Abstract
The isolation of genes from a given genomic region can be a rate-limiting step in the discovery of disease genes. We describe an approach to the isolation of cDNAs that have sequences in common with large genomic clones such as bacterial artificial chromosomes. We applied this method to loci both amplified and deleted in cancer, illustrating its usage in the identification of both oncogenes and tumor suppressor genes, respectively. The method, called rapid isolation of cDNAs by hybridization (RICH), depends on solution hybridization, enzymatic modification, and amplification/selection of sequences present in both cDNA populations and the genomic clones. The method should facilitate the development of transcription maps for large genomic clones, possibly even yeast artificial chromosomes.
Keywords: gene, bacterial artificial chromosome, exon trapping, sequencing
Powerful methods have facilitated the localization of disease genes to regions of the genome. Typically, candidate regions are contained on large yeast or bacterial cloning vectors, and these vectors must be searched assiduously by various means for candidate genes. This step has often proved to be an obstacle in gene discovery. The problem has been attacked in roughly three ways: by hybridization (1–3), sequence analysis (4, 5), and exon trapping methods (6–8). Each method has its own particular advantages, but no current method is without serious problems.
We present herein a method we call rapid isolation of cDNAs by hybridization (RICH) based on the identification of sequences in common between a cDNA library and a large clone of genomic DNA. The method selects and amplifies those restriction endonuclease fragments of cDNAs that hybridize precisely at one end to the end of a similarly cleaved genomic DNA fragment. Before hybridization, the cDNA and genomic fragments are modified with different adaptors. Those cDNAs that form hybrids with genomic DNA at at least one end are ligated to a “selection adaptor” that is complementary to the genomic adaptor and contains an additional sequence complementary to an RNA polymerase site. Such cDNAs can be selectively amplified by successive treatments with RNA and DNA polymerases.
We illustrate the basic method with two series of experiments: (i) a search for transcripts from the c-MYC locus in cDNAs from a breast cancer cell line and (ii) a search for transcripts from the PTEN tumor suppressor locus in cDNAs from normal breast tissue. Although the method is complex, in that many different enzymes (restriction endonucleases, various DNA ligases, RNA polymerase, various DNA endo- and exonucleases, reverse transcriptase, and various DNA polymerases) are used, they are all robust enzymes that are readily available. Only 3 days are required to yield candidates for further study.
MATERIALS AND METHODS
Materials.
The oligonucleotides which were synthesized for this research are listed in Table 1 and obtained from Biosynthesis (Lewisville, TX). P1 clone 8001 (Genome Systems, St. Louis) is an 80-kb genomic clone that contains the exons 1–3 of the c-MYC gene. A bacterial artificial chromosome (BAC) clone, 60C5, containing exons 4–9 of PTEN (containing nucleotides 1,244–2,246 of the published cDNA, GenBank accession no. U92436) was obtained from Genome Systems. The plasmid pUC18, digested with BamHI and treated with bacterial alkaline phosphatase was supplied by Amersham. pCR-Script SK(+) and Epicurian Coli XL2-Blue cells were obtained from Stratagene. SKBr3 is a breast cancer cell line from which the poly(A)+ RNA was extracted with the FastTrack kit (Invitrogen). cDNA was synthesized from the poly(A)+ RNA of SKBr3 and commercially available poly(A)+ RNA of mammary gland tissue (CLONTECH) by using the Copy kit (Invitrogen). The Megascript kit was from Ambion (Austin, TX).
Table 1.
Oligonucleotide sequences for RICH
| Oligo | Sequence | 5′ end |
|---|---|---|
| N-12 | GATCTTCCCTCG | Dephosphorylated |
| R-12 | GATCTGCGGTGA | Dephosphorylated |
| pN-24 | AGGCAACTGTGCTATCCGAGGGAA | Phosphorylated |
| pR-24 | AGCACTCTCCAGCCTCTCACCGCA | Phosphorylated |
| pR(+)SP6 | CTGCGGTGAGAGGCTGGAGAGTGCTCTATAGTGTCACCTAAAT | Phosphorylated |
| SP6 | TATTTAGGTGACACTATAGAGCA | Dephosphorylated |
The enzymes and their specific buffers used in RICH were obtained from the following suppliers: Sau3AI and T4 DNA ligase from New England Biolabs; Stoffel fragments, AmpliTaq, and AmpliTaq Gold from Perkin–Elmer; Ampligase from Epicentre Technologies (Madison, WI); Pfu DNA Polymerase and RNase-free Dnase from Stratagene; and λ exonuclease from Amersham.
Glycogen was obtained from Boeringer Mannheim. GeneQuant G-50, S-300HR, and S-400HR columns and Sephaglas BandPrep kit were obtained from Amersham. RNase-free water was supplied by Ambion. Maxi prep kit was supplied by Qiagen (Hilden, Germany). Phenol was prepared as described elsewhere (9).
RICH Standard Protocol.
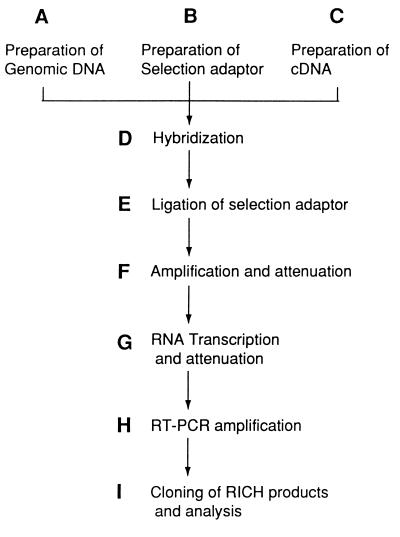
To facilitate description of the RICH protocol, we have broken up the procedure into discrete units, labeled those units with an alphabetic letter, and assigned that letter both to the procedure and its final product (Fig. 1).
Figure 1.
Flowchart of RICH. Each boldface letter represents a unit of the RICH procedure described in the standard RICH protocol.
A. Preparation of genomic DNA.
For the preparation of genomic DNA, 1 μg of BAC DNA is digested with 20 units of Sau3AI. The digests are purified by phenol/chloroform extraction and ethanol precipitation. The whole digests are mixed with 100 pmol of pR-24 and 100 pmol of R-12 oligonucleotides (Table 1) in 20 μl of 1× T4 DNA ligase buffer. The double-strand cDNA and the oligonucleotides are heated at 65°C for 5 min, annealed by cooling down the mixture to 4°C gradually, and then ligated by overnight incubation with 2,000 units of T4 DNA ligase at 11°C. The excess oligonucleotides are removed from the ligated fragments with S-400HR columns.
B. Preparation of selection adaptor.
The selection adaptor is synthesized, phosphorylated at its 5′end, and purified by PAGE. The working concentration of the adaptor is 50 nM.
C. Preparation of cDNA.
Double-stranded cDNA is synthesized from 5 μg of poly(A)+ RNA with the Copy kit. One microgram of cDNA is digested with 20 units of Sau 3AI and ligated to 100 pmol of pN-24 and 100 pmol of N-12 oligonucleotides in a 20-μl reaction volume as described above. A partial fill-in mixture is made by combining 21 μl of water, 5 μl of 10× Stoffel buffer, 1 μl of 10 mM dATP, 1 μl of 10 mM dGTP, and 1 μl of 10 mM dTTP and is added to the ligation mixture. After incubation at 72°C for 5 min, 2 units of Stoffel fragment are added to the mixture and incubated at 72°C for additional 5 min. The fragments are extracted twice with 50 μl of phenol/chloroform. The aqueous phase is transferred to a new tube.
D. Hybridization.
Ten microliters of genomic fragments (components A), 5 μl of R(+)SP6 (components B), and 25 μl of cDNA fragments (components C) are mixed and then extracted twice with 40 μl of phenol/chloroform. Ten microliters of 10 M ammonium acetate, 1 μl of glycogen (20 mg/ml), and 92 μl of ethanol are added. After centrifugation and washing with 70% ethanol, the DNA is dissolved in 4 μl of 3× EE buffer (10). The solution is overlaid with 30 μl of mineral oil and denatured by incubation in boiling water for 5 min. One microliter of 5 M NaCl is added and the DNA is reannealed for 16 h at 67°C.
E. Ligation of selection adaptor.
To ligate the selection adaptor to the cDNA fragments, 6 μl of 10× Ampligase buffer and 1 μl of Ampligase (100 units/μl) are added to 48 μl of water, and this ligation mixture is incubated at 67°C for 3 min before adding to the solution (component D). The total mixture is then incubated for 4 h at 67°C. After ligation, the DNA is extracted with 60 μl of phenol/chloroform twice, precipitated by ethanol as above, and dissolved in 50 μl of TE buffer (10 mM Tris⋅HCl, pH 7.5/1 mM EDTA). Excess R(+)SP6 is removed by S-400HR after denaturation.
F. Amplification and attenuation.
Five microliters of the DNA solution (component E) is mixed with 50 μl of reaction mixture [1× AmpliTaq buffer/all four dNTPs (each at 200 nM)/1 μM pN-24/1 μM SP6/1 unit of AmpliTaq Gold]. The DNA is amplified by a PCR program that specifies an incubation for 10 min at 94°C followed by 10 cycles of 1 min at 94°C, 1 min at 60°C, and 3 min at 72°C. After purification with S-300 HR columns, the DNA is digested at 37°C with 10 units of λ exonuclease. Eighty microliters of TE is added to the reaction mixture and incubated in boiling water for 5 min. The 100-μl PCR mixture contains 8 μl of the DNA, all four dNTPs (each at 200 μM), 1 μM pN-24, 1 μM SP6, and Pfu buffer at a 1× final concentration. After initial denaturation of 94°C for 5 min, 2.5 units of Pfu polymerase is added to the mixture and 20 cycles at 94°C for 1 min and 70°C for 4 min are performed, followed by a final extension at 72°C for 10 min. The PCR products are purified with phenol/chloroform extraction and ethanol precipitation. The DNA is dissolved in 25 μl of RNase-free water after washing with 70% ethanol.
G. RNA transcription and attenuation.
RNA is transcribed from 8 μl of the DNA template (component F) with the Megascript kit. After a 6-h incubation at 37°C, 20 units of RNase-free Dnase I is added and the incubation is continued for an additional 10 min. The RNA is purified with phenol/chloroform extraction and dissolved in 15 μl of RNase-free water after ethanol precipitation.
H. Reverse transcription-coupled PCR amplification.
To synthesize cDNA, 1.3 μl of 50 μM pN-24 is added to the mixture (component G), incubated at 65°C for 10 min, and then placed at room temperature for 5 min. Double-stranded cDNA is synthesized by the Copy kit following the supplier’s protocol. After the products are denatured by heating to 94°C for 10 min, they are purified with S-300HR columns. The 200-μl PCR mixture contains 10 μl of the cDNA, all four dNTPs (each at 200 μM), 1 μM pN-24, 1 μM pR-24, 5 units of AmpliTaq Gold, and AmpliTaq buffer at a 1× final concentration. After initial denaturation of 94°C for 10 min, 30 cycles at 94°C for 1 min and 72°C for 3 min were performed, followed by a final extension at 72°C for 10 min.
I. Cloning and sequencing.
The RICH products were purified with GeneQuant G-50 columns and digested with 40 units of Sau3AI in a 120-μl reaction volume at 37°C for 4 h. The digests were size-fractionated by electrophoresis through an agarose gel for Southern blot analysis and for cloning. The DNA bands were excised, purified with the Sephaglas BandPrep kit, and cloned into pUC18 digested with BamHI and treated with bacterial alkaline phosphatase. Epicurian Coli XL2-Blue cells were transformed with the plasmid. The plasmid DNA was purified with the Maxi prep kit and sequenced by the Cycle Sequencing system (Perkin–Elmer).
J. Southern blot analysis.
A part of RICH product was electrophoresed through a 4% agarose gel and transferred to Hybond N+ (Amersham). Hybridization with the PTEN cDNA probe was detected by the ECL system (Amersham).
K. Making representations of genomic clones.
In one variation of the basic method, we used representations of genomic clones. To make the representation, 100 ng of BAC DNA is digested with 5 units of Sau3AI. The digests are purified by phenol/chloroform extraction and ethanol precipitation. The whole digests are mixed with 100 pmol of pR-24 and 100 pmol of R-12 oligonucleotides (Table 1) in 20 μl of 1× T4 DNA ligase buffer. The double-strand cDNA and the oligonucleotides are heated at 65°C for 5 min, annealed by cooling the mixture to 4°C gradually, and then ligated by overnight incubation with 2,000 units of T4 DNA ligase at 11°C.
RESULTS
The Basic Method.
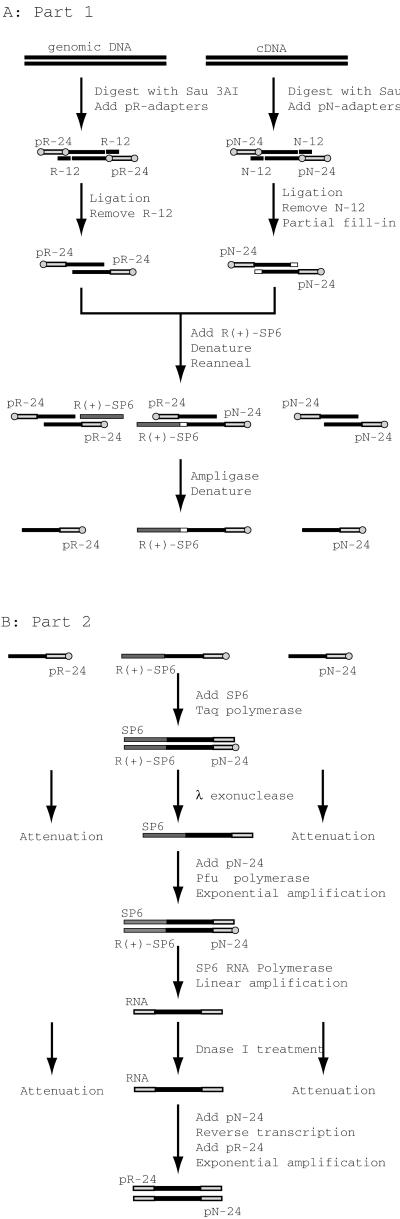
For convenience, we break the method into two parts. In part one, we add a selection adaptor to only those cDNA fragments that hybridize at one end to an end of a genomic fragment. In part two, we selectively amplify only those modified cDNA fragments. The complexity of the method arises from the need to suppress the amplification of self-annealed cDNA and genomic homoduplexes. Fig. 2 outlines the basic schema.
Figure 2.
RICH procedure. (A) Part 1. (B) Part 2. A circle represents a phosphorylated 5′ end. Open boxes represent filled-in bases. Oligonucleotides and their antisense sequences are drawn as dark and light shaded boxes.
Part One.
First, both the large genomic clone and cDNA are separately cleaved with Sau3AI and modified with T4 DNA ligase by the addition of oligonucleotide adaptors (pR-24 for genomic DNA and pN-24 for cDNA) at their 5′ ends only. These adaptors have phosphate groups at their 5′ ends, for reasons that will become apparent in part two.
Next, the 3′ ends of the adapted cDNA molecules are partially filled-in with the three nucleotides guanosine, adenosine, and thymidine by using the Stoffel fragment (a Taq DNA polymerase lacking both 3′ and 5′ exonuclease activity). This step enables us to distinguish later the homoduplexes of genomic DNA from the cDNA–genomic heteroduplexes. An alternative to partial filling is presented later.
The cDNA and genomic DNAs are mixed at equal mass ratios, and the selection adaptor pR(+)SP6 is added. pR(+)SP6 is complementary to pR-24 but has an extra cytidine at its 5′ end and a sequence complementary to the SP6 RNA polymerase promoter sequence at its 3′ end.
The mixture is heat-denatured and allowed to reanneal. During reannealing, two and three part structures will form, as indicated in Fig. 2A. The reannealed mixture is treated with a thermostable DNA ligase, Ampligase, at 68°C to ensure that perfect matches are preferentially ligated.
Ligation of pR(+)SP6 to cDNA will occur only when the latter is annealed to a matching genomic fragment, forming a perfect contiguous substrate for the ligase. The extra cytidine at the 5′ end of pR(+)SP6 is needed to fill the gap of the partially filled-in cDNA fragment. pR(+)SP6 will not be ligated to genomic homoduplexes because those three part structures will have a 3-nucleotide gap. Neither will pR(+)SP6 be ligated to cDNA homoduplex because the latter, lacking pR-24 and having only a 1-base overhang, will not base-pair to pR(+)SP6.
Thus only cDNAs with homology to genomic fragments at one end will have the pN-24 adaptors at their 5′ ends and the pR(+)SP6 selection adaptor at their 3′ ends.
Part Two.
The selectively modified cDNA fragments are amplified, in the following manner. First, after ligation, we denature the mixture and use Taq DNA polymerase and an oligonucleotide primer containing the SP6 promoter sequence to make the selectively modified cDNAs double-stranded. These SP6 primers are not phosphorylated at their 5′ ends. This step is repeated 10 times to give an arithmetic increase in the number of complementary strands (see Fig. 2B).
Next, the entire mixture is treated with λ exonuclease, which degrades double-stranded fragments from their phosphorylated 5′ ends. The strands synthesized from the SP6 primers are thus protected. A small number of duplexes will be formed during the reannealing of the previous step and, when filled-in by Taq polymerase, would contaminate the subsequent reactions. These contaminants are destroyed by λ exonuclease because the adaptors pR-24 and pN-24 have phosphorylated 5′ ends.
To amplify the SP6-primed strand, we perform a PCR with Pfu DNA polymerase and SP6 and pN-24 oligonucleotides as primers. We use Pfu polymerase because its products are blunt-ended, whereas Taq polymerase sometimes adds an extra nucleotide to the 3′ end of its product, and the SP6 RNA polymerase used in the next step does not work well on substrates with 3′ protruding ends.
In this amplification step, it is likely that cDNA homoduplexes that escaped λ exonuclease treatment will be amplified. Moreover, a significant amount of genomic homoduplexes would be amplified in later steps. Therefore, we use SP6 RNA polymerase (Ambion) to create RNA transcripts of those molecules that contain the SP6 promoter sequence and digest any surviving DNA molecules with DNase I.
Finally, reverse transcription with avian myeloblastosis virus reverse transcriptase (Invitrogen) using pN-24 as primer, followed by PCR with Taq DNA polymerase and pN-24 and pR-24 as primers, yields products for cloning and analysis.
Testing the c-MYC Locus.
Our first mock experiments, not shown, used serial dilutions of the bacterial plasmid pUC18 into a cDNA library, and DNA from a 160-kbp BAC (60C5) to which was added an equimolar amount of the same plasmid. These experiments demonstrated to our satisfaction that the procedure would work well with cDNA species containing as little as 0.01% of a cDNA library. Success of the method was not evident when the pUC18 was present in cDNA at the 0.0001% level. We proceeded next with actual test cases.
In our first test case, we used an 80-kb P1 bacterial cloning vector containing the entire c-MYC gene and used randomly primed double-stranded cDNA that was prepared from the poly(A)+ RNA of the breast cancer cell line SKBr3, in which the c-MYC locus is amplified. Northern blot analysis indicated that the level of c-MYC expression from SKBr3 was in the middle of the range for a panel of breast cancer cell lines, including BT20, Du4475, HS578T, MDA134, MDA231, MDA436, SKBr3, UACC812, UACC893, ZR75–1, and ZR75–30. We estimated that c-MYC expression was 3-fold higher in SKBR3 than in normal breast tissue and 3-fold lower than in the highest expressing tumor cell line tested, ZR75–30.
Clones were prepared from the RICH products by either blunt-ended ligation to pCR-Script SK(+) or cleavage with Sau3AI followed by ligation to pUC18. Vector inserts were sequenced. All blunt-ended inserts contained both the pN-24 and pR-24 sequences and the adjoining GATC sequences, indicating that they all derived from cDNA–genomic heteroduplexes.
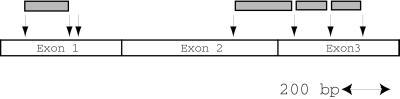
cDNA insert sizes varied from 61 to 259 bp. Roughly one-quarter of clones either had no matches in the data bases or were repeat sequences, mainly of the alu family. The remaining three-quarters had identity to sequences found on c-MYC mRNA, as indicated in Fig. 3. One type of RICH product derived from exon 1 of c-MYC entirely, one spanned exons 2 and 3, and two derived entirely from exon 3. Shorter fragments, less than 100 bp, and a larger fragment, 880 bp spanning exon 1 and 2, were not obtained in the first 81 clones examined. No clones from either the 3′ or 5′ ends of c-MYC were obtained, and this is predicted by theory because a cDNA fragment would need to have Sau3AI sites at both ends to be found.
Figure 3.
RICH products and the c-MYC gene. Arrows indicate Sau3AI recognition sites. Shadowed boxes represent RICH products, the sizes of which are 150, 250, 166, and 135 bp from the left. The large restriction fragment that we failed to isolate lies between third and forth arrows from the left and is 880 bp.
Testing the PTEN Locus.
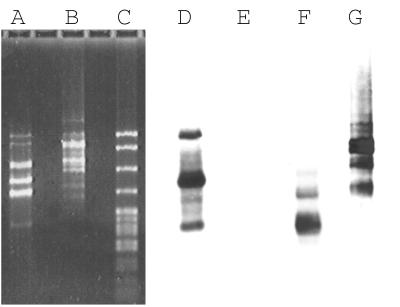
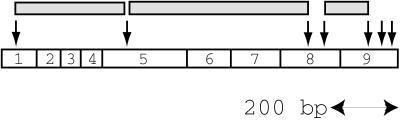
In a second test of the method, we used a 160-kb BAC, 60C5, containing PTEN, a tumor suppressor gene that encodes a mixed-specificity protein phosphatase expressed ubiquitously but at a low level in normal tissues (11–13). We prepared double-stranded cDNA from normal human breast tissue by random priming of poly(A)+ RNA. As a control we used another BAC from a different region of the genome. By following the same protocol as above, we obtained RICH products from each BAC and probed the products with PTEN cDNA by Southern blotting. Only RICH products from the PTEN BAC yielded fragments that hybridized with PTEN sequences (Fig. 4). In total at least three PTEN products were observed, about 200, 350, and 570 bp long. We expected fragment sizes of 153, 298, and 520 bp without adaptors and 201, 346, and 568 bp with adaptors, all of which span exons. We failed to observe a range of smaller fragments (Fig. 5 and see Discussion).
Figure 4.
RICH products from the PTEN gene. Lanes: A, RICH products obtained from the PTEN BAC, 60C5; B, RICH products from an unrelated BAC; C, pBR322 DNA digested with MspI. Lanes A and B were transferred to a nylon membrane and probed with PTEN cDNA (lanes D and E) or total human DNA (lanes F and G). Lanes: D and F, RICH products from the PTEN BAC; E and G, RICH products from the unrelated BAC.
Figure 5.
RICH products and the PTEN gene. Arrows indicate Sau3AI recognition sites. The shadowed boxes represent RICH products, the sizes of which are 298, 520, and 153 bp from the left. The restriction fragments in exon 8 and exon 9 that we failed to isolate are 81, 33, 27, and 29 bp from the left.
To estimate the ratio of the products that derived from PTEN cDNA to products deriving from repeat sequences, a collection of clones were analyzed by filter hybridization with both PTEN cDNA and total human DNA as probes. Six of 120 clones hybridized to PTEN cDNA and about 60 hybridized to total human genomic sequences.
To extend the utility of RICH, we tested a variation in the method: we used the PCR product from a large insert cloning vector as our source of genomic DNA. For this purpose, we gel-purified PTEN BAC and cleaved it with Sau3AI, ligated pR-24 to the 5′ ends, filled-in the 3′ ends, and used PCR to amplify fragments with pR-24 as primer. We used this amplified material in an otherwise identical protocol and obtained results that, by Southern blotting, appeared as satisfactory as when we began with restriction endonuclease-cleaved BAC DNA.
We analyzed in greater depth the RICH products of this experiment by blunt-ended cloning into pCR-Script SK(+). Ninety-six individual clones were analyzed. They were tested for the presence of inserts by PCR using primers derived from the vector cloning sites and for the presence of the Sau3AI sites by cleavage of the PCR product. By this simple test, only 17 had Sau3AI sites at the expected sites within the inserts. Sequence analysis indicated that all 17 of these contained both the pN-24 and pR-24 primers at the proper sites (this could also have been determined by PCR). Of the 17, five derived from PTEN cDNA, representing the 153-bp (three times) and the 298-bp (twice) Sau3AI fragments. The larger fragment, 520 bp long, was not obtained in this group. The remaining 12 inserts contain a Sau3AI site within human repeat sequences. Two of these contained repeats we have not yet found in the sequence of the PTEN BAC, the sequence of which is now 90% complete.
DISCUSSION
Finding genes in large chromosomal regions has been approached in three ways: exon trapping, DNA sequencing analysis, and direct hybridization selection. Exon trapping works when the gene in question contains splicing sites that are efficiently recognized by the host cell (6–8). But it fails when introns are absent or the intron–exon borders are not recognized; and exon trapping yields a background of false candidates derived from cryptic splice sites. DNA sequencing is effective when the gene in question has homology to a known expressed sequence. Even without homology, computational methods for predicting genes also have promise (4). But DNA sequencing on a massive scale is still costly. Direct hybridization selection (1) has also found use, but it diminishes in usefulness with rare messages and suffers from the vagaries of physical selection methods and background problems with repetitive sequences. We have described an additional approach to this problem: an effective protocol for selecting cDNA fragments that are homologous at one of their ends to one of the ends from a collection of genomic fragments. This method should work whenever a cDNA population is available that contains transcripts from the gene in question.
A protocol (end ligation coincident sequence cloning, EL-CSC) similar to ours, has been presented by Brookes et al. (14). Like ours, their protocol is based upon heteroduplex formation between DNA fragments made from two populations, and the use of ligation (with what they call “capture oligonucleotides”) to distinguish heteroduplex from homoduplex. Unlike our procedure, their method requires heteroduplex formation at both ends between cDNA and genomic fragments, because both ends of the heteroduplex must be ligated. Thus, cloning of cDNA fragments that span introns is much less likely. In our procedure, we use a selection adaptor that allows us to generate an RNA intermediate, and so we can isolate heteroduplexes that have formed at only one end. Moreover, EL-CSC uses physical trapping through biotin–avidin complex formation to enrich for products. We have experienced difficulty with protocols incorporating such methods and have avoided them in RICH. The report of Brookes et al. (14) does not contain sufficient information to enable us to make quantitative comparisons of our methods nor have we found their procedure used in the published literature.
Although the yield of c-MYC fragments in the RICH products was very satisfactory, the yield of PTEN products was less so. An additional prescreening of RICH products from PTEN was needed: namely, the verification of the adaptors and the Sau3AI sites that should be present upon proper priming and ligation. We believe that this is due to the reduced level of expression of the PTEN gene compared with c-MYC. Moreover, an analysis of our products from the PTEN BAC revealed a higher proportion of repeat-containing sequences than were found for c-MYC, presumably for the same reason.
Not all fragments from the same cDNA will have the same yield. For example, the 880-bp fragment from c-MYC was not obtained as a RICH product. We speculate this may be due to the difficulty of obtaining long transcripts from the SP6 polymerase. Also, we did not clone the 520-bp fragment of PTEN that was seen upon Southern blotting to be present in reduced amount. Most strikingly, we do not observe very short fragments in the RICH products. Possibly, this deficit is partially due to the slower kinetics of hybridization of shorter fragments (15). Gel fractionation before cloning might overcome some of these problems of underrepresentation. Alternatively, these limitations can be overcome by using different restriction endonucleases during the protocol.
We have introduced one variation in our method: amplifying genomic DNA fragments before use. This step was incorporated for two reasons. (i) If genomic fragments are amplified and not cleaved, it should be unnecessary to partially fill-in the ends of restriction endonuclease cleaved cDNA, because the selection adaptor cannot be ligated to PCR-amplified genomic DNA. Thus, with this variation, any restriction endonuclease can be chosen, if used both for cDNA cleavage and amplification of genomic fragments, overcoming limitations in the discovery of cDNAs that might result from the use of Sau3AI discussed above. (ii) With genomic amplification the user can initiate the search for transcripts with very small amounts of genomic DNA. In fact, we have performed RICH starting from small amounts of gel-purified BAC DNA. It may be possible to extend this method to gel-purified yeast artificial chromosome DNAs.
We cannot obtain the 3′ or 5′ ends of cDNA transcripts with RICH because only cDNA fragments with restriction endonuclease cleavage sites at both ends can be selectively amplified. However, RICH can be used in reverse (rapid isolation of genes by hybridization to transcripts, RIGHT) to identify genomic clones with homology to cDNAs. The directional modifications of genomic fragments and cDNAs used in RICH can be essentially reversed, rendering genomic fragments that form heteroduplex with cDNA the only selectable RIGHT products. When unamplified cDNAs are used, the RIGHT protocol should yield the 3′ and 5′ ends of a transcription unit. In addition, RIGHT might facilitate the confirmation of cDNAs found by RICH and aid in the determination of intron-exon boundaries.
Acknowledgments
We thank Robert Lucito for useful discussions; Eric Green, Raju Kucherlapati, and Susan Naylor for critical reading of the manuscript; Scott Powers for providing RNA; Mike Riggs for DNA sequencing; Jim Duffy for artwork; and Patricia Bird for secretarial assistance. This work was supported by grants to M.H.W. from the National Institutes of Health (OIG-CA39829 and 5P50-CA-68425); the U.S. Army (DAMD17-94-J-4247); 1 in 9: The Long Island Breast Cancer Action Coalition; and Tularik. M.H.W. is an American Cancer Society Research Professor.
ABBREVIATIONS
- RICH
rapid isolation of cDNA by hybridization
- BAC
bacterial artificial chromosome
- RIGHT
rapid isolation of genes by hybridization to transcripts
References
- 1.Lovett M, Kere J, Hinton L M. Proc Natl Acad Sci USA. 1991;88:9628–9632. doi: 10.1073/pnas.88.21.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mancini M, Sala C, Rivella S, Toniolo D. Genomics. 1996;38:149–154. doi: 10.1006/geno.1996.0610. [DOI] [PubMed] [Google Scholar]
- 3.Parimoo S, Patanjali S R, Shukla H, Chaplin D D, Weissman S M. Proc Natl Acad Sci USA. 1991;88:9623–9627. doi: 10.1073/pnas.88.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uberbacher E C, Mural R J. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M Q. Proc Natl Acad Sci USA. 1997;94:565–568. doi: 10.1073/pnas.94.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duyk G M, Kim S W, Myers R M, Cox D, R. Proc Natl Acad Sci USA. 1990;87:8995–8999. doi: 10.1073/pnas.87.22.8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckler A J, Chang D D, Graw S L, Brook J D, Haber D A, Sharp P A, Housman D E. Proc Natl Acad Sci USA. 1991;88:4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamaguchi M, Sakamoto H, Tsuruta H, Sasaki H, Muto T, Sugimura T, Terada M. Proc Natl Acad Sci USA. 1992;89:9779–9783. doi: 10.1073/pnas.89.20.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. B.4. [Google Scholar]
- 10.Straus D, Ausubel F M. Proc Natl Acad Sci USA. 1990;87:1889–1893. doi: 10.1073/pnas.87.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 12.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 13.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brookes A J, Slorach E M, Morrison E, Qureshi A J, Blake D, Davies K, Porteous D J. Hum Mol Genet. 1994;11:2011–2017. doi: 10.1093/hmg/3.11.2011. [DOI] [PubMed] [Google Scholar]
- 15.Wetmur J G, Davidson N. J Mol Biol. 1968;31:349– 370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]