Abstract
Molecular dynamics simulations of Staphylococcal nuclease and of 10 variants with internal polar or ionizable groups were performed to investigate systematically the molecular determinants of hydration of internal cavities and pockets in proteins. In contrast to apolar cavities in rigid carbon structures, such as nanotubes or buckeyballs, internal cavities in proteins that are large enough to house a few water molecules will most likely be dehydrated unless they contain a source of polarity. The water content in the protein interior can be modulated by the flexibility of protein elements that interact with water, which can impart positional disorder to water molecules, or bias the pattern of internal hydration that is stabilized. This might explain differences in the patterns of hydration observed in crystal structures obtained at cryogenic and room temperature conditions. The ability of molecular dynamics simulations to determine the most likely sites of water binding in internal pockets and cavities depends on its efficiency in sampling the hydration of internal sites and alternative protein and water conformations. This can be enhanced significantly by performing multiple molecular dynamics simulations as well as simulations started from different initial hydration states.
INTRODUCTION
Internal water molecules in proteins can play essential structural and functional roles. They can satisfy the hydrogen-bonding potential of backbone and side chains, and they can act as integral structural struts (1–6). They can influence the stability of proteins (7), their flexibility (8,9), and modulate the pKa values of internal ionizable groups (10,11). Internal water molecules can participate actively in many fundamental processes, including proton transfer reactions (12–14), ligand binding (15,16), catalysis (14,17,18), folding (19,20), and redox processes (21). In ion channels, water can play a role in gating (22,23) and in ion selectivity (24–26).
Despite their important structural and functional roles, the number, location, and dynamical character of internal water molecules is still a matter of debate. Internal water molecules that are observed in x-ray structures are necessarily restricted to well-defined locations where they form stable hydrogen bonds among themselves and with polar atoms of the protein. Internal water molecules that are structurally disordered or that occupy a certain site with less than full occupancy are often not observed in crystal structures (27), as they do not contribute significantly to the electron density. Such water molecules are also invisible in many NMR spectroscopy experiments because water molecules have to have relatively long residence times and be rotationally hindered to be visible in these experiments (28,29). Furthermore, the number and locations of internal water molecules resolved in crystal structures may be affected by the temperature at which the diffraction data are collected (30,31).
Molecular dynamics (MD) simulations have proved to be useful for studying protein hydration (32). They can provide microscopic details of hydration dynamics that are inaccessible experimentally, including the description of individual events of water penetration and exit from the protein (33–37). Thermodynamic information derived from MD simulations can be used to determine the occupancies of internal cavities by water (16,38–40). The structural, dynamic, and kinetic properties of water molecules described with MD simulations, such as mean-square displacements (MSD), rotational flexibility, or lifetimes, can also be compared directly with data from crystallography or NMR spectroscopy (32,34,41–43). One limitation of MD simulations in describing hydration is that they can fail to identify deeply buried water molecules. This problem is related to the limited simulation time that is practical, which is not always sufficient to sample rare or slow events (44).
Staphylococcal nuclease (SN) and its variants with internal ionizable groups have been extremely useful to describe internal hydration of proteins. The pKa values of Asp, Glu, and Lys residues buried at internal positions 66 and 92 in SN are shifted relative to their normal pKa values in water (10,45–47). When analyzed with continuum electrostatic methods, these pKa values reflect unusually high polarizability in the protein interior, comparable to that of a material with a dielectric constant of ∼10. Several explanations have been offered to rationalize this high apparent polarizability in the interior of this protein, including structural rearrangements concomitant with ionization of the internal groups, and the presence of internal water molecules (10,11,32,48). A possible role for internal water molecules as a source of polarity in the protein interior was first suggested by x-ray crystal structures of variants of SN with internal Glu and Asp residues. In structures obtained at cryogenic temperatures, internal water molecules are usually observed hydrating the polar moieties of internal Glu and Asp side chain that are buried deeply inside the protein. Internal water molecules have also been observed recently in structures of variants with internal Asn, Gln, and Tyr residues (J. L. Schlessman and B. García-Moreno E., unpublished).
The role of internal water molecules as determinants of the pKa values of internal ionizable groups was addressed previously by MD simulations of wild-type SN and two variants, one with Glu-66 and one with Lys-66 (32). These simulations suggested that internal water molecules were not likely to be as important a source of internal polarity in proteins as previously thought. The penetration of water into the protein, as observed in MD simulations, was fully consistent with internal water molecules observed in crystal structures: water molecules were observed in the interior near Glu-66, but none were observed in the interior in the presence of Lys-66. The MD simulation of wild-type SN identified an internal water molecule that has not been seen previously in any of the x-ray structures of SN. This water molecule makes two hydrogen bonds with the backbone atoms of a protein loop and one with another water molecule. The flexibility of the loop imparts significant disorder to the water molecule, rendering it invisible to NMR and x-ray crystallography, if this molecule is, indeed, present. The effects of the protein flexibility were quantified through calculations of the chemical potential. The chemical potential of a water molecule in the wild-type simulation shifted by 3 kcal/mol when the hydrogen-bonding partner of this water molecule in the neighboring loop changed. Similarly, in the simulation of the V66E variant the chemical potential changed by 3 kcal/mol when this water molecule exchanged with a bulk water molecule, and Glu-66 moved slightly away from it.
Crystal structures of many variants of SN with internal polar or ionizable groups at positions 66 and 92 have become available recently (J. L. Schlessman and B. García-Moreno E., unpublished; (45–47,49)). The side chains at positions 66 and 92 are located in a microcavity that is lined with nonpolar atoms. The focus of this study is on the state of hydration of this microcavity, thus it will be referred to as the cavity. In the wild-type protein, the cavity is sufficiently large to house as many as four water molecules, but it is sufficiently small to be undetectable when the molecular surface is calculated with a water probe radius of 1.4 Å. Crystallographic structures for many of the relevant variants of SN have been obtained separately under room temperature and cryogenic conditions. In most variants, the crystallographic structures obtained at different temperatures display different hydration patterns. In this article, we describe the results of MD simulations of these proteins. One goal of these simulations was to examine possible causes behind the effects of temperature on the patterns of internal hydration, and to investigate systematically the molecular determinants of hydration of pockets and internal cavities in proteins. These simulations also constitute a useful benchmark for MD-based protocols for artificial hydration of internal pockets and cavities in proteins.
METHODS
Simulated systems
The proteins that were simulated are the wild-type SN and the variants PHS/V66E, Δ+PHS/V66K, PHS/V66D, PHS/V66N, PHS/V66Q, PHS/V66Y, V66W, Δ+PHS/I92D, Δ+PHS/I92E, and Δ+PHS/I92K. PHS refers to a hyperstable form of SN with three substitutions (P117G, H124L, S128A). Δ+PHS is an even more stable protein consisting of PHS with two additional substitutions (G50F, V51N) and a 44–49 deletion.
The structure of the wild-type (WT) Staphylococcal nuclease that was used has the PDB accession number 1STN. Table 1 describes details of simulation setups and runs in terms of the exact number of amino acids in each system, the initial number of water molecules in the cavity, the protonation states of histidine residues, the number of counterions, the total number of water molecules, and the length of each simulation. Residues that were not resolved in the crystal structures were not included in the simulation. In some cases, the simulated systems contained additional amino-acid mutations, as described in Table 1.
TABLE 1.
Simulated systems
| Protein | IHS | Cavity H2O | His tautomeric state | Cl− | Na+ | H2O |
|---|---|---|---|---|---|---|
| WT (6-141) | crystal | 0 | His-8,-121,-124 (δ), His-46 (ɛ) | 27 | 17 | 5961 |
| WT (6-141) | full | 4 | His-8,-46,-121,-124 (δ) | 27 | 17 | 5895 |
| PHS/V66E (7-141) | crystal | 2 | His-8,-121 (δ), His-46 (ɛ) | 26 | 17 | 5495 |
| PHS/V66E (7-141) | full | 4 | His-8,-46,-121 (δ) | 22 | 13 | 5464 |
| PHS/V66E (7-141) | empty | 0 | His-8,-121 (δ), His-46 (ɛ) | 26 | 17 | 5484 |
| Δ+PHS/V66K (7-141)* | crystal | 0 | His-8,-121 (δ) | 25 | 19 | 5248 |
| Δ+PHS/V66K (7-141)* | full | 3 | His-8,-121 (δ) | 20 | 14 | 5055 |
| PHS/V66D (7-141) | crystal | 2 | His-8,-46,-121 (δ) | 26 | 17 | 5666 |
| PHS/V66D (7-141) | full | 7 | His-8,-46,-121 (δ) | 23 | 14 | 5692 |
| PHS/V66D (7-141) | empty | 0 | His-8,-46,-121 (δ) | 23 | 14 | 5684 |
| PHS/V66N (6-141)† | crystal | 2 | His-8,-46,-121 (δ) | 24 | 17 | 6106 |
| PHS/V66N (6-141)† | full | 7 | His-8,-46,-121 (δ) | 22 | 15 | 6071 |
| PHS/V66N (6-141)† | empty | 0 | His-8,-46,-121 (δ) | 22 | 15 | 6020 |
| PHS/V66Q (6-141) | crystal | 1 | His-8,-46,-121 (δ) | 26 | 16 | 6059 |
| PHS/V66Q (6-141) | full | 5 | His-8,-46,-121 (δ) | 22 | 12 | 6015 |
| PHS/V66Q (6-141) | empty | 0 | His-8,-46,-121 (δ) | 22 | 12 | 6006 |
| PHS/V66Y (7-141) | crystal | 2 | His-8,-46,-121 (δ) | 26 | 17 | 5507 |
| PHS/V66Y (7-141) | full | 3 | His-8,-46,-121 (δ) | 23 | 14 | 5506 |
| PHS/V66Y (7-141) | empty | 0 | His-8,-46,-121 (δ) | 23 | 14 | 5489 |
| V66W (7-141) | crystal | 2 | His-8,-121,-124 (δ), His-46 (ɛ) | 26 | 17 | 5926 |
| V66W (7-141) | full | 3 | His-8,-46,-121,-124 (δ) | 23 | 14 | 5448 |
| V66W (7-141) | empty | 2 | His-8,-121,-124 (δ), His-46 (ɛ) | 26 | 17 | 5445 |
| Δ+PHS/I92D (6-141)‡ | crystal | 2 | His-8,-121 (δ) | 22 | 17 | 5664 |
| Δ+PHS/I92D (6-141)§ | full | 6 | His-8,-121 (δ) | 22 | 21 | 5140 |
| Δ+PHS/I92D (6-141)‡ | empty | 0 | His-8,-121 (δ) | 22 | 17 | 5304 |
| Δ+PHS/I92E (7-143) | crystal | 4 | His-8,-121 (δ) | 21 | 17 | 5664 |
| Δ+PHS/I92E (7-143) | full | 5 | His-8,-121 (δ) | 20 | 16 | 5559 |
| Δ+PHS/I92E (7-143) | empty | 0 | His-8,-121 (δ) | 21 | 17 | 6033 |
| Δ+PHS/I92K (6-141) | crystal | 0 | His-8,-121 (δ) | 24 | 17 | 5978 |
| Δ+PHS/I92K (6-141) | full | 5 | His-8,-121 (δ) | 24 | 17 | 5832 |
| Δ+PHS/I92L (6-141)¶ | full | 6 | His-8,-121 (δ) | 22 | 21 | 5140 |
The first and last residue observed in the crystal structure is identified. IHS is the abbreviation for the initial hydration state. Cavity H2O refers to the number of water molecules in the cavity. Cl−, Na+, and H2O refer to the number of Cl−, Na+ ions, and water molecules. See footnotes for substitutions made. The following structures are available in the Protein Data Bank: 1U9R for PHS/V66E (room temperature), 2OXP for PHS/V66D (cryo temperature), 2SNM for V66K, 2PZU for PHS/V66N (cryo temperature), 2PZW for PHS/V66N (room temperature), 2PZT for PHS/V66Q (cryo temperature), 2PYK for PHS/V66Q (room temperature), 2PW7 for PHS/V66Y (cryo temperature), 2PW5 for PHS/V66Y (room temperature), 1TQO for Δ+PHS/I92E (cryo temperature), 1TR5 for Δ+PHS/I92E (room temperature), and 1TT2 for Δ+PHS/I92K (cryo temperature).
F50G, N51G.
K6A, K53A, K110S.
K6A, K110A.
K6A, K110A, K64A, K97A, K127A, K116S.
This protein was constructed artificially by mutating D into L.
As a first step in the preparation of the simulated systems, hydrogen atoms were added to the crystal structures. The protonation states of the histidine residues are listed in Table 1. All of the ionizable groups in positions 66 or 92 were set to the neutral state, which is the case at pH 7 (10,45,50). An acetylated N-terminus and an N-methylated C-terminus were added to all simulated structures. Missing hydrogen atoms and terminal heavy atoms were generated with the PSFGEN feature of the molecular dynamics program NAMD2 (51). The systems were first subjected to energy minimizations and then immersed in a preequilibrated water box with TIP3 water molecules. The position of the proteins in the box was such that in each Cartesian direction there were 10 Å between the minimal and maximal protein coordinate and the edge of the water box. All water molecules within 2.4 Å of a protein atom were removed (except for the crystallographic water molecules, and water molecules inserted with DOWSER, as described below). The systems contained, on average, an excess of 39 Na+ and Cl− ions, corresponding approximately to 380 mM salt. The ions were placed at random in the simulation box, but at least 6 Å away from the protein or from each other.
Initial state of hydration of the microcavity
Three different types of simulations were performed for most systems, differing in the initial hydration state of the cavity. The initial number of water molecules in the cavity is listed in Table 1. Systems labeled crystal initially contained all the water molecules that were resolved crystallographically under cryogenic conditions. In systems labeled empty, the cavity was initially empty. Sometimes, during the immersion of the protein into the box of water, water molecules are placed in interior sites in the protein. In the systems labeled empty, we ascertained that the cavity was devoid of water. Systems labeled full included the water molecules in the internal positions that were identified by the program DOWSER (52) as sites of both low and high potential for water binding. DOWSER identifies water molecules in the protein interior using the following protocol:
Step 1. The solvent-accessible surface of the protein is generated and a probe is used to identify interior surface points.
Step 2. Water molecules are placed in the interior pockets and cavities, their energies are minimized, and their positions refined and verified.
Step 3. The interaction energies of all interior water molecules are calculated. Water molecules with interaction energies <−10 kcal/mol are considered to occupy internal positions fully (52). Water in binding sites of lower energy are considered unoccupied.
Our modified procedure for identifying internal sites of hydration with DOWSER avoided Step 3 in the normal DOWSER procedure.
Molecular dynamics simulations
Energy minimization and molecular dynamics simulations were performed with NAMD2 (51). The force field used for the simulations was CHARMM (53), Ver. 27. The systems were simulated with periodic boundary conditions using the particle-mesh Ewald method with a real space interaction cutoff of 10 Å. Calculations were done using particle-mesh Ewald grid spacing of ∼1 Å (60 × 60 × 60 grid dimensions), and interpolation order of 4. The systems were first energy-minimized and then equilibrated at 300 K for 500 ps.
MD simulations were performed in an NPT ensemble at a pressure of 1 atm and a temperature of 300 K. The Langevin piston method (54) was used for pressure control, together with Langevin dynamics for temperature control. The following parameters were used: Langevin damping of 1/ps, and Langevin piston period and decay time of 200 fs and 100 fs, respectively. A 2-fs timestep was used. The default simulation time for each system in the crystal initial hydration state was 10 ns. The simulation times for systems started in the empty and full hydration states vary, as specified in Table 1. Coordinates from MD simulations were recorded every ps.
Analysis of MD trajectories
The simulations started from the crystal hydration state were analyzed in terms of the water molecules bound to the protein for longer than 1 ns, which were identified based on their coordination number (50). The average locations of such water molecules were determined and their residence times and mean-square deviations were measured (43). Additional details about the analysis procedure have been published previously (32). The trajectories resulting from simulations started from the full and empty hydration states were only inspected visually, using the program VMD (55).
RESULTS
Locations of internal water molecules and routes of water penetration
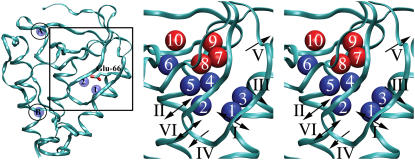
The crystal structure of the PHS/V66E variant of SN (45) shown in Fig. 1 illustrates the overall topology of the protein and the location of the microcavity of interest (highlighted with a box). The two crystallographic water molecules that interact with the buried carboxylic oxygens of Glu-66 are labeled in this figure as 1 and 2. Crystal structures of the wild-type protein and of variants with Q, N, D, W, Y at position 66, and with E and D at position 92, revealed that a total of six distinct sites can be occupied by water molecules in this cavity. These six sites are shown as blue spheres in Fig. 1 (right). Four additional, putative water-binding sites shown as red spheres in Fig. 1 were predicted by the DOWSER calculations. The labels used in Fig. 1 to identify the different water sites in the cavity will be used throughout this article: the average position of internal water molecules with residence time longer than one nanosecond will be approximated to be at either one of those 10 sites, or between two such sites.
FIGURE 1.
(Left) Crystal structure of the PHS/V66E variant showing the locations of the crystallographically resolved water molecules in the microcavity near position 66 and at two other sites, labeled A and B. (Right) Simultaneous representation of water-binding sites in the microcavity for all of simulated variants. Water molecules that are observed in crystal structures are shown in blue. Those that are only observed in DOWSER calculations are shown in red. Routes of water penetration and exit observed in MD simulations are indicated with arrows and labeled with Roman numerals I–VI.
Six routes of water penetration were identified by visual inspection of the MD trajectories. These routes are labeled with Roman numerals in Fig. 1. The directions of the arrows used to show the routes indicate whether a route is used for penetration, exit, or both. Route I is the most traveled route. A water molecule passing through this route travels a passage formed by Ala-17, Ile-18, Asp-19, Ser-59, and Lys-63. The internal water molecule found closest to this route is water molecule 1. Route II is formed by Thr-22, Val-23, Asp-19, and Phe-34. Its closest crystallographic water molecules are water molecules 2 and 5. Route III is formed by Leu-14, Ile-15, Lys-16, Ala-17, Lys-63, and Glu-67. Its closest crystallographic water molecule is water molecule 3. Route IV is formed by Gly-20, Leu-36, Thr-62, and Leu-103. Its closest crystallographic water molecules are water molecules 1 and 2. Route V is formed by Ala-12, Thr-13, Leu-14, and Ile-72. Its closest crystallographic water molecules are water molecules 7 and 9. Route VI is formed by Ile-18, Asp-19, Gly-20, and Thr-22. It involves a passage through a β-turn. Its closest crystallographic water molecules are water molecules 1 and 2.
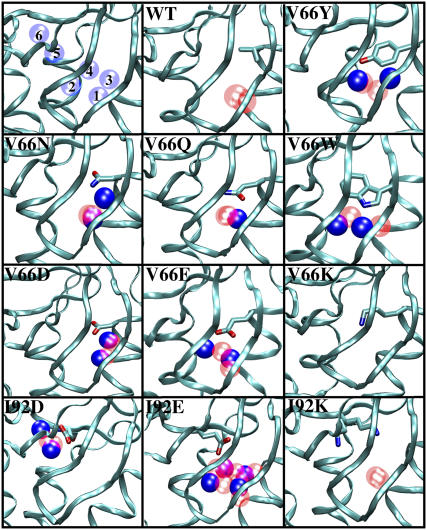
Results from simulations with the wild-type SN and with 10 of its variants were compared with each other and with crystallographic observations. Fig. 2 shows, for all variants, a closeup of the microcavity as observed in the crystal structure, the internal crystallographic water molecules, and the water molecules with residence times larger than 1 ns obtained from a single 10-ns-long MD simulation started from crystal hydration state. The lifetimes and mean-square deviations of the MD water molecules shown in Fig. 2 are given in Table 3. Table 2 summarizes hydration patterns observed through cryo and room-temperature crystallography, and through MD simulations started from crystal, full, and empty hydration states. For simulations started from the full hydration state, we list only the final hydration patterns. In addition, Table 2 lists simulation times and RMSD values between crystallographic and final MD structures. These RMSD values were calculated using the MMTSB Tool Set (56). To gain insight into whether alternative hydration patterns are coupled to distortions of the protein, RMSD values of residues 16–25 (which are in vicinity of residue 66) and residues 66 or 92 are listed. In the sections below, the hydration patterns and the observed pathways of water exit/penetration are described in detail.
FIGURE 2.
Locations of internal water molecules observed in crystallographic structures (blue), or MD simulations (red) started from the crystal hydration states. Only water molecules that have residence times longer than 1 ns are shown. Only the results of a single MD simulation for each variant are represented in this figure.
TABLE 3.
Residence times (tin-tout) and mean-square deviations (MSD) for water molecules that have residence times longer than 1 ns in MD simulations started from crystal hydration state (results for only one set of 10-ns-long simulations are shown)
| Water | tin-tout (ns) | MSD (Å2) |
|---|---|---|
| WT (1) | 1.2–2.9 | 2.35 |
| WT (1) | 3.6–8 | 2.25 |
| V66E (1) | 1.3–3.6 | 4.2 |
| V66E (1) | 5.1–9.4 | 1.60 |
| V66D (1) | 0–3.2 | 1.28 |
| V66D (3) | 0–3.7 | 1.83 |
| V66N (1) | 0–3.5 | 1.01 |
| V66N (1) | 3.5–10 | 0.60 |
| V66Q (1) | 0–1.8 | 3.4 |
| V66Q (1) | 2.9–10 | 0.59 |
| V66Y (1) | 0–3.9 | 4.64 |
| V66Y (2) | 0–10 | 1.67 |
| V66W (1) | 0.9–8.8 | 2.7 |
| V66W (2) | 0–10 | 1.78 |
| I92D (5) | 0–10 | 3.87 |
| I92D (7) | 0–10 | 2.52 |
| I92E (1) | 0–1.5 | 5.7 |
| I92E (1) | 1.8–3.3 | 5.75 |
| I92E (2) | 1–10 | 6.25 |
| I92E (1–3) | 0–1 | 4.65 |
| I92E (3) | 4.5–10 | 3.7 |
| I92E (3) | 7–9.5 | 4.22 |
| I92E (4) | 0–3.8 | 1.75 |
| I92K (1) | 3–7 | 2.42 |
| I92K (1) | 7.5–10 | 0.72 |
The numbers in parentheses correspond to water binding sites shown in Fig. 2.
TABLE 2.
Hydration sites observed in crystal structures obtained under cryogenic or room temperature conditions, and in MD simulations
| Protein | IHS | Observed water sites | τ (ns) | RMSD (Å) | ||||
|---|---|---|---|---|---|---|---|---|
| Cryo x-ray | Room T x-ray | MD | bb (all) | bb (16:25) | side (66 or 92) | |||
| WT | crystal | 0 | N/A | 1 | 10 | 1.83 | 0.29 | 66: 0.27 |
| WT | full | 0 | N/A | 0 | 10 | 2.80 | 0.36 | 66: 0.20 |
| PHS/V66E | crystal | 1,2 | 1 | 0; 1 | 10 | 1.41 | 0.54 | 66: 0.57 |
| PHS/V66E | full | 1,2 | 1 | 2 | 6 | 1.69 | 1.25 | 66: 1.45 |
| PHS/V66E | empty | 1,2 | 1 | 0; 1,2 | 50 | 1.92 | 0.59 | 66: 1.51 |
| PHS/V66E | empty | 1,2 | 1 | 0; 1,2,3 | 10 | 2.58 | 1.02 | 66: 1.29 |
| PHS/V66E | empty | 1,2 | 1 | 0; misc | 10 | 1.65 | 0.89 | 66: 0.90 |
| PHS/V66E | empty | 1,2 | 1 | 1; misc | 10 | 2.16 | 1.54 | 66: 1.56 |
| PHS/V66E | empty | 1,2 | 1 | 1,2; 2 | 10 | 1.76 | 0.78 | 66: 1.40 |
| PHS/V66E | empty | 1,2 | 1 | 0 | 10 | 2.11 | 0.67 | 66: 2.37 |
| PHS/V66E | empty | 1,2 | 1 | 0 | 10 | 1.82 | 0.46 | 66: 1.40 |
| PHS/V66E | empty | 1,2 | 1 | 1 | 10 | 2.05 | 0.76 | 66: 0.56 |
| PHS/V66E | empty | 1,2 | 1 | 1,2 | 10 | 1.91 | 0.65 | 66: 1.45 |
| Δ+PHS/V66K | crystal | 0 | 0 | 0 | 10 | 1.80 | 0.98 | 66: 1.32 |
| Δ+PHS/V66K | full | 0 | 0 | 0 | 3 | 2.47 | 1.10 | 66: 1.47 |
| PHS/V66D | crystal | 1,3 | 0 | 1; 1,3 | 10 | 1.81 | 0.78 | 66: 0.52 |
| PHS/V66D | crystal | 1,3 | 0 | 1 | 10 | 2.04 | 1.20 | 66: 0.63 |
| PHS/V66D | full | 1,3 | 0 | 2, 4 | 10 | 2.14 | 0.66 | 66: 0.38 |
| PHS/V66D | empty | 1,3 | 0 | 5 | 7 | 1.70 | 1.08 | 66: 0.41 |
| PHS/V66N | crystal | 1,3 | 0 | 1 | 10 | 1.65 | 0.61 | 66: 0.85 |
| PHS/V66N | crystal | 1,3 | 0 | 1 | 2 | 1.93 | 0.58 | 66: 0.09 |
| PHS/V66N | full | 1,3 | 0 | 1 | 10 | 1.48 | 1.95 | 66: 0.47 |
| PHS/V66N | empty | 1,3 | 0 | 1 | 2 | 1.09 | 1.31 | 66: 1.08 |
| PHS/V66Q | crystal | 1 | 0 | 1; | 10 | 1.77 | 0.45 | 66: 1.35 |
| PHS/V66Q | crystal | 1 | 0 | 1; 1,2; 1,2,4 | 10 | 1.87 | 0.70 | 66: 1.36 |
| PHS/V66Q | full | 1 | 0 | 1 | 4 | 1.74 | 0.82 | 66: 1.40 |
| PHS/V66Q | empty | 1 | 0 | 1 | E | 0.97 | 0.59 | 66: 1.43 |
| PHS/V66Y | crystal | 1,2 | 1,2 | 1,2 | 10 | 1.52 | 0.52 | 66: 0.55 |
| PHS/V66Y | full | 1,2 | 1,2 | 1,2 | 2 | 1.12 | 0.66 | 66: 1.12 |
| PHS/V66Y | empty | 1,2 | 1,2 | 1,2 | E | 1.10 | 0.63 | 66: 0.59 |
| V66W | crystal | 1,2 | N/A | 1,2 | 10 | 1.83 | 0.40 | 66: 0.92 |
| V66W | full | 1,2 | N/A | 6 | 10 | 2.04 | 0.37 | 66: 3.21 |
| V66W | empty | 1,2 | N/A | 1 | 10 | 1.85 | 1.33 | 66: 0.55 |
| Δ+PHS/I92D | crystal | 5,6 | N/A | 5,6 | 10 | 1.47 | 0.44 | 92: 0.43 |
| Δ+PHS/I92D | full | 5,6 | N/A | 5 | 20 | 1.91 | 1.28 | 92: 0.61 |
| Δ+PHS/I92D | empty | 5,6 | N/A | 5 | 10 | 1.94 | 1.18 | 92: 0.12 |
| Δ+PHS/I92E | crystal | 1,2,3,4 | 0 | 1,2; 3,4 | 10 | 1.95 | 1.19 | 92: 3.23 |
| Δ+PHS/I92E | full | 1,2,3,4 | 0 | 1,2 | 3 | 1.56 | 1.06 | 92: 2.40 |
| Δ+PHS/I92E | empty | 1,2,3,4 | 0 | 1,4 | E | 0.96 | 0.44 | 92: 1.44 |
| Δ+PHS/I92K | crystal | 0 | N/A | 0, 1 | 10 | 1.77 | 0.36 | 92: 1.38 |
| Δ+PHS/I92K | full | 0 | N/A | 0 | 10 | 2.62 | 0.89 | 92: 3.64 |
| Δ+PHS/I92L | full | N/A | N/A | 1 | 4 | 1.08 | 0.69 | 92: 2.69 |
Simulation time (τ) in nanoseconds. E indicates that only an equilibration run was performed. RMSD values between the crystallographic structures and final MD snapshots are shown. The three RMSD columns stand for: backbone atoms of all protein residues, backbone atoms of residues 16:25 (which are in vicinity of residue 66), and the side chain of residue 66 or 92.
Wild-type SN
The crystal structure of the wild-type protein contains no water molecules in the cavity. In this case the empty and the crystal hydration states of the cavity are the same, thus simulations were only performed with the crystal hydration state. The results of the simulation are shown in Fig. 2 and have also been discussed elsewhere (32,57). During the simulation, a single water molecule entered the protein and stayed in site 1. This site remained occupied during the entire simulation, although the individual water molecules exchanged. The longest recorded residence time in this site was 4.4 ns.
The structure with the full hydration state initially contained water molecules in sites 2, 4, 5, and 6. The cluster of four water molecules rearranged into positions 1, 2, 3, and 4 and was stable during the entire course of the equilibration stage. Soon into the production run, three out of four water molecules left the pocket via route I. The remaining water molecule stayed trapped close to site 2 during the first seven nanoseconds of the production run. Eventually, it left the cavity through route I. No water molecules were found in the interior thereafter.
V66E
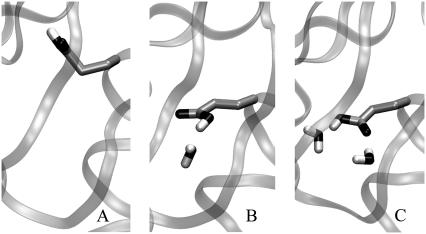
The crystal structure of the V66E variant obtained at cryogenic temperatures contains water molecules in sites 1 and 2. In the simulation with the crystal hydration state one water molecule diffused out during equilibration, through route I. Two conformations of Glu-66 were recorded during the simulation, a straight one with one internal water molecule, and a twisted one without any internal water molecules. Snapshots from the MD run with Glu-66 in different conformations are shown in Fig. 3. The lifetime of the twisted conformation was 2 ns. The hydration patterns and conformations of Glu-66 are discussed in more detail ahead.
FIGURE 3.
Snapshots representing the three most common hydration patterns sampled during MD simulations of the V66E variant.
The structure with the full hydration state initially contained water molecules in positions 1, 2, 6, and 7. One water molecule left the cavity within 100 ps into equilibration through route II. Shortly into the production run, the side chain of Glu-66 changed into the twisted conformation. This was followed by the release of two internal water molecules. The remaining water molecule stayed trapped by interactions with the polar groups along exit route II. This caused a slight separation between strands β2 and β3. In this position the water molecule did not interact with the carboxylic moiety of Glu-66. The twisted conformation was stable for ∼2.5 ns. Since it was devoid of water, Glu-66 started alternating between the twisted and the straight conformation during the next 1.5 ns. During one of these shifts, it pulled the water molecule that was trapped in route II into the interior site 2. This was followed by penetration of an additional water molecule through route II. During the rest of the simulation, Glu-66 was found mostly in a straight conformation, interacting with the water molecule in site 2.
V66K
No internal water molecules have been observed in the microcavity in any of the crystal structures of the V66K variant (50). Therefore, the simulation with the empty hydration state was not performed. During the course of the MD run started from the crystal hydration state, no water molecules penetrated because route I was blocked by the backbone carbonyl group of Asp-19. A more detailed discussion of this simulation has been presented elsewhere (32).
The structure with the full hydration state contained initially water molecules in sites 2, 3, and 6. The water molecule in site 3 left the interior through route III soon after the equilibration was started. A hydrogen bond between Lys-66 and water molecule in site 2 did not hold the Lys in place as it started shifting into different rotameric states. The water molecule in site 2 left the interior within 800 ps into the production run, through route I. At 1 ns into the production run, the third water molecule, initially in site 6, was attracted by Lys-66 toward site 5. It stayed hydrogen-bonded to Lys-66 for another 1.5 ns and eventually left the cavity through route II.
V66D
The crystal structure of the V66D variant obtained at cryogenic temperatures contains water molecules in sites 1 and 3. Within ∼3 ns into the production run started from the crystal hydration state, water molecule 1 left the protein interior using route I. The water molecule in site 3 left within the next 500 ps, also through route I. Approximately 7 ns into the production run, site 1 became reoccupied briefly, while site 3 remained empty. To increase sampling, an additional 10-ns-long MD run was performed starting from the crystal hydration state. The water molecule in site 3 diffused out through route III, 100 ps after the production run was started, while the water molecule in site 1 stayed bound for another 2 ns into the production run before leaving the cavity. None of the sites were reoccupied.
Approximately 2 ns into the production run, with the empty hydration state a water molecule diffused through route II into a location close to site 2. This water molecule was followed immediately by another water molecule that took its place, and pushed the first one into a location close to site. By the end of the 7-ns production run one water molecule left through route II, leaving one water molecule located close to site 5.
The structure with the full hydration state initially contained water molecules in sites 1, 2, 3, 4, 6, 7, and 8. Within 350 ps into the equilibration run, four water molecules left the protein interior, three using route I, and one using route IV. The remaining three water molecules were stable in positions 5, 6, and 8 for 4.5 ns. After a water molecule left through route V, two remaining water molecules rearranged themselves into positions 2 and 4. This conformation was kept until the end of the 10-ns-long production run.
V66N
The crystal structure of the V66N variant obtained at cryogenic temperatures contains water molecules in sites 1 and 3. Within a few hundred picoseconds into the production run of the simulation with the crystal hydration state, the water molecule in site 3 diffused out through route I. The water molecule in site 1 stayed bound for an additional 3.5 ns and diffused out through route I. It was replaced by another water molecule that stayed bound until the end of the simulation. Site 3 was never reoccupied.
An additional 2-ns-long MD run started from the crystal hydration state was performed to increase sampling. During equilibration, both internal water molecules diffused out. The water molecule in site 1 was replaced by another water molecule, while site 3 was not occupied again. In the simulation with the empty hydration state, one water molecule diffused into site 1 through route I during the equilibration period.
The structure with the full hydration state initially contained water molecules in sites 1, 2, 3, 6, 7, 8, and 9. During equilibration three water molecules left the cavity through route I, and one through route III. The remaining three water molecules were stabilized at sites 1, 2, and 4 for ∼5 ns. One water molecule eventually left through route I. The remaining two water molecules were stabilized at sites 1 and 2 for the rest of the 10-ns-long run. Positions 3 and 4 were occupied occasionally during the simulation.
V66Q
The crystal structure of the V66Q variant obtained at cryogenic temperatures contains a water molecule in site 1. On average, this site was occupied during the entire 10-ns-long MD run started from the crystal hydration state. The original water molecule stayed bound for 1.8 ns before exiting through route VI. Site 1 was filled again, temporarily, by a water molecule that used route VI for both penetration and exit. This water molecule was in turn replaced by another one, which used route I and stayed in the protein interior for >7 ns, until the end of the simulation. In all simulations of the V66Q variant, Gln-66 is rotated about its axis, with positions of O and N atoms exchanged as compared to the crystal structure.
An additional 10-ns-long MD run started from the crystal hydration state was performed to improve sampling. The crystallographic water molecule remained inside the protein during the entire run. From 0 to 1.5 ns, it was in position 1. After that, a second water molecule diffused into site 1 and pushed the first water molecule into position 2. This situation was preserved until 8 ns into the production run, when another water molecule diffused in through route I. From 8 to 10 ns, three water molecules were found in the protein interior, approximately in sites 1, 2, and 4. In contrast, in the simulation started with the empty hydration state, one water molecule diffused into site 1 through route I during the equilibration period.
The structure with the full hydration state initially contained water molecules in sites 1, 2, 6, 7, and 9. During equilibration, one water molecule left the protein through route V. Also during equilibration, Gln-66 rotated to a position where the NE2 atom was hydrogen-bonded with Thr-62:O. Soon into the production run, one water molecule left through route I. A cluster of three water molecules was stabilized close to sites 1, 2, and 4 for ∼2.5 ns. At ∼2.5 ns, one water molecule left through route I, immediately followed by another one—thus recovering the hydration pattern of the crystal hydration state.
V66Y
The crystal structure of the V66Y variant obtained at cryogenic temperatures contains water molecules in sites 1 and 2. Both sites remained occupied during the entire 10-ns-long MD run started from the crystal hydration state. Water molecule in site 1 exchanged with bulk water molecules. The water molecule in site 2 never exchanged. Similarly, in the simulation started from the empty hydration state, both water sites were filled up quickly during equilibration. Penetration was via route I.
The structure with the full hydration state initially contained water molecules in the cavity, in sites 1, 2, and 6. One-hundred-and-fifty picoseconds ps into the equilibration, the water molecule in position 6 explored several alternative positions. When the water molecule from position 1 left the interior through route I, the two remaining water molecules settled in sites 1 and 2.
V66W
The crystal structure of the V66W variant obtained at cryogenic temperatures also contains water molecules in sites 1 and 2. Both sites were occupied during the entire 10-ns-long MD run started from the crystal hydration state. The water molecule in site 1 exchanged with bulk water molecules through route I. The water molecule in site 2 did not exchange.
In the simulation started from the empty hydration state, the observed hydration pattern (one water molecule in position 1) differs from that of the crystal structure. This is an issue related to the starting structure. Because internal water molecules are absent initially, Trp-66 made a hydrogen bond with the carbonyl of Gly-20. This bond was not broken throughout the run. This was not the case in the simulation started from the crystal hydration state, in which a water molecule was found in between Trp-66 and Gly-20. It is likely that this slight difference in hydrogen-bonding behavior would be eliminated with longer simulation times that allowed Trp-66 to sample non-hydrogen-bonded conformations.
The structure with the full hydration state initially contained water molecules in sites 1, 2, and 6. At ∼5 ns into the production run, the Trp residue flipped about the Cβ-Cγ bond into an alternative rotameric state, and stayed in this state until the end of the run. Immediately after the flip, the water molecules from positions 1 and 2 left through route I. These water-binding sites were never reoccupied. The water molecule in site 6 remained bound during the entire 10-ns-long run, far away from other water molecules or polar groups that might have helped it leave the protein. The chemical potential for this water molecule was calculated as described in Damjanović et al. (32). The chemical potential was ∼3 kcal/mol higher than the chemical potential of a bulk water molecule, suggesting that this water molecule would have probably exited the protein interior if it were able to interact with other polar groups.
I92D
The crystal structure of the I92D variant obtained at cryogenic temperatures contains water molecules in sites 5 and 6. The two water molecules are hydrogen-bonded to Asp-92 and to each other. They were present during the entire 10-ns-long MD run started from the crystal hydration state. In the 10-ns-long simulation started from the empty hydration state, one water molecule entered the cavity through route I within 1.5 ns and stayed bound to Asp-92 until the end of the simulation.
The structure with the full hydration state initially contained six water molecules in the cavity, in sites 5, 6, 7, 8, 9, and 10. During the equilibration, these six water molecules moved to sites 1, 2, 4, 5, 7, and 9. Within 300 ps into the production run, one water molecule left through route I. During the next 700 ps, the remaining five water molecules stayed in positions 2, 4, 5, 8, and 9. Then they rearranged into positions 4, 7, 8, 9, and 10, where they remained for 1.6 ns. Approximately 2.6 ns into the production run, one water molecule left through route I. The remaining four water molecules stayed in sites 4, 5, 8, and 9, surrounding the carboxylic group of Asp-92. This configuration was stable for ∼2.5 ns. Approximately 5 ns into the production run, the four remaining water molecules rearranged. At 1.5 ns later, one of them diffused out through route I while another one diffused in. These four water molecules were stabilized in different sites while another water molecule was present mostly in site 1 without interacting directly with the rest of the water cluster. Approximately 9.3 ns into the production run, one water molecule diffused out through a route between routes II and IV. At 11.8 ns, another water molecule diffused in through route II. At 13.6 ns, one water molecule left through route I, leaving three water molecules inside. At 14.3 ns, another water molecule left through route II. Until the end of the 20 ns run, the number of water molecules in the cavity varied between zero and two; the exchange occurred through route II. During that time period, water molecules were actively changing locations. Site 5 had the largest occupancy.
I92E
The crystal structure of the I92E variant obtained at cryogenic temperatures contains water molecules in sites 1, 2, 3, and 4. During the simulation started from the crystal hydration state, the number of internal water molecules varied between two and four. However, most of the time only two water molecules were present in the microcavity. Water molecules were seen mostly in positions 1, 2, 3, 4, and only rarely in position 5. Route I was used for water exit and penetration. During the equilibration of the simulation with empty hydration state, two water molecules diffused into the cavity through route I.
The structure with the full hydration state initially contained water molecules in sites 1, 2, 3, 4, and 6. Four-hundred ps into the production run, one water molecule left through route I, followed by another one at 700 ps. Three remaining water molecules occupied various positions within the protein. At 2.8 ns into the production run, the hydration patterns became similar to the hydration patterns observed in the simulation started from the crystal hydration state, which had two water molecules within the cavity.
I92K
The crystal structure of the I92K variant obtained at cryogenic temperatures contains no internal water molecules. This was reproduced by the simulation started from the crystal hydration state. A water molecule can occupy site 1; however, this water molecule does not interact with the Lys side chain. The MD simulations showed that the Lys-92 side chain changed its rotameric state very often. This is fully consistent with the weak electron density for the middle atoms of the side chain of this group in the x-ray crystallographic study (46).
The structure with the full hydration state initially contained water molecules in sites 4, 5, 6, 9, and 10. During equilibration stage, one water molecule left through route V and one through route IV. The remaining three water molecules were stabilized in sites 2, 5, and 6. At 2.6 ns into the production run, they all exited through route II. Occasionally one water molecule penetrated and exited through the same route. Until the end of the 10-ns-long MD run, a water molecule resided in the exit pathway II, making occasional interactions with Lys-92.
I92L
A simulation was performed with the I92L variant to identify the origin of the long lifetime of the water clusters observed in the simulation of the I92D variant started from the full hydration state. The side chains of Leu and Asp are comparable in size but different in polarity. The setup for the simulation of the I92L variant with the full hydration state was the same as for the I92D variant. The full hydration state contained water molecules in sites 5, 6, 7, 8, 9, and 10. During the equilibration stage, two water molecules left the cavity via route I and two via route II. During the first 3 ns of the production run, some of the remaining water molecules exchanged with bulk water through routes I–III. During the last nanoseconds of the production run, only one internal water molecule was found, in site 1.
Multiple simulations of the V66E variant
To investigate how sampling affects the description of water penetration and occupancy of internal pockets and cavities with MD simulations, nine additional 10-ns-long MD simulations were performed with the PHS/V66E variant, starting from the empty hydration state. One goal of these simulations was to investigate whether the water-binding sites observed in crystal structures were reproduced through MD simulations that started from an empty hydration state.
In the nine MD runs, the initial system configurations were identical. The difference was in the assignment of initial velocities. In addition, five of the nine simulations used a slow heatup process. Instead of heating up directly to 300 K in the equilibration procedure, the system was heated up in increments of 25 K. At each temperature the system was equilibrated for 50 ps, except when 300 K were reached, when the system was equilibrated for 450 ps.
In addition to examining the populations of water molecules observed in the microcavity, the populations of two crystallographically resolved water molecules that have long residence times according to both MD (32) and NMRD (49) were analyzed in detail. These two water molecules make three very stable hydrogen bonds with protein residues and are almost completely isolated from the rest of the solvent. The location of these water molecules is shown in Fig. 1. The molecule labeled A was observed in all nine simulations and the molecule labeled as B was observed in six of nine simulations.
In the crystal structure of the PHS/V66E variant obtained at cryogenic temperature, water molecules are observed in sites 1 and 2. Multiple MD simulations started from the empty hydration state identified three dominant hydration patterns. The hydration pattern characterized by the absence of interior water molecules is labeled A in Fig. 3. In this case the Glu-66 side chain is found deeply buried in the protein, usually in a conformation that is either slightly twisted or fully twisted and pointed toward the backbone of Tyr-93 (Fig. 3, left). In the hydration pattern labeled B, a water molecule is found in site 1. In this case, Glu-66 is usually in an extended conformation, with the carboxylic group hydrogen-bonded to the water molecule in site 1 (Fig. 3, middle). In the hydration pattern labeled C, water molecules are found in sites 1 and 2. In this case, Glu-66 is also usually in the extended conformation, with the carboxylic group hydrogen-bonded to water molecules in site 2 (Fig. 3, right). Occasionally, additional hydration patterns were observed, such as when water molecules are located in sites 1, 2, and 3, or when they are randomly clustered around Glu-66, which is in an extended conformation.
In several simulations, a single hydration pattern was predominant. The A hydration pattern was predominant in two simulations, and hydration patterns B, and C were predominant in one simulation each. The remaining MD runs featured a mixture of two or sometimes three hydration patterns. An additional 40-ns-long MD simulation was performed for one of the runs (totaling 50 ns of simulation time). During this long simulation, the A hydration pattern was predominant during the first 25 ns, and the C hydration pattern was predominant during the last 25 ns.
DISCUSSION
Comparison of water binding sites observed by MD and crystallography
The locations of water molecules observed in x-ray structures obtained at cryogenic conditions were compared with the locations of water molecules that have residence times >1 ns in a single 10-ns-long MD simulation started from the crystal hydration state (Fig. 2). The sites where water was observed by the two methods are in agreement except in the case of the wild-type protein, and the I92K, V66N, and V66E variants.
In the wild-type and in the I92K variant the MD simulations report more internal water molecules than in the crystal structure, where no internal water molecules are observed. MD simulations report a water molecule at site 1, with two hydrogen bonds to backbone atoms of the neighboring loop (comprised of residues Asp-19, Gly-20, and Asp-21), and one to an external water molecule (32). This water molecule can interact with several backbone atoms of the loop, thus it is positionally disordered. The flexibility of the loop prevents the water molecule from becoming ordered, thus preventing its detection in the crystal studies.
In the case of the V66E and V66N variants, the MD simulations report fewer internal water molecules than in the crystal structures. However, in both cases there is a discrepancy between the patterns of hydration observed in structures obtained at cryogenic and at room temperatures (see Table 2). More water molecules are almost always observed in the structures obtained at cryogenic than at room temperatures. Room temperature structures have been obtained recently for several variants: V66E, V66Q, V66D, V66N, V66K, V66Y, and I92E (J. L. Schlessman and B. García-Moreno E., unpublished; also (47,49)). Out of these nine variants, only two display internal water molecules: V66Y in position 1 and 2, V66E in position 1.
MD simulations offer insight into the discrepancy between hydration patterns observed in x-ray structures obtained at different temperatures. For seven out of 10 variants (V66E, V66D, V66Q, V66W, I92D, I92E, and I92K), more than one hydration pattern is observed in the MD simulations (Table 2). Four of seven display a discrepancy between the hydration patterns observed through cryo and room temperature crystallography. Furthermore, most of the internal water molecules display rather large MSD values (Table 3). Water motions are much larger at room temperatures than at cryogenic temperatures. This might explain why such water molecules are absent from room temperature structures. However, in the case of the water molecule in site 1 in the V66Y variant, the large MSD value observed in the MD simulations (4.64 Å2) is in disagreement with the presence of this water molecule in the room temperature structure. This could be an issue related to either incomplete sampling, or inaccurate force field.
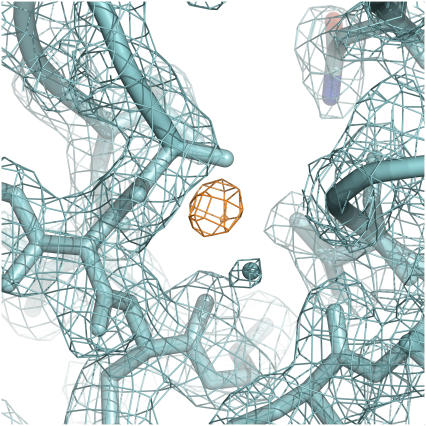
It is worth stating that the number of water molecules resolved in crystallographic structures may depend on the selection criteria used by individual crystallographers during the model building and refinement process. The selection criteria used for the room temperature structures relevant for this study (J. L. Schlessman and B. García-Moreno E., unpublished) were: 1), spherical electron density must be observed in both the 2Fo-Fc and 1Fo-Fc electron density maps; 2), at least one protein hydrogen-bonding partner must be identified; and 3), the B-factor for the water should remain <50 Å in subsequent rounds of refinement. Following these criteria, in the case of V66N, a water molecule was not observed in position 1 in the room temperature structure (Fig. 4). Some electron density was observed in the 1Fo-Fc map at position 1; however, no electron density was observed in the 2Fo-Fc electron density map, even when that map contour was lowered to noise levels (Fig. 4). This suggests that a disordered water molecule, or a water molecule with partial occupancy may be present in this position, even though no water molecule in this position is reported in the final structure. A water molecule may have been reported in the x-ray structure, in full agreement with what the MD simulations suggest, had a less stringent set of criteria been used.
FIGURE 4.
Region near Asn-66 in the crystal structure of PHS/V66N variant obtained at room temperature. 2Fo-Fc electron density map is in cyan, and 1Fo-Fc electron density map is in orange.
Simulations with the empty hydration state
The MD simulations started from the empty hydration state were designed to test the ability of MD simulations to identify sites where internal water molecules might exist but which are not observed in crystal structures for the reasons discussed above. A single simulation was performed for each variant with the exception of the V66E variant, for which eight 10-ns-long simulations and one 50-ns-long simulation were performed.
Multiple simulations of the V66E variant
Two internal water molecules at the sites other than those in the microcavity are present in all of the crystal structures of SN (sites A and B as shown in Fig. 3). These water molecules display long residence times in both the NMRD experiments (49) and MD simulations (32). The locations of these two water molecules are reproduced well by the nine simulations of the V66E variant. The water molecule in site A is observed in all of the simulations whereas the water molecule in site B is reproduced in 67% of the simulations. Inspection of the trajectories revealed that the timescale for water penetration into site B ranges between a few hundred picoseconds and 5 ns. This suggests that this water molecule could have been observed in all simulations if the simulations were longer than 10 ns.
In terms of the state of hydration of the microcavity, multiple MD simulations of the V66E variant revealed multiple hydration patterns, with three occurring most frequently. In four simulations, a single hydration pattern was predominant; the remaining six simulations showed a mixture of mostly two hydration patterns. Only two of the three hydration patterns observed in MD simulations were observed in crystal structures: the structure obtained at cryogenic temperature shows two internal water molecules and the structure obtained at room temperature shows only one internal water molecule (49).
The hydration pattern characterized by the absence of water molecules in the microcavity has not been observed crystallographically. When water is absent from the protein, the side chain of Glu-66 is found deeply buried inside the protein either in an extended, in a slightly twisted, or in a fully twisted conformation in which its polar group interacts with the backbone of Tyr-93. The twist of Glu-66 could thus be caused by the tendency of the polar group of Glu-66 to interact with another polar group. Furthermore, the starting structures that lack water molecules in the protein interior could be influencing the results and driving Glu-66 into the twisted state. We note, however, that the twisted state of Glu-66 was also observed in simulations started from the crystal hydration state.
The nine simulations of the V66E variant show the importance of performing multiple MD runs to enhance sampling. Multiple simulations enhance the probability of observing water penetration events. Multiple MD simulations can also reveal multiple rotameric states of side chains and multiple hydration patterns that are sometimes not observed in a single 10-ns-long simulation.
Simulations of all other variants
In most of the simulations started from the empty hydration state, the hydration pattern that was recovered was similar to the one observed in the simulations started from crystal hydration state. The hydration patterns were usually established during the equilibration phase, on the timescale of a few hundred picoseconds. In three cases, V66W, V66D, and I92D, hydration patterns different from those observed in simulations started from crystal hydration state were observed. In the case of the V66W variant, the observed hydration pattern is most likely an artifact that results from a lack of water molecules in the starting MD structures, and sampling insufficient to allow the breaking of a hydrogen bond formed at the beginning of the simulation.
Simulations with the full hydration state
The simulations started from the full hydration state were designed to test if there are regions inside the protein that could be populated by water but where penetration cannot occur in the 10-ns timescale. In seven of eleven simulations performed in this way, the hydration pattern observed in the crystal structures, or in the simulations started from the crystal hydration state, were recovered by the MD simulations. The exiting of water from the protein was achieved on a nanosecond timescale (except for I92D which took more than 10 ns).
Hydration patterns alternative to those observed in the crystal structures or in the simulations started from the crystal hydration state were found in four cases. One water molecule stayed trapped in a potential minimum in site 6 in V66W. The remaining three cases occurred in simulations of V66E, V66D, and I92D variants.
Local flexibility and water content
Protein flexibility appears to be one of the factors responsible for multiple hydration patterns, disorder, and partial occupancies of water binding sites. In simulations with the WT protein, the flexible loop elements provided a dynamical environment that led to positionally disordered internal water molecules.
In the case of simulations with the V66E variant, the three different rotameric states of Glu-66 were dominated by a unique hydration pattern. The same was true in simulations with the I92E variant. Four water molecules were observed in the crystal structure of I92E obtained at low temperature (46). However, in the simulations predominantly only two of those sites are populated at a single moment. In one of the conformations of Glu-92, water molecules are present in sites 1 and 2. In a slightly different conformation of Glu-92, sites 3 and 4 are the ones that are occupied.
The simulations of V66K and I92K variants offer even more dramatic examples of how flexibility affects the patterns of hydration observed in MD simulations. The long side chain of Lys, which is inherently flexible, is partially disordered and invisible in the crystal structures. According to the MD simulations, the reason that internal water molecules are not observed in crystal structures of variants with internal Lys is that water is not stable in the presence of internal Lys residues owing to fast fluctuations between different rotameric states of the side chain.
Are hydrophobic microcavities in other proteins hydrated?
Recent theoretical studies on nanotubes (58) and small hydrophobic cavities in carbon spheres (59) suggest that water molecules can exist in an ordered state in confined hydrophobic environments. These environments provide rigid shells in which water molecules can form stable hydrogen-bonded networks among themselves. This is a situation similar to the case of large protein cavities, such as those found in the β-barrel of the fatty-acid binding protein (35), where large clusters of water molecules can be stabilized.
The MD simulations of the wild-type SN, and the I92D and I92L variants started from the full hydration state suggest that small nonpolar cavities in proteins are likely to be dry. The cavity in the structure of I92D and I92L can house as many as six water molecules. The simulation of the variant containing the polar Asp-92 shows that during the MD simulation clusters of five, then four, then three, and then two water molecules are stabilized for a few nanoseconds. During the last few nanoseconds, between zero and two water molecules reside in the interior and interact with Asp-92. The size and shape of the microcavity of the variant with nonpolar Leu-92 is approximately the same as for I92D. Five out of six internal water molecules placed initially in the cavity in the I92L variant by DOWSER leave the protein. One water molecule remains in polar site 1. In the case of the wild-type SN started from the full hydration state (i.e., four interior water molecules), the cavity, which contains only hydrophobic residues, also becomes devoid of water. These simulations provide a strong indication that flexible, small, apolar pockets and cavities in proteins will not house ordered water molecules unless a source of polarity is present.
CONCLUSIONS
The patterns of internal hydration observed in crystal structures obtained under cryogenic conditions are almost always sampled during MD simulations. The number of water molecules observed in simulations can be larger than the number reported in crystal structures when water molecules are disordered or when they have partial occupancies. The simulations can sometimes appear to underestimate the number of internal water molecules when flash-freezing induces water penetration or ordering of disordered water molecules in crystal structures (31). In the crystallographic structures of SN variants with internal polar groups, more water molecules are always observed under cryogenic than under room-temperature conditions. The simulations suggest that room-temperature structures contain less water molecules because multiple hydration patterns are feasible, and because observed water molecules have large amplitudes of motion. It thus appears that in the crystallographic structures of SN variants with internal polar groups, cryogenic conditions organize internal water molecules by minimizing disorder.
There are two scenarios under which MD simulations can fail to detect water penetration. One case is when the simulations are started without any internal water molecules, and polar residues hydrogen-bond to the backbone or other polar groups to compensate for the lack of water. The other scenario occurs when sampling in a single 10-ns-long simulation is simply insufficient. Both deficiencies can be decreased by performing multiple simulations as well as simulations started from different initial states of hydration. The comparison of simulations started from three different initial states of hydration indicates that water penetration occurs on timescales of hundreds of ps, and water exiting occurs on timescales of a few nanoseconds. To increase the usefulness of MD simulations to hydrate internal pockets and cavities or active sites artificially, it is advisable to compare simulations started from different initial hydration states.
The results of nine separate simulations with the V66E variant, started from nine sets of initial velocities, suggest that multiple simulations increase the likelihood of observing proper structures, of realistic sampling of alternative conformational states, and of hydration of internal sites with partial occupancies. Given a fixed number of simulation timesteps, multiple MD simulations are advantageous not only in terms of efficient parallelization and decreased simulation time, but more importantly in terms of increased ability of sampling of hydration patterns.
The simulations show that multiple patterns of hydration can be found in the interior of proteins, and that the patterns are linked to the flexibility and dynamics of the protein backbone and of internal polar side chains. The inherent flexibility of loops can alter hydration patterns of internal sites, but an even more significant observation is that internal polar side chains such as Lys and Glu can themselves exhibit significant flexibility, even in their buried, internal sites. The different conformational substates of Glu-66, for example, stabilize different patterns of internal hydration. Similarly, the high flexibility of internal Lys side chains appears to be the main reason that these internal side chains are dehydrated in crystal structures.
In contrast to rigid hydrophobic cavities, such as those found in buckeyballs, apolar, flexible protein cavities initially overstuffed with water become dehydrated during molecular dynamics simulations. This is consistent with a recent study (60) in which the increase in flexibility of apolar pores was found to result in expulsion of water from the pore. When a source of polarity, such as an internal polar or ionizable group is present, internal water molecules cluster around such groups. It appears unlikely that water molecules, alone or in small clusters, can be stabilized in small, flexible hydrophobic cavities in the protein interior unless there is a source of polarity.
Acknowledgments
We thank Carlos Castañeda for performing DOWSER calculations. We are also grateful to Apostolos Gittis and Wesley Stites for coordinates of crystal structures of variants with Asp-92 and Trp-66. Figs. 1–3 were generated with program VMD (55), while Fig. 4 was generated with PyMOL (61).
This work was supported by a National Institutes of Health grant (No. R01 GM061597) to B.G.M.E., and a National Science Foundation grant (Nos. MCB 0543769 and DMR 0117792) to A.E.G. A.D. was supported by the Burroughs Wellcome Fund. J.S. was supported by a John F. Crowley fellowship from the United States Naval Academy. Research Opportunity Award support for J.S. through National Science Foundation grant (No. MCB-0212414) to B.G.M.E. is also gratefully acknowledged.
Editor: Klaus Schulten.
References
- 1.Rose, G. D., and W. B. Young. 1983. Interior turns in globular proteins. Nature. 304:655–657. [DOI] [PubMed] [Google Scholar]
- 2.Sreenivasan, U., and P. H. Axelsen. 1992. Buried water in homologous serine proteases. Biochemistry. 31:12785–12791. [DOI] [PubMed] [Google Scholar]
- 3.Langhorst, U., R. Loris, V. P. Denisov, J. Doumen, P. Roose, D. Maes, B. Halle, and J. Steyaert. 1999. Dissection of the structural and functional role of a conserved hydration site in RNase T1. Protein Sci. 8:722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Likic, V. A., N. Juranic, S. Macura, and F. G. Prendergast. 2000. Structural water molecules in the family of fatty acid binding proteins. Protein Sci. 9:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covalt, J. C., M. Roy, and P. A. Jennings. 2001. Core and surface mutations affect folding kinetics, stability, and cooperativity I IL-1b: does alteration in buried water play a role? J. Mol. Biol. 307:657–669. [DOI] [PubMed] [Google Scholar]
- 6.Park, S., and J. G. Saven. 2005. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins Struct. Funct. Gen. 60:450–463. [DOI] [PubMed] [Google Scholar]
- 7.Takano, K., J. Funahashi, Y. Yamagata, S. Fujii, and K. Yutani. 1997. Contribution of water molecules in the interior of a protein to the conformational stability. J. Mol. Biol. 274:132–142. [DOI] [PubMed] [Google Scholar]
- 8.Mao, Y., M. A. Ratner, and M. F. Jarrold. 2000. One water molecule stiffens a protein. J. Am. Chem. Soc. 122:2950–2951. [Google Scholar]
- 9.Fischer, S., and C. S. Verma. 1999. Binding of buried structural water increases the flexibility of proteins. Proc. Natl. Acad. Sci. USA. 96:9613–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch, C. A., D. Karp, K. Lee, W. Stites, E. Lattman, and B. García-Moreno E. 2002. Experimental pKa values of buried residues: analysis with continuum methods and role of water penetration. Biophys. J. 82:3289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schutz, C. N., and A. Warshel. 2002. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins Struct. Funct. Gen. 44:400–417. [DOI] [PubMed] [Google Scholar]
- 12.Hofacker, I., and K. Schulten. 1998. Oxygen and proton pathways in cytochrome c oxidase. Proteins Struct. Funct. Gen. 30:100–107. [PubMed] [Google Scholar]
- 13.Pomès, R., and B. Roux. 2002. Molecular mechanism of H+ conduction in the single-file water chain of the gramicidin channel. Biophys. J. 82:2304–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decoursey, T. E. 2003. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83:465–579. [DOI] [PubMed] [Google Scholar]
- 15.Cao, W., J. F. Christian, P. M. Champion, F. Rosca, and J. T. Sage. 2001. Water penetration and binding to ferric myoglobin. Biochemistry. 40:5728–5737. [DOI] [PubMed] [Google Scholar]
- 16.Petrone, P., and A. E. García. 2004. MHC-peptide binding is assisted by bound water molecules. J. Mol. Biol. 338:419–435. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, E. 1992. Internal water molecules and H-bonding in biological macro-molecules: a review of structural features with functional implications. Protein Sci. 1:1543–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denisov, V. P., B. H. Jonsson, and B. Halle. 1999. Dynamics of functional water in the active site of native carbonic anhydrase from 17O magnetic relaxation dispersion. J. Am. Chem. Soc. 121:2327–2328. [Google Scholar]
- 19.Cheung, M. S., A. García, and J. N. Onuchic. 2002. Protein folding mediated by solvation: water expelling and formation of the hydrophobic core occur after the structural collapse. Proc. Natl. Acad. Sci. USA. 99:685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García, A., and J. Onuchic. 2003. Folding a protein in a computer; an atomic description of the folding/unfolding of protein A. Proc. Natl. Acad. Sci. USA. 100:13898–13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garau, G., S. Geremia, and L. Randaccio. 2002. Relationship between hydrogen-bonding network and reduction potential in c-type cytochromes. FEBS Lett. 516:285–286. [DOI] [PubMed] [Google Scholar]
- 22.de Groot, B. L., D. P. Tieleman, P. Pohl, and H. Grubmüller. 2002. Water permeation through gramicidin A desformylation and the double helix: a molecular dynamics study. Biophys. J. 82:2934–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anishkin, A., and S. Sukharev. 2004. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys. J. 86:2883–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]
- 25.Tajkhorshid, E., P. Nollert, M. Ø. Jensen, L. J. W. Miercke, J. O'Connell, R. M. Stroud, and K. Schulten. 2002. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 296:525–530. [DOI] [PubMed] [Google Scholar]
- 26.Roux, B., and K. Schulten. 2004. Computational studies of membrane channels. Structure. 12:1343–1351. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, X.-J., and B. W. Matthews. 1994. Conservation of solvent-binding sites in 10 crystal forms of T4 lysozyme. Protein Sci. 3:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denisov, V. P., and B. Halle. 1994. Dynamics of the internal and external hydration of globular proteins. J. Am. Chem. Soc. 116:10324–10325. [Google Scholar]
- 29.Denisov, V. P., G. Carlstrom, K. Venu, and B. Halle. 1997. Kinetics of DNA hydration. J. Mol. Biol. 268:118–136. [DOI] [PubMed] [Google Scholar]
- 30.Juers, D. H., and B. W. Matthews. 2001. Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions. J. Mol. Biol. 311:851–862. [DOI] [PubMed] [Google Scholar]
- 31.Halle, B. 2004. Biomolecular cryocrystallography: structural changes during flash-cooling. Proc. Natl. Acad. Sci. USA. 101:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damjanović, A., B. García-Moreno E., E. E. Lattman, and A. E. García. 2005. Molecular dynamics study of water penetration in staphylococcal nuclease. Proteins Struct. Funct. Gen. 60:433–449. [DOI] [PubMed] [Google Scholar]
- 33.Oprea, T. I., G. Hummer, and A. E. García. 1997. Identification of a functional water channel in cytochrome P45 enzymes. Proc. Natl. Acad. Sci. USA. 94:2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.García, A. E., and G. Hummer. 2000. Water penetration and escape in proteins. Proteins Struct. Funct. Gen. 38:261–272. [PubMed] [Google Scholar]
- 35.Bakowies, D., and W. F. van Gunsteren. 2002. Water in protein cavities: a procedure to identify internal water and exchange pathways and application to fatty acid-binding protein. Proteins Struct. Funct. Gen. 47:534–545. [DOI] [PubMed] [Google Scholar]
- 36.Bakowies, D., and W. F. van Gunsteren. 2002. Simulations of apo and holo-fatty acid binding protein: structure and dynamics of protein, ligand and internal water. J. Mol. Biol. 315:713–736. [DOI] [PubMed] [Google Scholar]
- 37.Damjanović, A., I. Kosztin, U. Kleinekathoefer, and K. Schulten. 2002. Excitons in a photosynthetic light-harvesting system: a combined molecular dynamics, quantum chemistry and polaron model study. Phys. Rev. E. 65:031919. [DOI] [PubMed] [Google Scholar]
- 38.Wade, R. C., M. Mazor, J. A. McCammon, and F. A. Quiocho. 1991. A molecular dynamics study of thermodynamic and structural aspects of the hydration of cavities in proteins. Biopolymers. 31:919–931. [DOI] [PubMed] [Google Scholar]
- 39.Roux, B., M. Nina, R. Pomes, and J. Smith. 1996. Thermodynamics stability of water molecules in the bacteriorhodopsin proton channel—a molecular dynamics free energy perturbation study. Biophys. J. 71:670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olano, L. R., and S. W. Rick. 2004. Hydration free energies and entropies for water in protein interiors. J. Am. Chem. Soc. 126:7991–8000. [DOI] [PubMed] [Google Scholar]
- 41.McDowell, R. S., and A. A. Kossiakoff. 1995. A comparison of neutron diffraction and molecular dynamics structures: hydroxyl group and water molecule orientations in trypsin. J. Mol. Biol. 250:553–570. [DOI] [PubMed] [Google Scholar]
- 42.Sterpone, F., M. Ceccarelli, and M. Marchi. 2001. Dynamics of hydration in hen egg white lysozyme. J. Mol. Biol. 311:409–419. [DOI] [PubMed] [Google Scholar]
- 43.García, A. E., and L. Stiller. 1993. Computation of the mean residence time of water in the hydration shells of biomolecules. J. Comput. Chem. 14:1396–1406. [Google Scholar]
- 44.Imai, T., A. Kovalenko, and F. Hirata. 2004. Solvation thermodynamics of protein studied by the 3D-RISM theory. Chem. Phys. Lett. 395:1–6. [Google Scholar]
- 45.Dwyer, J. J., A. G. Gittis, D. A. Karp, E. E. Lattman, D. S. Spencer, W. E. Stites, and B. García-Moreno E. 2000. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 79:1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen, D. M., R. L. Reynald, A. G. Gittis, and E. E. Lattman. 2004. X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior. J. Mol. Biol. 341:565–574. [DOI] [PubMed] [Google Scholar]
- 47.Karp, D. A., A. G. Gittis, M. A. Stahley, C. A. Fitch, W. E. Stites, E. E. Lattman, and B. García-Moreno E. 2007. Molecular determinants of the abnormal pKa value of an internal aspartic acid: contributions by local unfolding and water penetration. Biophys. J. 92:2041–2053.17172297 [Google Scholar]
- 48.Kato, M., and A. Warshel. 2006. Using a charging coordinate in studies of ionization induced partial unfolding. J. Phys. Chem. B. 110:11566–11570. [DOI] [PubMed] [Google Scholar]
- 49.Denisov, V. P., J. L. Schlessman, B. García-Moreno E., and B. Halle. 2004. Stabilization of internal charges in a protein: water penetration or conformational change? Biophys. J. 87:3982–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Moreno E., B., J. J. Dwyer, A. G. Gittis, E. E. Lattman, D. S. Spencer, and W. E. Stites. 1997. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys. Chem. 64:211–224. [DOI] [PubMed] [Google Scholar]
- 51.Kalé, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151:283–312. [Google Scholar]
- 52.Zhang, L., and J. Hermans. 1996. Hydrophilicity of cavities in proteins. Proteins Struct. Funct. Gen.. 24:433–438. [DOI] [PubMed] [Google Scholar]
- 53.MacKerell, A. D., Jr., D. Bashford, M. Bellott, R. L. Dunbrack, Jr., J. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, I. W. E. Reiher, B. Roux, M. Schlenkrich, J. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-hydrogen empirical potential for molecular modeling and dynamics studies of proteins using the CHARMM22 force field. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 54.Feller, S. E., Y. H. Zhang, R. W. Pastor, and B. R. Brooks. 1995. Constant pressure molecular dynamics simulation—the Langevin piston method. J. Chem. Phys. 103:4613–4621. [Google Scholar]
- 55.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 56.Feig, M., J. Karanicolas, and C. L. Brooks III. 2001. MMTSB tool set. MMTSB NIH Research Resource, The Scripps Research Institute, La Jolla, CA.
- 57.Damjanović, A., B. García-Moreno E., E. E. Lattman, and A. E. García. 2005. Molecular dynamics study of hydration of the protein interior. Comput. Phys. Commun. 169:126–129. [Google Scholar]
- 58.Hummer, G., J. C. Rasaiah, and J. P. Noworyta. 2001. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 414:188–190. [DOI] [PubMed] [Google Scholar]
- 59.Vaitheeswaran, S., H. Yin, J. C. Rasaiah, and G. Hummer. 2004. Water clusters in nonpolar cavities. Proc. Natl. Acad. Sci. USA. 101:17002–17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreev, S., D. Reichman, and G. Hummer. 2005. Effect of flexibility on hydrophobic behavior of nanotube water channels. J. Chem. Phys. 123:194502. [DOI] [PubMed] [Google Scholar]
- 61.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. http://www.pymol.org.