Abstract
Large granular lymphocyte (LGL) leukemia is characterized by the expansion of antigen-activated cytotoxic T lymphocytes. These leukemic cells are resistant to Fas-mediated apoptosis despite expressing high levels of Fas. We found that leukemic LGL from 19 patients displayed high levels of activated STAT3. Treatment of leukemic LGL with the JAK-selective tyrosine kinase inhibitor AG-490 induced apoptosis with a corresponding decrease in STAT-DNA binding activity. Moreover, using an antisense oligonucleotide approach to diminish STAT3 expression, we found that Fas sensitivity was restored in leukemic LGL. AG-490–induced apoptosis in leukemic LGL was independent of Bcl-xL or Bcl-2 expression. However, we found that the Bcl-2–family protein Mcl-1 was significantly reduced by AG-490 treatment. Activated STAT3 was shown to bind an SIE-related element in the murine mcl-1 promoter. Using a luciferase reporter assay, we demonstrated that v-src overexpression in NIH3T3 induced STAT3-dependent transcriptional activity from the mcl-1 promoter and increased endogenous Mcl-1 protein levels. We conclude that STAT3 activation contributed to accumulation of the leukemic LGL clones. These findings suggest that investigation should focus on novel strategies targeting STAT3 in the treatment of LGL leukemia.
Introduction
Large granular lymphocyte (LGL) leukemia (CD3+) is a clonal T-cell lymphoproliferative disease with autoimmune features and chronic neutropenia (1). The disease is characterized by the expansion of CD3+, CD8+, CD57+ LGLs, with many similarities to antigen-activated cytotoxic T lymphocytes (CTLs) (1, 2). For example, characteristic of CTLs, the leukemic cells constitutively express perforin and Fas ligand (FasL) (3). After repeated antigenic stimulation, normal CTL are targeted for cell death through the Fas/FasL apoptotic pathway (FasR, CD95) (4, 5). Animal models of lymphoproliferative disorders with autoimmune features such as lpr/lpr and gld/gld mice are characterized by insensitivity to Fas-dependent cell death due to mutations in the FasR or FasL, respectively (6, 7). Our laboratory has shown that leukemic LGL are resistant to Fas-induced apoptosis despite high levels of Fas and FasL expression (8). No mutations in Fas were found in these patients. Treatment of leukemic LGLs with PHA + IL-2 or with ceramide reversed Fas resistance, indicating that the FasR and apoptotic machinery are intact.
The myeloma cell line, U266, is also resistant to Fas-mediated apoptosis despite expressing high levels of Fas (9). Recent data indicate that constitutive activation of STAT3 signaling leads to apoptotic resistance in U266 cells. STAT family proteins are latent cytoplasmic transcription factors that bind to sequence-specific transcriptional regulatory elements in the promoter of target genes (10–12). They are regulated by tyrosine phosphorylation, which leads to homodimerization or heterodimerization with other STAT proteins and translocation to the nucleus (13, 14). The Janus (JAK) family of tyrosine kinases that include JAK1, JAK2, JAK3, and TYK2 mediates tyrosine phosphorylation of STAT proteins (15–17). STAT activation is also induced by diverse oncoproteins including v-src, v-abl, v-fps, v-fes, and v-eyk (18–22). Importantly, many types of human tumors have been demonstrated to express constitutively activated STAT3, STAT5, or STAT1 proteins (23–27). The v-src transformation of NIH3T3 cells was inhibited by a STAT3-dominant interfering protein (STAT3β) that blocks transcription but not dimerization, yielding direct evidence of STAT3 involvement in cellular transformation (28–30).
In this study, we first assessed whether leukemic LGLs harbor activated STAT proteins. We discovered that STAT3 and/or STAT1 were constitutively activated in leukemic LGL. To elucidate the role of activated STAT1 and STAT3 proteins in the survival of leukemic LGL, we treated the patients’ PBMCs in vitro with AG-490, a selective inhibitor of JAK-family kinases. We also utilized STAT3 antisense oligonucleotides to inhibit STAT3 expression. Here, we demonstrate that inhibition of the JAK/STAT pathway resulted in apoptosis of CD8+ leukemic cells and reversal of Fas resistance in some leukemic cells. STAT3 transcriptional activation of an antiapoptotic protein, Bcl-xL, controlled resistance to Fas-mediated apoptosis and chemotherapeutic drug resistance in U266 cells (9). In contrast, we found that induction of apoptosis was independent of Bcl-xL regulation in LGL leukemia. Instead, AG-490 treatment resulted in reduced protein expression of another Bcl-2–family protein, Mcl-1. Here, we further establish that mcl-1, like Bcl-xL, is also a STAT3-regulated gene.
Methods
Patients.
All patients met the clinical criteria of T-cell (CD3+) LGL leukemia with increased LGL counts and clonal T-cell receptor (TCR) gene rearrangements. Leukemic LGL from all but two patients expressed TCR-αβ; these two patients (10122 and 10110) displayed TCR-γδ instead. All patients had chronic disease not requiring treatment at the time of analysis. Patient numbers were assigned after accrual into the national LGL leukemia registry initiated at the H. Lee Moffitt Cancer Center and Research Institute (www.moffitt.usf.edu/lgl-leukemia). Informed consents were signed by all patients and normal individuals to allow the use of their cells for these experiments (IRB no. 4451).
Cell lysate preparation and electrophoretic mobility shift assay.
Nuclear extracts were prepared from 2 × 107 PBMCs of 19 patients with LGL leukemia or from seven normal donors, as well as from U266 cells, as described previously (9, 19). PBMCs were separated by centrifugation on Ficoll-Hypaque (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). Some LGL leukemia cells were then quickly frozen in 10% DMSO containing RPMI and placed at –70°C before extract preparation. However, lysates prepared from frozen cells of three patients did not display any activated STAT proteins. Reanalysis using lysates prepared from freshly isolated cells in these patients demonstrated STAT activation. The loss of STAT activity with freeze thawing of cells has been described previously (24). Subsequently, all cell extracts were prepared from freshly isolated normal and patient cells. Aliquots of the extracts were stored at –80°C and used only once after thawing to prevent protein degradation.
Electrophoretic mobility shift assay (EMSA) was performed as described previously (19). To identify specific STAT family members, blocking or supershifting analyses were performed. For both STAT3 and STAT5 supershifting, antibodies were used. In such experiments, the presence of STAT3, for example, is indicated by decreased intensity of the original STAT3 complexes and by the appearance of a new higher band (supershift). For STAT1, a blocking antibody was used. In this case, identification of STAT1 is shown by elimination of the STAT1 complex. For these experiments, 1 μl of rabbit polyclonal anti-STAT1 blocking antibody (clone C136), STAT3 supershifting (clone H190), or STAT5B supershifting antibody (clone C17) (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) was added to the incubation reaction for 30 minutes at room temperature in a 10-μl reaction volume. Oligonucleotide probes (2 pmol) containing the hSIE sequence (5′-CTTCATTTCCCGTAAATCCCTA), Mcl-1 SIE (–103 to –81) (5′-TTTCCCCTTTTACGGGAAGTCCT), MGFe sequence (5′-AGATTTCTAGGAATTCAA-3′), and FIRE (5′-GTCCCCCGGCCGGGGAG GCGCT-3′) (all from Life Technologies Inc., Gaithersburg, Maryland, USA) were labeled with 32P-dATP and dCTP. The degree of binding was determined by electrophoretic analysis on a 5% native polyacrylamide gel as described previously (19, 31).
Drug and antisense oligonucleotide treatment.
Purified PBMCs and leukemic LGLs were placed in 24-well plates at a concentration of 2 × 106 per 0.5 ml of complete medium (RPMI-1640 medium supplemented with 10% fetal bovine, 2 mM L-glutamine, 10 U/ml penicillin, and 100 μg/ml streptomycin, all from Life Technologies Inc.). The U266 multiple myeloma cell line was obtained from the American Type Culture Collection (Rockville, Maryland, USA) maintained in complete medium, and placed at a concentration of 1 × 106 cells per milliliter in 24-well plates. Normal PBMCs were also activated with 1 μg/ml of phytohemagglutinin (PHA; Sigma Chemical Co., St. Louis, Missouri, USA) and IL-2 (500 U/ml) for 7–10 days to induce Fas sensitivity (8). These cells displayed CD25 in 98–100 % of the cells by FACS analysis at the time of apoptosis analysis (data not shown). All cells were treated with AG-490 at the indicated concentration or an equal volume of drug solvent (vehicle control), dimethyl sulfoxide (DMSO; Sigma Chemical Co.). Incubation proceeded at 37°C for 24 hours, and cells were retreated for an additional 18 hours with or without anti-Fas antibody, as described previously (32). To determine Fas sensitivity, 1 μg/ml of anti-Fas mAb (CH11; Kamiya Biomedical Corp., Seattle, Washington, USA), or in some experiments purified mouse IgM control protein (PharMingen, San Diego, California, USA), was added. Antisense STAT3 (5′-ACT CAA ACT GCC CTC CTG CT-3′) or control oligonucleotide (5′-TCT GGC AAA GTG TCA GTA TG-3′) (Isis Pharmaceuticals Inc., Carlsbad, California, USA) was added at the concentrations indicated. Oligonucleotides were synthesized using phosphorothioate chemistry, and 2′-O-methoxyethyl modification of the five terminal nucleotides (underlined) was used to increase stability, as described previously (33). After 6 hours without sera, 5% FBS and CH11 were added. Incubation continued for an additional 18 hours before collection for extract preparation and apoptosis analysis. For experiments with the caspase inhibitor (Ac-DEVD-fmk; Calbiochem-Novabiochem, San Diego, California, USA), cells were pretreated for 4 hours before the addition of AG-490.
Apoptosis assay.
To measure apoptosis, we performed two-color flow cytometry with 7-amino-actinomycin D (7-AAD; Calbiochem-Novabiochem) and annexin-V-FITC (5 μl; CLONTECH Laboratories Inc., Palo Alto, California, USA). Data acquisition and analysis were performed by the Flow Cytometry Core Facility at the H. Lee Moffitt Cancer Center. The peak emission for 7-AAD is approximately 685 nm, and a FL3 photomultiplier tube was used with a 670-nm-long pass filter. No distinction was made between intermediate and late apoptosis. Any 7-AAD single positive stained cells were considered necrotic and excluded from the calculation for apoptosis. Forward and side scatter analyses, as well as surface staining with anti-CD45 (data not shown), were used to identify the lymphocyte population in peripheral blood samples from leukemic LGLs and normal PBMCs. This lymphocyte gate was then used to examine the population that stained positive for annexin-V-FITC, and the percent specific apoptosis was calculated using the following formula:
[(% annexin-V-FITC positive in the assay well) – (% annexin-V–FITC positive in the control well) × 100] ÷ [100 – (% annexin-V-FITC positive in the control well)]
For three-color flow cytometry, leukemic LGLs were first stained with 5 μl of anti–CD8-PE (Coulter Corp., Miami, Florida, USA) for 30 minutes on ice in the dark, washed two times in 1 × PBS and stained with 7-AAD and annexin-V-FITC.
Western immunoblotting.
Cells were lysed in a buffer composed of 50 mM Tris-Cl (pH 7.6); 5 mM EDTA; 150 mM NaCl; 0.5% NP-40; 0.5% Triton-X-100 (RIPA) containing 1 μg/ml leupeptin, aprotinin, and antipain; 1 mM sodium orthovanadate; and 0.5 mM PMSF (all from Sigma Chemical Co.). Protein concentration was determined in all cell extracts using the Bradford assay (Bio-Rad Laboratories, Hercules, California, USA). Unless otherwise indicated, 25 μg of total protein was loaded per lane in Laemmli SDS-PAGE sample loading buffer and boiled for 5 minutes before separation by 10% SDS-PAGE. Twenty-five micrograms of total protein was found to give ECL signals within the linear range for β-actin in titration experiments. The proteins were then transferred to a membrane for Western blot analysis. Antibodies were obtained from the following sources and used at the dilutions recommended by the manufacturer: Bcl-2, Mcl-1, STAT1, and Bcl-xL antibodies, 1:1,000 dilution (Santa Cruz Biotechnology Inc.), anti-STAT3, 1:2,000 (Santa Cruz Biotechnology Inc.), and anti–β-actin 1:2,500 (Sigma Chemical Co.). Antibody detection was performed by standard ECL techniques as recommended by the manufacturer (Amersham Pharmacia Biotech). Protein quantification was performed by densitometry using the ImageQuant program (Molecular Dynamics, Sunnyvale, California, USA).
Transient transfection assay.
A luciferase reporter (p-203/+10mcl-1-luc) driven by the murine mcl-1 promoter region (–203 to +10) and an identical reporter with a mutated SIE site (p-203/+10mS) have been described previously (34, 35). The pLucSRE-luciferase was included as a reporter control. Transfection of the indicated plasmids into NIH3T3 cells was performed using CaP04, as described previously (28). pM-v-src and pSG5-STAT3β are plasmid vectors expressing v-src and a dominant negative mutant of STAT3, respectively. Control transfections were also performed using pSG5 to assess whether nonspecific repression occurred as a result of the vector alone. Transfection efficiency was normalized by expression of a CMV–β-galactosidase reporter gene.
Statistical analysis.
Statistical analysis was performed by A. Cantor (Biostatistic Core, H. Lee Moffitt Cancer Center) using the SAS statistical software program (SAS Institute Inc., Cary, North Carolina, USA).
Results
Leukemic LGLs display constitutively activated STAT3 and/or STAT1.
EMSA using the oligonucleotide probe containing a high-affinity mutant of the STAT-specific DNA sequence from the hSIE is used to detect homodimers and heterodimers of STAT3 and STAT1 (10). Nuclear extracts were prepared from the PBMCs of 19 LGL leukemia patients. The diagnosis was confirmed on all patients by TCR gene rearrangement studies, and the leukemic cells constituted 70–90% of lymphocytes by flow cytometry. We found that nuclear extracts from all 19 patients contained DNA-binding complexes that recognized the hSIE probe (results from 12 patients are shown in Figure 1a). In agreement with previously reported data, little or no SIE-DNA–binding activity was detected with nuclear extracts derived from five normal donors (Figure 1a, lanes 1–5) (36). Normal PBMCs treated for 7 days with PHA + IL-2 (activated normal), however, contained robust SIE-binding activity (Figure 1a, lane 6). Western blot analysis of STAT3 protein expression was also examined in leukemic LGLs from each sample and in the normal PBMCs (Figure 1a, top). In addition to the increased STAT3 DNA-binding activity observed in leukemic LGLs by EMSA, we found that the amount of STAT3 protein was higher in leukemic LGLs and activated normal PBMCs compared with the amount in unstimulated normal PBMCs, suggesting continued activation in leukemic cells (Figure 1, a and c).
Figure 1.
Activation of STAT3 and STAT1 in leukemic LGLs. (a) EMSA with nuclear extracts from PBMCs of normal donors (NL), from normal donors activated for 7 days with PHA + IL-2 (NL-AC), and from patients with LGL leukemia (LGL). Cytoplasmic extracts from each sample were run on an SDS-PAGE gel and blotted for expression of STAT3. (b) EMSA supershift or blocking experiments with untreated (lanes 1, 4, 7, 10, and 13), anti-STAT1–preincubated (lanes 2, 5, 8, 11, and 14), and anti-STAT3–preincubated (lanes 3, 6, 9, 12, and 15) reaction mixtures. (c) Western blot analysis of whole-cell extracts for phosphorylated STAT3, total STAT3, and β-actin. (d) EMSA for STAT5-DNA–binding activity with activated normal and leukemic LGLs (10024-AC) used as a positive control. The relative migration of STAT3:3 homodimers (ST3:3), STAT1:3 heterodimers (ST1:3), and STAT1:1 homodimers (ST1:1) is shown in a and b. (d) STAT5:5 homodimers (ST5:5) and STAT1:1 homodimers (ST1:1) are indicated. WB, Western blot.
To identify the STAT family members bound to the SIE probe in leukemic LGL, we performed blocking or supershift analyses with anti-STAT1 and 3 specific antibodies, respectively. Supershift data for five patients are shown in Figure 1b and demonstrate that the majority of the SIE-binding activity consisted of STAT3:3 homodimers and to a lesser extent STAT1:1 homodimers and STAT1:3 heterodimers. An anti-STAT1 blocking antibody completely eliminated the complex observed in extracts from one patient with LGL leukemia (no. 10104), suggesting the presence of activated STAT1:1 homodimers (Figure 1b, lanes 4–6). This patient’s cells displayed a slightly different phenotype, being double CD4/CD8+ rather than the usual CD8+/CD4– seen in leukemic LGLs from all other patients.
The dimerization and DNA binding activity of STAT3 are dependent on tyrosine phosphorylation (14). The phosphorylation status of STAT3 was examined by Western blot analysis using a phosphorylation-specific STAT3 antibody (Figure 1c). We found that leukemic LGL from seven patients, PHA+IL-2–activated PBMCs, and U266 cells displayed varying amounts of constitutively phosphorylated STAT3. In contrast, normal unactivated PBMCs displayed no detectable constitutively phosphorylated STAT3. These data further confirm the EMSA results suggesting that constitutively activated STAT3 is present in leukemic LGLs.
STAT5-DNA binding is induced after TCR stimulation of normal T lymphocytes (37). Because leukemic LGLs share many characteristics of activated T cells, we examined STAT5 DNA binding activity using an oligonucleotide probe containing the mammary gland factor element (MGFe) that recognizes STAT5 and STAT1 homodimers (31). We found that treatment with IL-2 + PHA resulted in strong activation of STAT5 in both normal activated PBMCs and inactivated leukemic LGLs (Figure 1d) but that constitutive STAT5 activity was detected in leukemic LGLs from only two of 12 patients (data shown for eight patients).
JAK-family kinase inhibitor induces apoptosis in leukemic LGL.
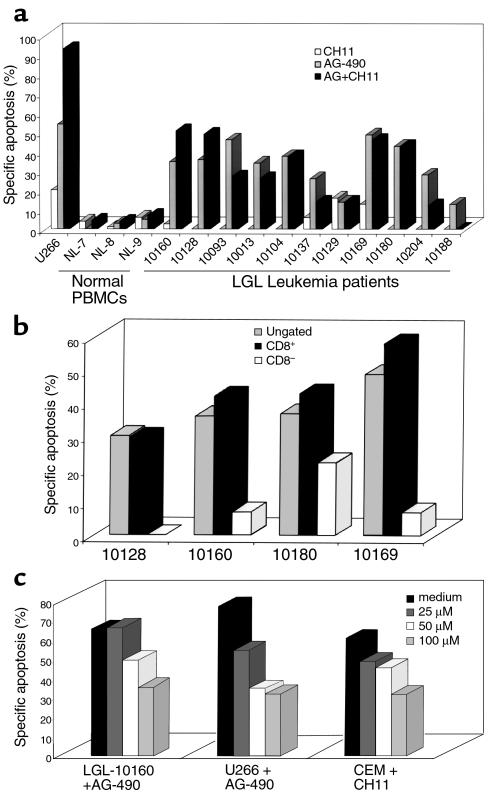
We previously demonstrated that leukemic LGLs display resistance to Fas-mediated apoptosis (8). The multiple myeloma cell line U266 expressed constitutively activated STAT3 and demonstrated Fas resistance that was reversed by the addition of a selective JAK inhibitor, AG-490 (9). We first examined the apoptotic inducing effects of AG-490 on leukemic LGLs (patient 10160), normal unactivated PBMCs, normal PHA+IL-2–activated PBMCs, and U266 cells. Normal unactivated PBMCs, leukemic LGLs, and U266 cells displayed inherent resistance to anti-Fas–mediated apoptosis (Figure 2a). AG-490 alone induced an increase in annexin-V-FITC binding in leukemic LGLs (patient 10160) after 48 hours (Figure 2a, row 1). The combination of CH11 and AG-490 further enhanced apoptosis in leukemic LGL from patient 10160, in normal activated PBMCs, and in U266 cells. In contrast to leukemic LGLs, normal unactivated PBMCs displayed no increase in apoptosis in response to either AG-490 alone or in combination with the anti-Fas mAb. Figure 2b demonstrates that increasing doses of AG-490 induced a dose-dependent increase in the percent specific apoptosis in leukemic LGLs but had little effect on normal PBMCs (Figure 2b). The 50 μM dose of AG-490 was then subsequently chosen for the remaining experiments owing to the greater differential effect between leukemic LGLs and normal PBMCs.
Figure 2.
AG-490 induction of apoptosis in leukemic LGLs. (a) Analysis of apoptosis was assessed by annexin-V-FITC and 7-AAD binding in PBMCs isolated from a patient with LGL leukemia (no. 10160; row 1), from a normal unactivated donor (NL-7; row 2), from activated normal PBMCs (NL-7AC; row 3) or from the U266 cell line (row 4). Cells were treated with DMSO + mouse IgM (drug solvent; column 1), anti-Fas agonistic antibody (CH11; column 2), 50 μM of AG-490 + mouse IgM (column 3), or AG-490 in combination with CH11 (column 4). The average percent of annexin-V-FTIC–positive cells and SD of two separate experiments is shown in the upper right corner of the histogram. (b) Annexin-V-FITC binding of leukemic LGL and normal PBMCs in response to DMSO (0) and AG-490 (25, 50, and 100 μM for 48 hours). The percent specific apoptosis is indicative of three separate experiments using the same patient. AStatistically significant; P < 0.05 using a Student’s t test. P value of normal unactivated PBMCs with 100 μM approached significance (P = 0.06).
We then examined leukemic LGL from 11 patients and PBMCs from three normal donors to determine whether AG-490 treatment consistently induced apoptosis and Fas sensitivity. We found that AG-490 treatment induced apoptosis in leukemic LGL from all 11 patients tested, in contrast to results with normal unactivated PBMCs (Figure 3a). The addition of anti-Fas agonistic mAb CH11, however, produced variable results. Similar to the results in U266 cells, leukemic LGLs from patients 10160 and 10128 displayed reversal of Fas resistance, as demonstrated by an additive induction of apoptosis after both AG-490 and CH11 treatment. Leukemic LGLs from the remaining nine patients demonstrated no effect or a decrease in apoptotic cells after the combination of AG-490 and CH11.
Figure 3.
AG-490 consistently induced annexin-V-FITC binding in leukemic LGLs but not sensitization of Fas. (a) The percent specific apoptosis was calculated for normal PBMCs (NL), leukemic LGLs (patient number), and U266 cells after treatment with DMSO. Open bars, anti-Fas (CH11); gray bars, 50 μM AG-490 (AG); filled bars, AG-490 with CH11. These results were representative of at least two experiments for each patient, and the assay was performed in duplicate. The only exception was patient 10129, for whom the procedure was done only once because therapy was soon initiated. AG-490–induced apoptosis in leukemic LGLs was statistically significant with P < 0.05 by ANOVA. (b) Three-color FACS analyses were performed on PBMCs from three patients with LGL leukemia. The patient’s cells were first stained with an isotype control antibody-PE or anti–CD8-PE before staining with annexin-V FITC and 7-AAD. A live gate was used to collect either the CD8+ cells or the CD8– cells, and then apoptosis was assessed (c). AG-490 was added to leukemic LGLs (patient 10160), and U266 cells or CH11 was added to CEM after pretreatment with medium (filled bars), or the caspase-3 inhibitor Ac-DEVD-fmk at doses of 25 μM (dark gray bars), 50 μM (open bars), and 100 μM (light gray bars). The data are shown as percent specific apoptosis and were performed in duplicate.
Approximately 70% of the circulating PBMCs in most patients with leukemic LGLs are CD8+ T lymphocytes as determined by flow cytometry (data not shown). To determine whether only the leukemic LGL cells (CD8+) undergo apoptosis and not the nonleukemic (CD8–) cells, we performed three-color flow cytometry analyses with annexin-V-FITC, 7-AAD, and a live gate to collect either CD8+ or CD8– cells. We found that AG-490–induced apoptosis in all cells (isotype control) was similar to that of CD8+-enriched leukemic population in all four patients examined, with little or no apoptosis induced in the CD8– population (Figure 3b). These results indicate that only the leukemic cells, and not nonleukemic cells, are sensitive to the apoptosis-inducing effects of AG-490.
Cleavage and activation of caspase proteases actuate the death machinery involved in apoptosis (38, 39). The cell-permeable competitive peptide inhibitor Ac-DEVD-fmk blocks the activity predominantly of caspase-3 (CPP32, YAMA). We added AG-490 to leukemic LGLs and U266 cells in the presence of increasing doses of the caspase-3 inhibitor. There was a dose-dependent decrease in AG-490–mediated apoptosis in both U266 and leukemic LGLs in response to coincubation with Ac-DEVD-fmk (Figure 3c). Incubation of a T-cell lymphoblastic leukemia cell line (CEM) with CH11 and Ac-DEVD-fmk also resulted in a considerable decrease in anti-Fas–mediated apoptosis. These results demonstrate that AG-490–dependent cell death is mediated by activation of caspase-3 that is indicative of apoptosis and not necrosis. Interestingly, Fas-dependent and AG-490–dependent apoptosis converge at the activation of the effector caspases.
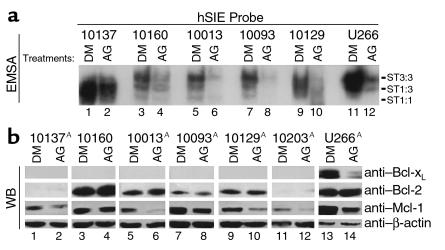
Effects of AG-490 on STAT3 DNA-binding activity and expression of antiapoptotic proteins in leukemic LGLs.
Nuclear extracts were prepared from leukemic LGLs of five patients after incubation for 48 hours in medium containing DMSO or AG-490. STAT-DNA binding activity using the hSIE probe was then determined by EMSA. Leukemic LGLs from all of these patients were sensitive to AG-490–mediated apoptosis (Figure 2a). There was a decrease in both STAT3 and STAT1 DNA-binding activity in response to AG-490 treatment in extracts from all five patients and from control U266 cells (Figure 4a).
Figure 4.
AG-490–reduced DNA-binding activity of STAT3 and expression of Mcl-1. (a) EMSA with leukemic LGLs (lanes 1–10) or U266 (lanes 11 and 12) treated with DMSO (DM; lanes 1, 3, 5, 7, 9, and 11) or AG-490 (lanes 2, 4, 6, 8, 10, and 12). The relative position of STAT3:3 homodimers (ST3:3), STAT1:3 heterodimers (ST1:3), and STAT1:1 homodimers (ST1:1) is indicated. The results are shown for five patients with LGL leukemia and are representative of three separate experiments. A statistically significant decrease in EMSA binding occurred in all AG-490–treated cells tested compared with DMSO (P < 0.05 using Student’s t test.) (b) Western blot analysis for expression of Bcl-xL, Bcl-2, Mcl-1, and β-actin. Ten micrograms of total protein was used for U266 cells and 25 μg for leukemic LGLs. Samples were analyzed on 10% SDS-PAGE gel. The results shown are representative of three separate experiments. AStatistically significant decrease in Mcl-1 expression after AG-490 treatment; P < 0.05 using Student’s t test.
In multiple-myeloma cells (U266 is an example), an IL-6–dependent autocrine or paracrine loop acts to stimulate the expression of the antiapoptotic protein Bcl-xL (9, 40, 41). STAT3 and STAT1 control bcl-x transcription (9, 42). Therefore, we examined the levels of Bcl-2–family antiapoptotic proteins by Western blot analysis. No Bcl-xL was observed in leukemic LGLs from six patients (Figure 4b). As described previously, the level of Bcl-xL decreased in AG-490–treated U266 cells (Figure 4b, top row, lanes 13 and 14) (9). Because of the strong relationship between STAT3-mediated Bcl-xL regulation, we wanted to examine further the role of Bcl-xL by RNase protection assay. We found that Bcl-xL mRNA was barely detectable in leukemic LGLs (data not shown). Bcl-2 mRNA and protein expression was unchanged by AG-490 treatment of leukemic LGLs and U266 cells (Figure 4b, second row; RNase protection data not shown).
AG-490 treatment downregulates Mcl-1 protein expression.
In contrast to Bcl-2 and β-actin expression, we observed a decrease in Mcl-1 protein expression after AG-490 treatment in U266 cells and in five of seven samples of leukemic LGLs (decreased expression was in the range of 55–98% after normalization by densitometry). Leukemic LGLs from two patients demonstrated no reduction in Mcl-1 protein expression in response to AG-490 (no. 10160, Figure 4b; and no. 10128, data not shown).
STAT3 binds a promoter element in the murine mcl-1 promoter.
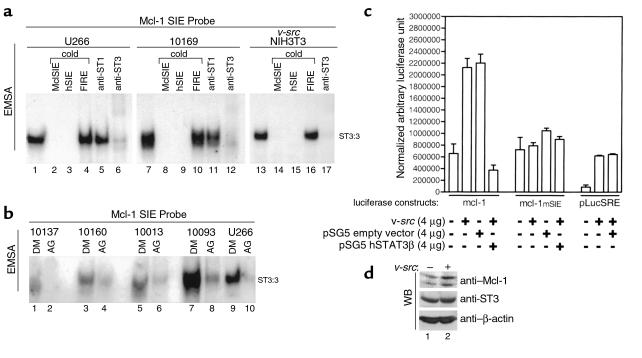
Decreased levels of Mcl-1 in the presence of AG-490 suggest that STATs may transcriptionally regulate the expression of Mcl-1. An SIE-like binding site has been identified in the mouse mcl-1 promoter and recently shown to contribute to IL-3-induced mcl-1 gene expression in IL-3-dependent Ba/F3 (34, 35). The STAT proteins that bound this element were not identified. To examine whether STAT3 could bind to the SIE element located at –103 to –87 of the mcl-1 promoter, we performed EMSA assays utilizing a synthetic oligonucleotide probe corresponding to this region and nuclear extracts known to contain activated STAT3; namely, those from U266, leukemic LGLs, and v-src–transformed NIH3T3 (9, 19). We found that a protein complex bound to this probe in all three extracts. Excess cold oligonucleotide competition (100×) with either the Mcl-1 SIE-DNA or authentic hSIE completely eliminated this binding activity (Figure 5a, lanes 2 and 3, 8 and 9, and 14 and 15). In contrast, the nonspecific FIRE sequence failed to eliminate the Mcl-1 SIE-DNA–binding activity. These results suggest that the Mcl-1 SIE-DNA–binding activity contained a STAT3 or STAT1 protein. We then performed supershift analysis with either anti-STAT1 or anti-STAT3 antibodies to positively identify the proteins contained within the DNA-binding complex. In U266 cells, leukemic LGLs, and v-src–transformed NIH3T3, supershift or elimination of the complex was observed with only the STAT3 antibody. These data demonstrate that STAT3 binds to the SIE-element within the murine mcl-1 promoter.
Figure 5.
STAT3 DNA-binding to an SIE-like element in the Mcl-1 promoter and transcription regulation. (a) EMSA was performed using nuclear extracts from U266 (lanes 1–6), leukemic LGLs (patient 10169; lanes 7–12), and v-src–transformed NIH3T3 (lanes 13–17) with the Mcl-1 SIE probe labeled with P32. Competition assays were performed by adding excess cold oligonucleotide (×100) for MclSIE (lanes 2, 8, and 14), hSIE (lanes 3, 9, and 15), and nonspecific competitor (FIRE, lanes 4, 10, and 16). Blocking or supershift analysis was performed by adding anti-STAT1 antibody (lanes 5 and 11) or anti-STAT3 antibody (lanes 6, 12, and 17), respectively. (b) EMSA analysis with mcl-1 SIE using nuclear extracts of leukemic LGLs (lanes 1–8) or U266 (lanes 9 and 10) treated with solvent control (DMSO, lanes 1, 3, 5, 7, and 9) or AG-490 (lanes 2, 4, 6, 8, and 10). Results shown are representative of two experiments. (c) Transcriptional analysis was performed by transfection of NIH3T3 with 4 μg of pGL2 mcl-1 (p-203/+10mcl-luc), pGL2 mSIE mcl-1 (p-203/+10mS), and pLucSRE-luciferase alone or in combination with 4 μg v-src, pSG5-STAT3β, and pSG5 vector control expression vectors. The values shown are normalized by cotransfection with CMV-β–galactosidase expression vectors. (d) Western blot analysis in NIH3T3 without (–) or with (+) v-src overexpression for Mcl-1, STAT3, and β-actin protein expression.
To examine whether nuclear extracts from leukemic LGL consistently bound the Mcl-1 SIE element, we performed EMSA assays with nuclear extracts from DMSO and AG-490–treated leukemic LGL. Mcl-1-SIE-DNA–binding activity was present in all extracts examined and was significantly reduced by AG-490 treatment (Figure 5b).
Mcl-1 gene regulation is induced by v-src expression.
To determine whether the murine mcl-1 gene is transcriptionally activated by STAT3, a transcriptional-reporter assay was performed with the murine mcl-1 promoter fused to a luciferase reporter gene by transient transfection, as described previously. (28, 34, 35). We found that the mcl-1 promoter was induced by coexpression of v-src in NIH3T3 (Figure 5c). Inducible expression was completely abolished by cotransfection of a dominant-negative pSG5-STAT3 (STAT3β), suggesting that induction of the mcl-1 promoter by v-src is STAT3-dependent. Control experiments with the pSG5 empty vector displayed no inhibition of reporter activity. An mcl-1 promoter construct harboring a point mutation in the SIE-binding element was used to confirm these results further (35). Figure 5c shows that an intact SIE-binding element is necessary for v-src inducibility. In contrast to results with the wild-type mcl-1 promoter, STAT3β displayed no reduction of mcl-1 basal transcriptional activity of the mutant reporter. As a control, we performed experiments with the pLucSRE-luciferase identified previously as v-src–inducible by a STAT3-independent mechanism. As demonstrated previously, the pLucSRE was inducible by v-src overexpression, but no inhibition was observed by coexpression of STAT3β (28). We concluded from these data that the mcl-1 gene is regulated by a STAT3 element in the murine mcl-1 promoter and that STAT3β overexpression does not interfere nonspecifically with v-src–induced promoter activity.
Endogenous levels of Mcl-1 protein induced by v-src overexpression.
The inducibility of Mcl-1 protein by v-src was examined in NIH3T3 without and with v-src overexpression. In comparison to STAT3 and β-actin, Mcl-1 expression was enhanced by v-src (Figure 5d). These data corroborate the results observed with in vitro reporter assays using the mcl-1-luciferase construct.
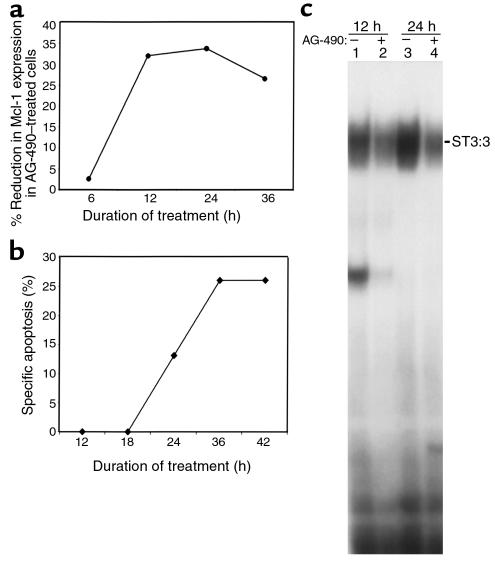
Kinetic analysis of AG-490 treatment.
Kinetic analysis was performed to determine whether the reduction in Mcl-1 protein expression and STAT3 activity preceded the induction of apoptosis induced by AG-490 in leukemic LGLs. Western blot analysis of Mcl-1, Bcl-2, and β-actin in DMSO and AG-490–treated leukemic LGLs was performed on extracts collected after 6, 12, 24, and 36 hours’ incubation. At each time point, cells were also studied for apoptosis. Nuclear extracts were prepared only after 12 and 24 hours owing to the limited number of cells available. We found that the expression of Mcl-1 in relation to β-actin had decreased in AG-490–treated cells by 12 hours (Figure 6a). Likewise, the reduction in STAT3 activation by EMSA analysis was also apparent after 12 hours (Figure 6c). Importantly, drug-induced apoptosis was not induced until 24 hours (Figure 6b). There was a rapid AG-490–mediated decline in Mcl-1 protein expression in leukemic LGLs. Rapid degradation of Mcl-1 has been previously linked to the presence of a PEST sequence in the protein (34). These data are suggestive but not conclusive for a role of Mcl-1 in AG-490–mediated apoptosis in leukemic LGLs.
Figure 6.
Kinetic analysis with AG-490 treatment. Leukemic LGLs were treated with DMSO or AG-490 for 6, 12, 24, and 36 hours. (a) Graphic representation of the percent reduction in Mcl-1 expression in AG-490–treated cells in comparison with DMSO-treated cells. Mcl-1 protein expression was normalized to β-actin by densitometry using the ImageQuant program (Molecular Dynamics). (b) Degree of annexin-V-FITC–positive binding with percent specific apoptosis, calculated as described in Methods, in leukemic LGLs treated for 12, 18, 24, 36, and 42 hours in DMSO and AG-490. (c) EMSA for STAT3 activity with DMSO-treated (–) or AG-490–treated (+) cells after 12 and 24 hours. Data shown are representative of results in two patients. The apoptosis assay was performed in duplicate in each patient.
Antisense STAT3 reverses Fas resistance in leukemic LGLs.
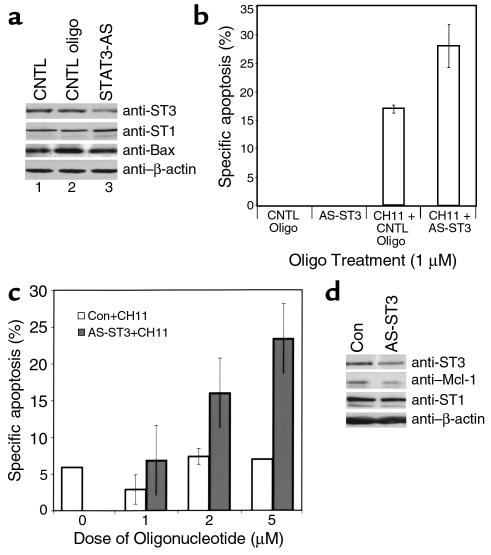
Because of the possibility that AG-490 impacts multiple signaling pathways mediated by JAK-tyrosine kinases, we used an antisense approach to reduce STAT3 levels (33). Figure 7a demonstrates that extracts obtained from leukemic LGLs treated with antisense STAT3 (AS-ST3) contained lower levels of STAT3 protein in comparison with cells incubated with medium and control oligonucleotide. These results were reproducible in experiments using leukemic LGLs from each of three patients, with the percent of reduction in STAT3 protein in the range of 25–45%. Sensitivity to Fas-mediated apoptosis was significantly increased in the AS-STAT3–treated cells (Figure 7b), an effect that was dose dependent (Figure 7c). We also found that Mcl-1 protein expression was decreased in response to AS-STAT3 (Figure 7d), supporting a direct role for STAT3 in Mcl-1 signaling in leukemic LGLs. To assess the specificity of the antisense for STAT3, STAT1 protein levels were also determined. In contrast to the STAT3 reduction, there was no effect of oligonucleotide treatment on STAT1 expression (Figure 7, a and d).
Figure 7.
Sensitivity to Fas and reduced expression of Mcl-1 in leukemic LGLs treated with antisense STAT3. Leukemic LGLs (patient 10160) were incubated with medium (CNTL), 1 μM control oligonucleotide (CNTL oligo), or 1 μM antisense STAT3 oligonucleotide. (a) Western blot analysis for STAT3, STAT1, Bax, and β-actin expression. (b) Annexin-V-FITC binding was determined in the presence (CH11+) or absence of agonistic anti-Fas antibody. Data shown are calculations of percent specific apoptosis of the average results obtained in duplicate reactions. (c) Specific apoptosis calculated for control (open bars) or antisense STAT3 oligonucleotide (filled bars) in the absence (0) or presence of 1, 2, and 5 μM oligonucleotides (patient 10205). (d) Western blot analysis in the presence of 2 μM control oligonucleotide (Con) or antisense STAT3 oligonucleotide (AS-ST3) for expression of STAT3, Mcl-1, STAT1, and β-actin protein.
Discussion
Here we report that leukemic LGL from all 19 patients tested constitutively expressed high levels of activated STAT3 and/or STAT1. Unlike normal TCR-stimulated T cells, STAT5 was distinctly absent in leukemic LGLs from all but two patients examined (36, 43, 44). IL-2 is not expressed constitutively or after anti-CD3 stimulation in leukemic LGLs (8, 45–47). However, the leukemic cells do express the intermediate affinity IL-2 receptor containing the β and γ subunits and become sensitive to Fas-mediated apoptosis after treatment with high concentrations of IL-2 in vitro, indicating that the signaling pathway mediated by IL-2 is intact (45–47). IL-2–deficient mice share some features in common with LGL leukemia, such as splenomegaly and autoantibody formation (44). Also similar to LGL leukemia, the IL-2–/– cells are phenotypically characteristic of antigen activated T cells but are resistant to Fas-mediated apoptosis despite expression of Fas (43). The lack of IL-2 production and the absence of STAT5 activation may prove to be an important factor in the growth and survival of leukemic LGLs.
We hypothesized that the dysregulated STAT activation was involved in cell survival. Therefore, we used a tyrphostin inhibitor (AG-490) selective for the JAK-family tyrosine kinases to determine whether inhibition of the pathway upstream of STAT activation would affect apoptosis in LGL leukemia cells (32). We found that PBMCs from these patients underwent apoptosis in response to AG-490 treatment, suggesting that a STAT3-regulated pathway may be involved in cell survival. However, reversal of the Fas-resistant phenotype was observed in leukemic LGLs from only two of ten patients. These results indicate that biochemical differences exist in the mechanism for cell survival and Fas resistance in leukemic LGLs. Proteolytic cleavage and activation of a cascade of caspases (death proteases) mediate the cleavage of cellular targets, resulting in programmed cell death (38, 39). To ascertain the role of the effector caspases in AG-490–mediated apoptosis, we performed experiments with the inhibitor Ac-DEVD-fmk. We found that the caspase inhibitor blocked AG-490–induced apoptosis in a dose-dependent fashion, in both leukemic LGLs and U266, as well as in CH11-treated CEMs. These data show that AG-490–induced death results from apoptosis rather than nonspecific activation of necrosis and that AG-490 and Fas-mediated cell death converge at the activation of the effector caspases.
The signaling pathway resulting in STAT3 activation in leukemic LGLs is not known. In myeloma, STAT activation has been attributed to an IL-6 feedback loop. Possible mechanisms in leukemic LGLs could include the following: (a) a survival-promoting autocrine or paracrine cytokine pathway; (b) dysregulated STAT activation due to the absence of IL-2 production; and (c) activation by retroviral infection. The sera of approximately 50% of LGL leukemia patients are reactive to an HTLV envelope protein, suggesting infection with an HTLV-related virus (48, 49).
STAT3 was found to positively regulate the transcription of the antiapoptotic protein Bcl-xL and control the IL-6–dependent survival of U266 cells (9). However, we failed to detect Bcl-xL protein in leukemic LGL. Our data suggested that a Bcl-xL–independent pathway was involved in the AG-490–induced cell death observed in leukemic LGLs. We then evaluated other members of the Bcl-2–family proteins as possible candidates. The mcl-1 gene was recently demonstrated to be controlled by both PI3-K–mediated CRE-2 activation and PI3-K–independent activation of an SIE (STAT3-like) sequence in response to IL-3 and GM-CSF (34, 35). It was also shown that elevation in Mcl-1 protein expression correlated with relapse in acute leukemias (50). Mcl-1, when overexpressed, can inhibit cell death induced by multiple apoptotic stimuli, suggesting that it plays a role in cell survival (51–53). We carefully examined whether STAT3 was capable of transcriptionally regulating the murine mcl-1 promoter in v-src–transformed NIH3T3. We demonstrated that the SIE-like element in the mcl-1 promoter was STAT3 responsive and v-src inducibility was dependent on this site. A correlation between transcriptional regulation of the human and murine mcl-1 promoter can not be made at this time because the human genomic sequence for mcl-1 is yet to be cloned. The mechanism of Mcl-1 antiapoptotic activity has not been established, but the protein binds to proapoptotic Bcl-2–family members Bax-α, hypophosphorylated BAD, Bak, Bok, Bik, and BOD (54–56). The ratio of anti- and proapoptotic proteins determines cell survival. Therefore, the reduction in Mcl-1 protein by inhibition of STAT signaling may contribute to apoptotic induction in leukemic LGLs. Further experiments are needed to define more clearly the role of Mcl-1 in abnormal cell survival. Because Mcl-1 has antiapoptotic activity, the regulation of Mcl-1 gene expression by oncogenic v-src–induced STAT3 has potentially important implications in mechanisms of tumorigenesis.
These studies provide an important demonstration that an AG-490–inhibitable pathway may contribute to the survival of primary tumor cells such as leukemia samples as well as malignant cell lines. Cell survival by the JAK/STAT pathway appears complex, involving control of antiapoptotic proteins and possibly other unidentified mechanisms. Recent data has demonstrated that AG-490 inhibits not only the JAK-STAT pathway but also mitogen-activated-protein kinase (MAPK), another well-known pathway involved in cell survival and proliferation (57). In leukemic LGLs, enhanced apoptosis after anti-Fas ligation was observed in cells treated with antisense STAT3. These data show that STAT3 activation contributes to Fas resistance and implicate this signaling pathway in abnormal survival of leukemic LGL. Results of these studies identify the STAT3 signaling pathway as molecular targets for drug discovery in LGL leukemia and possibly other chronic lymphoproliferative diseases (58).
Acknowledgments
This investigation was supported by a grant from the Veterans Administration, an institutional research grant from the American Cancer Society (1093–02), and grants awarded by the National Cancer Institute (CA-78724 and CA-55652). We thank William Dalton and Terry Landowski for their informative suggestion on the manuscript preparation and data analysis. We also thank the Flow Cytometry Core Facility at Moffitt Cancer Center for acquisition and analysis of flow cytometry data, and Alan Cantor of the Biostatistics Core for statistical analysis.
References
- 1.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993; 82:1–14. [PubMed] [Google Scholar]
- 2.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997; 89:256–260. [PubMed] [Google Scholar]
- 3.Oshimi K, Shinkai Y, Okumura K, Oshimi Y, Mizoguchi H. Perforin gene expression in granular lymphocyte proliferative disorders. Blood. 1990; 75:704–708. [PubMed] [Google Scholar]
- 4.Van Parijs L, Ibraghimov A, Abbas AK. The roles of costimulation and Fas in T cell apoptosis and peripheral tolerance. Immunity. 1996; 4:321–328. doi: 10.1016/s1074-7613(00)80440-9. [DOI] [PubMed] [Google Scholar]
- 5.Spaner D, Raju K, Radvanyi L, Lin Y, Miller RJ. A role for perforin in activation-induced cell death. J Immunol. 1998; 160:2655–2664. [PubMed] [Google Scholar]
- 6.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferative disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992; 356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, et al. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell. 1994; 76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 8.Lamy T, Liu JH, Landowski TH, Dalton WS, Loughran TP., Jr Dysregulation of CD95/CD95 ligand-apoptotic pathway in CD3+ large granular lymphocyte leukemia. Blood. 1998; 92:4771–4777. [PubMed] [Google Scholar]
- 9.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human myeloma tumor cells. Immunity. 1999; 10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 10.Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990; 9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidel HM, et al. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995; 92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milocco LH, Haslam JA, Rosen J, Seidel HM. Design of conditionally active STATs: insights into STAT activation and gene regulatory function. Mol Cell Biol. 1999; 19:2913–2920. doi: 10.1128/mcb.19.4.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank DA, Robertson MJ, Bonni A, Ritz J, Greenberg ME. Interleukin 2 signaling involves the phosphorylation of Stat proteins. Proc Natl Acad Sci USA. 1995; 92:7779–7783. doi: 10.1073/pnas.92.17.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenlund AC, et al. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995; 2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 15.Liu KD, Gaffen SL, Goldsmith MA. JAK/STAT signaling by cytokine receptors. Curr Opin Immunol. 1998; 10:271–278. doi: 10.1016/s0952-7915(98)80165-9. [DOI] [PubMed] [Google Scholar]
- 16.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998; 334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayajima A, Ito Y, Kinoshita T. Cytokine signaling for proliferation, survival, and death in hematopoietic cells. Int J Hematol. 1999; 69:137–146. [PubMed] [Google Scholar]
- 18.Garcia R, Jove R. Activation of STAT transcription factors in oncogenic tyrosine kinase signaling. J Biomed Sci. 1998; 5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
- 19.Yu C-L, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995; 269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 20.Danial NN, Pernis A, Rothman PB. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995; 269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 21.Nelson KL, Rogers JA, Bowman TL, Jove R, Smithgall TE. Activation of STAT3 by the c-Fes protein-tyrosine kinase. J Biol Chem. 1998; 273:7072–7077. doi: 10.1074/jbc.273.12.7072. [DOI] [PubMed] [Google Scholar]
- 22.Besser D, Bromberg FJ, Darnell JEJ, Hanafusa H. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol Cell Biol. 1999; 19:1401–1409. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia R, et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Diff. 1997; 8:1267–1276. [PubMed] [Google Scholar]
- 24.Gouilleux-Gruart V, et al. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996; 87:1692–1697. [PubMed] [Google Scholar]
- 25.Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997; 159:4720–4728. [PubMed] [Google Scholar]
- 26.Grundis JR, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998; 102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun WH, et al. Interferon-alpha resistance in a cutaneous T-cell lymphoma cell line is associated with lack of STAT1 expression. Blood. 1998; 91:570–576. [PubMed] [Google Scholar]
- 28.Turkson J, et al. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998; 18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999; 98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 30.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JEJ. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998; 18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1995; 14:854–855. doi: 10.1002/j.1460-2075.1995.tb07064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meydan N, et al. Inhibition of acute lymphoblastic leukemia by a Jak-2 inhibitor. Nature. 1996; 379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- 33.Karras JG, et al. STAT3 regulates the growth and immunoglobulin production of BCL (1) B cell lymphoma through control of cell cycle progression. Cell Immunol. 2000; 202:124–135. doi: 10.1006/cimm.2000.1661. [DOI] [PubMed] [Google Scholar]
- 34.Chao J-R, et al. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is a component of the GM-CSF viability response. Mol Cell Biol. 1998; 18:4883–4898. doi: 10.1128/mcb.18.8.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J-M, et al. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999; 19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin JX, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995; 4:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 37.Welte T, et al. STAT5 interaction with the T cell receptor complex and stimulation of T cell proliferation. Science. 1999; 8:222–225. doi: 10.1126/science.283.5399.222. [DOI] [PubMed] [Google Scholar]
- 38.Enari M, Hug H, Nagata S. Involvement of an ICE-like protease in Fas-mediated apoptosis. Nature. 1995; 375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson DW, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995; 376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 40.Ferlin-Bezombes M, et al. IFN-alpha is a survival factor for human myeloma cells and reduces dexamethasone-induced apoptosis. J Immunol. 1998; 161:2692–2699. [PubMed] [Google Scholar]
- 41.Treon SP, Anderson KC. Interleukin-6 in multiple myeloma and related plasma cell dyscrasias. Curr Opin Hematol. 1998; 5:42–48. [PubMed] [Google Scholar]
- 42.Fujio Y, Kunisada K, Hirota H, Yamauchi-Takihara K, Kishimoto T. Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J Clin Invest. 1997; 99:2898–2905. doi: 10.1172/JCI119484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berridge MJ. Lymphocyte activation in health and disease. Crit Rev Immunol. 1997; 17:155–178. doi: 10.1615/critrevimmunol.v17.i2.30. [DOI] [PubMed] [Google Scholar]
- 44.Kneitz B, Herrman T, Yonehara S, Schimpl A. Normal clonal expansion but impaired Fas-mediated cell death and anergy induction in interleukin-2-deficient mice. Eur J Immunol. 1995; 25:2572–2577. doi: 10.1002/eji.1830250925. [DOI] [PubMed] [Google Scholar]
- 45.Loughran TP, Jr, Aprile JA, Ruscetti FW. Anti-CD3 monoclonal antibody-mediated cytotoxicity occurs through an interleukin-2-independent pathway in CD3+ large granular lymphocytes. Blood. 1990; 4:935–940. [PubMed] [Google Scholar]
- 46.Yoon HJ, Sugamura K, Loughran TP., Jr Activation of leukemic large granular lymphocytes by interleukin-2 via the p75 interleukin-2 receptor. Leukemia. 1990; 4:848–850. [PubMed] [Google Scholar]
- 47.Zambello R, et al. Cell membrane expression and functional role of the p75 subunit of interleukin-2 receptor in lymphoproliferative disease of granular lymphocytes. Blood. 1990; 76:2080–2085. [PubMed] [Google Scholar]
- 48.Loughran TP, Jr, et al. Epitope mapping of HTLV envelope seroreactivity in LGL leukaemia. Br J Haematol. 1998; 101:318–324. doi: 10.1046/j.1365-2141.1998.00691.x. [DOI] [PubMed] [Google Scholar]
- 49.Loughran TP. HTLV infection and hematologic malignancies. Leuk Res. 1996; 20:457–458. doi: 10.1016/0145-2126(95)00121-2. [DOI] [PubMed] [Google Scholar]
- 50.Kaufmann SH, et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998; 91:991–1000. [PubMed] [Google Scholar]
- 51.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. Mcl-1, a gene expressed in programmed myeloid cell differentiation has sequence similarity to Bcl2. Proc Natl Acad Sci USA. 1993; 90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds JE, et al. Mcl-1 a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 1994; 54:6348–6352. [PubMed] [Google Scholar]
- 53.Zhou P, Quian L, Kozopas KM, Craig RW. Mcl-1, a bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 1997; 89:630–643. [PubMed] [Google Scholar]
- 54.Wang K, Gross A, Waksman G, Korsmeyer SJ. Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol Cell Biol. 1998; 18:6083–6089. doi: 10.1128/mcb.18.10.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu SY, Lin P, Hsueh AJ. BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Mol Endocrinol. 1998; 12:1432–1440. doi: 10.1210/mend.12.9.0166. [DOI] [PubMed] [Google Scholar]
- 56.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJ. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci USA. 1997; 94:12401–12406. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang LH, Kirken RA, Erwin RA, Yu CR, Farrar WL. JAK3, STAT, and MAPK signaling pathways as novel molecular targets for the tyrphostin AG-490 regulation of IL-2-mediated T cell response. J Immunol. 1999; 162:3897–3904. [PubMed] [Google Scholar]
- 58.Levitzki A. Protein tyrosine kinase inhibitors as novel therapeutic agents. Pharmacol Ther. 1999; 82:231–239. doi: 10.1016/s0163-7258(98)00066-7. [DOI] [PubMed] [Google Scholar]