | Score | Genbank ID 1 | t-stat 1 | Genbank ID 2 | t-stat 2 |
|---|---|---|---|---|---|
| Breast | 0.838 | X03453 | 4.39 | X82634 | 2.25 |
| Prostate | 0.902 | M84526 | 7.46 | M55914 | 4.13 |
| Leukemia | 0.979 | L11373 | 1.99 | X95735 | 10.92 |
| Leukemia | 0.979 | D86976 | 1.60 | X95735 | 10.92 |
| Leukemia | 0.979 | J05243 | 7.87 | M23197 | 6.62 |
Gene descriptions
X03453 Bacteriophage P1 cre gene for recombinase protein
X82634 Homo sapiens mRNA for hair keratin acidic 3-II
M84526 Human adipsin/complement factor D mRNA, complete cds
M55914 Homo sapiens c-myc binding protein (MBP-1) mRNA, complete cds
L11373 Human protocadherin 43 mRNA, complete cds for abbreviated PC43
X95735 Homo sapiens mRNA for zyxin
J05243 Human nonerythroid alpha-spectrin (SPTAN1) mRNA, complete cds
M23197 Human differentiation antigen (CD33) mRNA, complete cds
D86976 Human mRNA for KIAA0223 gene, partial cds
3.2 Score Significance
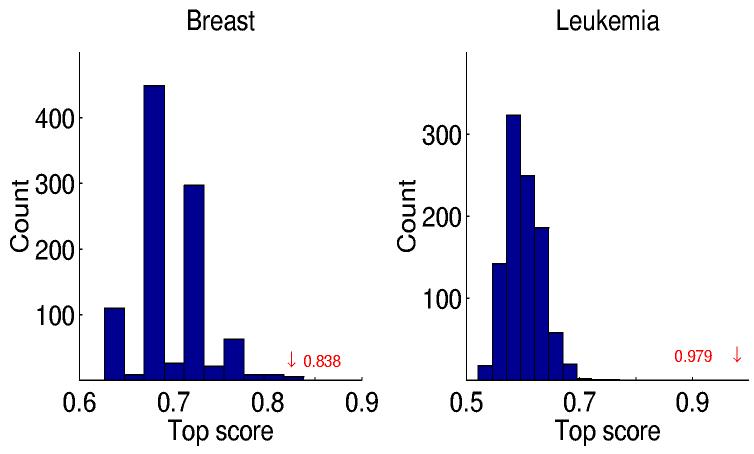
The significance of a score can be assessed by a permutation analysis. For any given study, artificial data sets can be constructed by randomly permuting the class labels, hence maintaining the sample sizes n1 and n2 of the two classes. The resulting top scores are then indicative of those obtained when attempting to classify based on profile labels which cannot be predicted from the expression values while maintaining the overall statistical dependency structure among the genes. In Figure 1, we display the histograms of top scores for the Breast and Leukemia studies based on 1000 permutations. In the latter case, for example, there is a top score for each of the 1000 random assignments of class labels to the n = 72 samples constrained by n1 = 47 and n2 = 25. From the permutation analysis we can compute a p-value associated with a given score obtained in the actual data by taking the fraction of permuted data sets in which a score at least as large is obtained. This p-value can be interpreted as the probability of observing such a large score under the null hypothesis that the pairs are non-informative for classification. No score among the 1000 permutation trials came near the top score actually observed (see Figure 1) on either the Leukemia or Prostate data, and hence the estimated p-values are virtually zero. For the Breast data the estimated p-value of the top score is 0.001.
Figure 1.
The distribution of top scores for a random class label permutation analysis. The locations of the top score on the real Breast and Leukemia data sets are shown in red; the estimated p-values are 0.001 and 0 respectively. The top-score histogram for the Prostate data looks qualitatively the same as the one for Leukemia, and the maximum score achieved among all of the artificial data sets is 0.586; the score observed on the real data is Δ = 0.902.
3.3 Classification Results
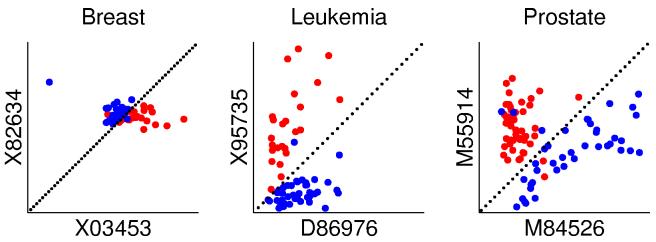
An intuitive appreciation of the nature of decision-making for the TSP classifier (i.e., predicting the class labels based entirely on the observed ordering among the pairs obtaining the top score) can be gleaned from Figure 2. For each study, there is a scatter plot of the expression levels for two genes – the unique top-scoring pair for the Breast and Prostate data and one of the three top-scoring pairs for the Leukemia data.
Figure 2.
Scatter plots for a top pair of genes for each study. The two classes are represented using red and blue, the axes represent the expression levels of the two genes and the dotted line y = x represents the decision boundary.
The estimated (correct) prediction rate of the TSP classifier for each study is displayed in Table 3 along with other reported results (and indicated references) for these data. All TSP results are based on leave-one-out cross-validation.
Table 3.
Some comparisons of performance between the TSP classifier and previously reported prediction rates. All TSP results are based on leave-one-out cross-validation. Breast: The range 41% – 88% (Dudoit and Fridlyand (2003)) covers both parameter settings, including the number of genes, and classification methods: k-nearest neighbors (8 – 26 errors in 49 samples), diagonal linear discriminant analysis (8 – 19 errors), diagonal quadratic discriminant analysis (11 – 26 errors), logitboost (9 – 21 errors), random forests (6 – 20 errors) and support vector machines (7 – 29 errors). More details appear in the text. Leukemia: In Golub et al. (1999), first the most “informative genes” are discovered by correlating profiles with ideal class identity vectors (a “signal-to-noise” variation on the t-statistic) and choosing the most significant ones based on a permutation test; these genes are then combined with a weighted voting scheme involving the correlations, class averages and a threshold for determining the “prediction strength” of a vote. The two stated rates, 85% and 95%, refer, respectively, to validation on the test set (see text) and leave-one-out cross-validation on the training set. Prostate: In Singh et al. (2002), a k-nearest neighbor classifier was applied to m genes (for selected values of m from 1 to 256) identified by measuring differential expression from normal to tumor samples using a variation of the signal-to-noise statistic (Golub et al. (1999)). For each m, prediction error was estimated using leave-one-out cross-validation; the range 86% – 92% corresponds to 4 ≤ m ≤ 256; the choice of k is not specified in Singh et al. (2002).
| Problem | Sample Size | TSP (# genes) | Previous Results (# genes) |
|---|---|---|---|
| Breast | 49 | 79% (2) | 41%-88% (10-7129) |
| Leukemia | 72 | 94% (5) | 85%,95% (50) |
| Prostate | 102 | 95% (2) | 86%-92% (4-256) |
3.3.1 Breast Study
In predicting the status of lymph nodes (affected or non-affected) in the Breast study, there are nine errors and three ties out of the 49 cross-validation loops; random tie-breaking then results in 10.5 errors on average which corresponds to an estimated classification rate of 79%. For comparison, estimated error rates for these data, also based on leave-one-out cross-validation and using a wide variety of common machine learning techniques, are summarized in Dudoit and Fridlyand (2003) for varying numbers of pre-filtered genes: m = 10, 50, 100, 200, 500, 1000, and m = 7129. Most parameter choices are external to the cross-validation in estimating the error rates listed in the main comparison in Dudoit and Fridlyand (2003); see the comprehensive discussion there. These external parameters include those which are method-specific as well as the choice of the number of genes that are pre-filtered.
For example, in the case of support vector machines, there are 48 experiments corresponding to choosing the kernel (linear or radial), the penalty, the filtering method and the number genes to be filtered; the number of errors varies considerably according to the protocol. Since the TSP classifier based on a unique top-scoring pair can be interpreted as a linear decision rule, albeit trivial, it can be seen as belonging to the same family of classifiers as a support vector machine with a linear kernel.
All of these methods are more complex than the TSP classifier and relatively few parameter choices yield better results. Moreover, it is not clear that some of these differences would remain after proper cross-validation of the other methods. One fully cross-validated experiment was performed in Dudoit and Fridlyand (2003) for k-NN and naive Bayes, resulting in 9 errors in both cases (and always using 10 genes).
3.3.2 Leukemia Study
The feasibility of cancer class prediction (as well as class discovery) based on gene expression monitoring was established in the pioneering work of Golub et al. (1999). The test case was separating AML from ALL leukemias (see Table 3 for a brief summary of the method in Golub et al. (1999)). Prediction rates are reported for a classifier based on fifty genes using cross-validation to measure accuracy on the initial data set of n = 38 acute leukemia samples (two samples are labeled “uncertain” and remaining 36 are correctly classified) as well as on an independent sample of 34 samples (making “strong predictions” for 29, all correct). Following the same protocol for training and testing, the TSP classifier correctly classifies 31 of the 34 samples (all three mistakes on ALL samples). On the combined data sets (see the Appendix), the TSP classifier uses five genes (the three pairs listed in Table 2) and classifies 68 samples correctly out of 72.
Biological Context
The first two pairs in Table 2 compare expression levels of L11373 (protocadherin gamma subfamily c,3; PCDHGC3) and D86976 (minor histocompatibility antigen; HA-1) with that of X95735 (zyxin; ZYX). ZYX is a member of the LIM protein family, co-localizes with integrins at sites of cell-substratum adhesion and is postulated to serve as a docking site for the assembly of protein complexes involved in regulating cell motility. The expression level of ZYX is up-regulated substantially in AML (382 in ALL versus 3258 in AML). Average expression levels of PCDHGC3 and HA-1 were (respectively) 1349 and 1396 in ALL versus 1063 and 1057 in AML. Differences between expression level in ALL and AML were not significant. Thus, PCDHGC3 and HA-1 may function in the TSP classifier as reference genes against which up-regulation of ZYX becomes highly discriminatory. Many other machine learning methods have also identified ZYX as a highly discriminating marker of ALL versus AML (Soukop and Lee (2003); Hwang et al. (2002); Siedow (2001); Golub et al. (1999)). The third pair is J05243 (the human nonerythroid -spectrin SPTAN1) and M23197 (the myeloid differentiation CD33 antigen). CD33 is well recognized as being a cell surface marker for AML (Griffin et al. (1983); Bradstock et al. (1989)), as it is generally not expressed in ALL (average expression level 174) but is expressed in most myeloid leukemias (average expression level 861). Expression of SPTAN1, which is involved in receptor binding and actin crosslinking, is elevated in ALL (average expression level 868) relative to AML (average expression level 162). This differential expression pattern produce a highly discriminating inversion of class probabilities.
3.3.3 Prostate Study
The results in Singh et al. (2002) confirmed a strong correlation between patterns of gene expression obtained at the time of diagnosis of prostate cancer and various clinical and pathological aspects of the disease. Specifically, it was established that “sufficiently robust” gene expression differences could be found to predict the identity of prostate samples. Results are reported for a k–nearest neighbor classifier (with Euclidean distance) applied to m genes for selected values of m (see Table 3). Prediction rates are estimated separately for each m using leave-one-out cross-validation, yielding a range from 86% to 92%, with top performance for m ≥ 4. The choice of k does not appear to be cross-validated, in which case the estimated prediction rates may be biased upwards.
Biological Context
The top scoring gene pair using the TSP classifier is M84526 (human adipsin complement factor D; DF) and M55914 (human c-myc binding protein; MBP-1). C-myc is a DNA-binding phosphoprotein protooncogene involved in the regulation of cell growth and differentiation and binding of MBP-1 to c-myc leads to tumor suppression (Pancholi (2001)). DF is a serine protease secreted by adipocytes into the bloodstream and functions as part of the alternative complement pathway of the innate immune system (Walport (2001)). Adipsin was identified as one of the top 50 marker genes in Singh et al. (2002), however, c-myc was not. Nonetheless, the joint behavior of c-myc and adipsin is highly discriminative of non-tumor versus prostate tumor samples, yielding a prediction rate of 95%.
3.3.4 A Very Small Sample Case
We applied the TSP classifier to one extreme case in terms of the ratio of sample size to the number of genes: We considered classifying gene expression profiles consisting of 22, 283 probes for 22 samples of myocardial tissue samples of patients diagnosed with idiopathic cardiomyopathy (IDCM) (12 samples) versus control (10 samples); these data are described in the Appendix. Previous studies have shown that virtually perfect discrimination between IDCM and controls can be achieved by methods including hierarchical clustering [36] and multidimensional scaling (unpublished results). Not surprisingly, many pairs provide perfect discrimination between IDCM and controls, so that best observed score is maximal (Δ = 1.0), and this score is realized by a large number (2,460) of gene pairs. Some of these top-scoring pairs are surely spurious due to the very small sample size and the very large space of possible gene pairs. Despite this, the cross-validated 100% prediction rate demonstrates that the discriminating power of the entire family of pairs estimated to achieve the maximal score is not due to chance. (Were the pairs detected purely by chance, then, for each loop of CV, each such pair would vote correctly with probability one-half, which is inconsistent with the cross-validated estimate. Consequently, many of the high-scoring pairs are genuinely informative.) Thus accuracy is maintained but at the possible expense of transparency. However, further analysis in this study (e.g., determining which pairs are most discriminating) is limited by the very small sample size.
4 Discussion
We have introduced a new classification methodology for microarray data based entirely on pairwise comparison of relative gene expression levels. Basing prediction on ratios of concentrations provides a natural link with biochemical activity which can only become stronger – more biologically meaningful – when mRNA abundance is replaced by actual protein expression data. Indeed, the full potential of this method may not be realized until high-throughput protein comparisons become practical.
Moreover, concrete hypotheses about the predictive significance of specific mRNA comparisons are generated naturally by the method, and follow-up studies could be focused on the corresponding list of gene pairs. Examples were provided in the cases of separating leukemia subtypes and detecting prostate cancer.
We have chosen leave-one-out (“n-fold”) CV to estimate the error rate of the TSP classifier in order to provide an “apples-to-apples” comparison with the other work we cite. In addition, this method is well-known to have low bias. On the other hand, methods such as k-fold CV and bootstrap resampling techniques have been asserted to have smaller variance (see, e.g., Efron (1983); Efron and Tibshirani (1997)) and be more appropriate for microarray analysis in many cases (Braga-Neto and Dougherty (2004)). For instance, with 10-fold CV, the estimated error rates should be unbiased for a training set of size .9n (rather than of size n) although sensitivity to the training set may be smaller than with n-fold CV. Of course, performance is expected to degrade somewhat due to the smaller number of training samples for constructing the classifier.
For the three data sets presented here the estimated prediction rate with 10-fold CV is consistent with the rates estimated with leave-one-out CV. Earlier, we had included another study – the benchmark Golub data (Pomeroy et al. (2002)). These data consist of gene expression profiles measured in embryonal tumors of the central nervous system. One objective is to predict the outcome of treatment for medulloblastoma tumors, with tissue samples labeled as “non-survivor” (n1 = 21 samples) or “survivor” (n2 = 39 samples). The prediction rate of the TSP classifier as measured by leave-one-out CV is 83% (corresponding to 11 errors in 60 loops), which is substantially higher than the rates reported in Dudoit and Fridlyand (2003) as part of the same large comparison study cited earlier in connection with breast cancer. In fact, there is a unique top-scoring pair (the human polyposis locus (DP1) gene (Genbank ID M73547) and the human E2 ubiquitin conjugating enzyme UBE2D2 (Genbank ID U39317) which appears to have biological interest; moreover, neither of these genes was identified as being important to prediction of treatment outcome using prior classification methods (Pomeroy et al. (2002)) and neither turns up significant when tested for differential expression. Nonetheless, in this case the estimated prediction rate with 10-fold CV is much lower than with leave-one-out CV, more in line with the results using other methods. Due to this ambiguity, we decided to remove this experiment.
We have focused our study on the TSP classifier in which predictions are based entirely on the top-scoring pairs. In most of the cases we have encountered there is in fact a unique top-scoring pair. However, there may be many pairs of genes whose relative expression values is informative. Moreover, the top-scoring pair may change when the training data is even slightly perturbed by adding or deleting a few samples. One avenue of future work is to find a more stable, comparison-based signature than the top-scoring pair or pairs. For example, one may also consider a k – TSP classifier based on all pairs achieving the k best scores. In this case, k is a parameter that should be estimated using cross-validation, hence requiring a double loop of cross-validation to estimate the generalization error. An investigation of the k – TSP classifier, and other extensions of the method introduced here, will be reported elsewhere.
The results already provide strong evidence that discriminating comparisons among expression levels can be discovered even under conditions of small sample size. Given the large number of variables (genes), we regard n = 100 as “small.” With somewhat larger samples, say several hundreds, the induction of modest-depth decision trees, based on successive entropy reduction and using only comparison questions, becomes feasible, thereby maintaining results which are both easy to interpret and invariant to normalization. The corresponding decision rules would then be based on more complex mRNA comparisons involving more than two genes. The methodology extends almost without modification to more complex and heterogeneous data sets, for example consisting of mixed mRNA and protein abundances.
Finally, one could also envision modeling the statistical dependency structure among families of genes and proteins, for example regulatory pathways, based on observed order statistics. With small amounts of data, it may only be possible to collect reliable estimates of pairwise comparisons among expression levels. More data could lead to estimating the order statistics of triplets, and so forth. This provides a natural, hierarchical family of models which can be adapted to the amount of data.
5 Appendix: Data Sets
The first three sets of data, the main ones used to test the classifier, are publicly available from the Kent Ridge Bio-medical Data Set Repository (http://sdmc.lit.org.sg/GEDatasets/Datasets.html).
Breast: Determining Lymph Node Status in Breast Tumor Samples: The data (West et al. (2001)) consist of gene expression profiles measured in breast tumor samples. Profiles were obtained using Affymetrix HuGeneFL arrays comprised of G = 7, 129 human probe sequences. One objective was to improve predictions about the future course of disease by accurately determining the status of the lymph nodes. Tissue samples were labeled as “positive” (affected node present, n1 = 25 samples) or “negative” (affected node absent, n2 = 24 samples).
Leukemia: Classifying Profiles from Different Leukemia Subtypes: The data (Golub et al. (1999)) consist of gene expression profiles with G = 7, 129 probes (6, 187 human genes) from 27 bone marrow samples of ALL (acute lymphoblastic leukemia) and from 11 samples of AML (acute myeloid leukemia), also obtained with Affymetrix HuGeneFL arrays. There is also a test set consisting of 34 samples (20 ALL and 14 AML). In order to utilize the same method of error estimation (namely, leave-one-out cross-validation) on all studies, we combined these two data sets into one of size n = 72 with n1 = 47 ALL samples and n2 = 25 AML samples.
Prostate: Distinguishing Tumors from Normal Profiles: The data is drawn from the study of prostate cancer reported in Singh et al. (2002), where three separate class prediction problems are investigated based on expression values for G = 12, 600 genes and ESTs derived from Affymetrix HU95Av2 microarray chips. One problem is to assign profiles to either tumor or normal tissue class (the others involve predicting clinical outcome and pathological features). There are n1 = 52 prostate tumor samples and n2 = 50 non-tumor samples, selected from among several hundred radical prostatectomy patients.
Cardiac: Classifying Gene Expression Profiles from Control Versus Failing Human Heart Tissue. Recently, we have used the Affymetrix GeneChip HG-U133A oligonucleotide array with G = 22, 283 probes to identify genes that are differentially regulated in left ventricular midmyocardial tissue isolated from patients diagnosed with end-stage idiopathic dilated cardiomyopathy (IDCM) versus that from patients in which cause of death is unrelated to heart disease (Yung et al. (2004)). Experiments were conducted on n = 22 preparations of which n1 = 10 were tissue samples obtained from control tissue and n2 = 12 were from patients diagnosed with IDCM (all data are available at www.ccbm.jhu.edu). The goal is to correctly classify gene expression profiles into control versus IDCM categories.
Footnotes
The authors wish to express their sincere thanks to Christina Yung for processing some of the raw data related to this study and to Arnaud Zeboulon for his insightful comments throughout this project. In addition, the referees and the Editor made several helpful suggestions. Daniel Naiman has been supported during the period of this research while on sabbatical at the Inherited Disease Research Branch of NHGRI and by NSF Grant ATM-0222238. This work was also supported by the Falk Medical Trust and NIH RO1-HL72488.
Publisher's Disclaimer: No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior written permission of the publisher, bepress, which has been given certain exclusive rights by the author. Statistical Applications in Genetics and Molecular Biology is produced by The Berkeley Electronic Press (bepress). http://www.bepress.com/sagmb
References
- Bicciato S, Pandin M, Didone G, Di Bello C. Pattern identification and classification in gene expression data using an autoassociative neural network model. Biotechnol. Bioeng. 2003;81(5):594–606. doi: 10.1002/bit.10505. [DOI] [PubMed] [Google Scholar]
- Bloom G, Tang IV, Boulware D, Kwong KY, Coppola D, Eschrich S, Quackenbush J, Yeatman TJ. Multi-platform, multi-site, microarray-based human tumor classification. Am. J. Pathol. 2004;164(1):9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bø TH, Jonassen I. New feature subset selection procedures for classification of expression profiles. Genome Biology. 2002;3(4):research0017.1–0017.11. doi: 10.1186/gb-2002-3-4-research0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulestiex AL, Tutz G, Strimmer K. A CART-based approach to discover emerging patterns in microarray data. Bioinformatics. 2003;19(18):2465–2472. doi: 10.1093/bioinformatics/btg361. [DOI] [PubMed] [Google Scholar]
- Bradstock KF, Kirk J, Grimsley PG, Kabral A, Hughes WG. Unusual immunophenotypes in acute leukemias: incidence and clinical correlations. Br. J. Haematol. 1989;72(4):512–518. doi: 10.1111/j.1365-2141.1989.tb04315.x. [DOI] [PubMed] [Google Scholar]
- Braga-Neto UM, Dougherty ER. Is cross-validation valid for small-sample microarray classification? Bioinformatics. 2004;20:374–380. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- Dettling M, Buhlmann P. Boosting for tumor classification with gene expression data. Bioinformatics. 2003;19(9):1061–1069. doi: 10.1093/bioinformatics/btf867. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J. Classification in microarray experiments. In: Speed T, editor. Statistical Analysis of Gene Expression Microarray Data. Chapman and Hall; 2003. [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statist. Sinica. 2002;12:111–139. [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nature Genetics Supplement. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Efron B. Estimating the error rate of a prediction rule: Improvement on cross-validation. J. Amer. Statist. Assoc. 1983;78(382):316–331. [Google Scholar]
- Efron B, Tibshirani R. Improvements on cross-validation: The .632+ boostrap method. J. Amer. Statist. Assoc. 1997;92(438):548–560. [Google Scholar]
- Geman S, Bienenstock E, Doursat R. Neural networks and the bias/variance dilemma. Neural Computation. 1992;4:1–58. [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Collier H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Griffin JD, Mayer RJ, Weinstein HJ, Rosenthal DS, Coral FS, Beveridge RP, Schlossman SF. Surface marker analysis of acute myoblastic leukemia: identification of differentiation-associated phenotypes. Blood. 1983;62(3):557–563. [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning. Springer-Verlag; 2001. [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Hwang KB, Cho DY, Park SW, Kim SD, Zhang BT. Methods of Microarray Data Analysis. Kluwer Academic Publishers; 2002. Applying machine learning techniques to analysis of gene expression data: cancer diagnosis. [Google Scholar]
- Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artifical neural networks. Nat. Med. 2001;7(6):659–659. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. Genome-wide coexpression dynamics: Theory and application. Proc. Natl. Acad. Sci. USA. 2002;99(26):16875–16880. doi: 10.1073/pnas.252466999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SPA, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nature Genetics Supplement. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Pancholi V. Multifunctional a-enolase: its role in diseases. Cell Mol. Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Ling XB, Peng X, Du W, Chen L. Molecular classification of cancer types from microarray data using the combination of genetic algorithms and support vector machines. FEBS Lett. 2003;555(2):358–362. doi: 10.1016/s0014-5793(03)01275-4. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JYH, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(4):436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Gussoni E, Kohane IS, Ramoni MF. Statistical challenges in functional genomics. Statistical Science. 2003;18(1):33–70. [Google Scholar]
- Siedow J. Making sense of microarrays. Genome Biology. 2001;2(2):reports4003.1–4003.2. doi: 10.1186/gb-2001-2-2-reports4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. Journal of the National Cancer Institute. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- Singh D, Febbo PG, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- Soukop M, Lee JK. Developing optimal prediction models for cancer classification using gene expression data. J. Bioinfo. Comp. Biol. 2003;1(4):681–694. doi: 10.1142/s0219720004000351. [DOI] [PubMed] [Google Scholar]
- Stolovitzky G. Gene selection in microarray data: the elephant, the blind men and our algorithms. Curr. Opin. Struct. Biol. 2003;13:370–376. doi: 10.1016/s0959-440x(03)00078-2. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport MJ. Complement: First of two parts. N. Engl. J. Med. 2001;344(14):1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr., Marks JR, Nevins JR. Predicting the clinical status of human breast cancer using gene expression profiles. Proc. Natl. Acad. Sci. USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngal J, Speed TP. Normalization for cDNA microarray data. Microarrays: Optical Technologies and Informatics, Proc. SPIE. 4266. 2001:141–152. [Google Scholar]
- Yeang CH, Ramaswany S, Tamayo P, Mukherjee S, Rifkin RM, Angelo M, Reich M, Lander E, Mesirov J, Golub T. Molecular classification of multiple tumor types. Bioinformatics. 2001;17(Suppl 1):S316–322. doi: 10.1093/bioinformatics/17.suppl_1.s316. [DOI] [PubMed] [Google Scholar]
- Yung CK, Halperin VL, Tomaselli GF, Winslow RL. Gene expression profiles in end-stage human idiopathic dilated cardiomyopathy: altered expression of apoptotic and cytoskeletal genes. Genomics. 2004;83(2):281–297. doi: 10.1016/j.ygeno.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yu CY, Singer B. Cell and tumor classification using gene expression data: construction of forests. Proc. Natl. Acad. Sci. USA. 2003;100(7):4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]