Abstract
It has unambiguously been shown that genetic, environmental, demographic, and technical factors may have substantial effects on gene expression levels. In addition to the measured variable(s) of interest, there will tend to be sources of signal due to factors that are unknown, unmeasured, or too complicated to capture through simple models. We show that failing to incorporate these sources of heterogeneity into an analysis can have widespread and detrimental effects on the study. Not only can this reduce power or induce unwanted dependence across genes, but it can also introduce sources of spurious signal to many genes. This phenomenon is true even for well-designed, randomized studies. We introduce “surrogate variable analysis” (SVA) to overcome the problems caused by heterogeneity in expression studies. SVA can be applied in conjunction with standard analysis techniques to accurately capture the relationship between expression and any modeled variables of interest. We apply SVA to disease class, time course, and genetics of gene expression studies. We show that SVA increases the biological accuracy and reproducibility of analyses in genome-wide expression studies.
Author Summary
In scientific and medical studies, great care must be taken when collecting data to understand the relationship between two variables, such as a drug and its effect on a disease. In any given study there will be many other variables at play, such as the effects of age and sex on the disease. We show that in studies where the expression levels of thousands of genes are measured at once, these issues become surprisingly critical. Due to the complexity of our genomes, environment, and demographic features, there are many sources of variation when analyzing gene expression levels. In any given study, it is impossible to measure every single variable that may be influencing how our genes are expressed. Despite this, we show that by considering all expression levels simultaneously, one can actually recover the effects of these important missed variables and essentially produce an analysis as if all relevant variables were included. As opposed to traditional studies, the massive amount of data available in this setting is what makes the method, called surrogate variable analysis, possible. We hypothesize that surrogate variable analysis will be useful in many large-scale gene expression studies.
Introduction
Large-scale gene expression studies allow one to characterize transcriptional variation with respect to measured variables of interest, such as differing environments, treatments, time points, phenotypes, or clinical outcomes. However, a number of unmeasured or unmodeled factors may also influence the expression of any particular gene. Besides inducing widespread dependence in measurements across genes [1,2], these influential factors create additional sources of differential expression, which, unlike gene-specific fluctuations, represent common sources of variation in gene expression that can be observed among multiple genes.
We call “primary measured variables” (or primary variables) those variables that are explicitly modeled in the analysis of an expression study. These variables may or may not be associated with any given gene's expression variation. We classify all the remaining sources of expression variation into three basic types. “Unmodeled factors” are sources of variation explained by measured variables, but are not explicitly included in the statistical model (e.g., because their relationship to expression is intractable or the relevant measured variables were excluded because of sample size restrictions). “Unmeasured factors” are sources of expression variation that are not measured in the course of the study, so we also call these unmodeled factors. Finally, “gene-specific noise” refers to random fluctuations in gene expression independently realized from gene to gene.
As a simple example meant only for illustrative purposes, consider a human expression study where disease state on a particular tissue type is the primary variable. Suppose that in addition to changes in expression being associated with disease state, the age of the individuals also has a substantial influence on expression. Thus, some genes exhibit differential expression with respect to disease state, some with respect to age, and some with respect to both. If age is not included in the model when identifying differential expression with respect to disease state, we show that this may (a) induce extra variability in the expression levels due to the effect of age, decreasing our power to detect associations with disease state, (b) introduce spurious signal due to the fact that the effect of age on expression may be confounded with disease state, or (c) induce long-range dependence in the apparent “noise” of the expression data, complicating any assessment of statistical significance for differential expression. In practice, even if age were known, it may be one of dozens of available measured factors, making it statistically intractable to determine which to include in the model. Furthermore, even measured factors such as age may act on distinct sets of genes in different ways, or may interact with an unobserved factor, making the effect of age on expression difficult to model. “Expression heterogeneity” (EH) is used here to describe patterns of variation due to any unmodeled factor.
Major sources of expression variation are due to technical [3,4], environmental [5,6], demographic [7,8], or genetic [9–11] factors. It is well known that sources of variation due to experimental design or large-scale systematic sources of signal may be present in expression data [3,4,12,13], sometimes even after normalization has been applied [14]. Genetic factors can also have a large-scale impact on gene expression levels. Specific genetic loci have been shown to influence the expression of hundreds or thousands of genes in several organisms [10,11,15]. Expression heterogeneity is particularly pronounced in human expression data, especially in the study of complex systems, such as cancer or responses to stress [16–18]. Recently, Lamb et al. proposed the “Connectivity Map” for identifying functional connections between cancer subtypes, genetic background, and drug action [19]. Lamb et al. noted EH (e.g., due to cell type and batch effects) presented a major hurdle for extracting relevant biological signal from the Connectivity Map.
In each of these studies, expression variation with respect to one or at most a handful of variables is explored. However, it is likely that in each study multiple sources of EH will act on distinct, but possibly overlapping, sets of genes. Normalization techniques are commonly used to detect and adjust for systematic expression variation due to well-characterized laboratory and technical sources [12,13,20]. However, to date there has been no approach for identifying and accounting for all sources of systematic expression variation, including variation due to unmeasured or unmodeled factors of both biological and technical sources. We show here that biological sources of variation not modeled in the analysis can be just as problematic as technical sources of variation.
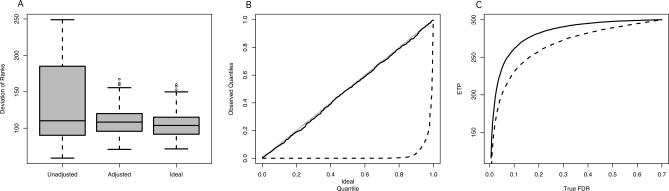
Here, we introduce “surrogate variable analysis” (SVA) to identify, estimate, and utilize the components of EH. Figure 1 shows the effects of failing to account for unmodeled factors in a differential expression analysis, and the potential benefits of the SVA approach. EH causes drastic increases in the variability of the ranking of genes for differential expression (Figure 1A), distorts the null distribution potentially causing highly conservative or anticonservative significance estimates (Figure 1B), and reduces the power to distinguish true associations between a measured variable of interest and gene expression (Figure 1C). However, employing SVA in these studies produces operating characteristics nearly equivalent to what one would obtain with no EH at all.
Figure 1. Impact of Expression Heterogeneity.
One thousand gene expression datasets containing EH were simulated, tested, and ranked for differential expression as detailed in Simulated Examples.
(A) A boxplot of the standard deviation of the ranks of each gene for differential expression over repeated simulated studies. Results are shown for analyses that ignore expression heterogeneity (Unadjusted), take expression heterogeneity into account by SVA (Adjusted), and for simulated data unaffected by expression heterogeneity (Ideal).
(B) For each simulated dataset, a Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution (Text S1). A quantile–quantile plot of the 1,000 Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the unadjusted analysis (dashed line). It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the unadjusted analysis does not due to EH.
(C) A plot of expected true positives versus FDR for the SVA-adjusted (solid) and -unadjusted (dashed) analyses. The SVA-adjusted analysis shows increased power to detect true differential expression.
We apply SVA to three distinct expression studies [7,21,22], where each study contains clear patterns of EH (Figure S1). These studies represent major classes of gene expression studies performed in practice: genetic dissection of expression variation, differential expression analysis between disease classes, and differential expression over time. We show that SVA is able to accurately identify and estimate the impact of unmodeled factors in each type of study, using only the expression data itself. We further show that SVA improves accuracy and consistency in detecting differential expression. SVA orders the significant gene lists to more accurately and reproducibly reflect the ordering of the genes with respect to their true differential expression signal. SVA is particularly useful in producing reproducible results in microarray studies, because adjusting for surrogate variables reduces differential expression due to sources other than the primary variables. These results indicate that EH is prevalent across a range of studies and that SVA can be used to capture and account for these patterns to improve the characterization of biological signal in expression analyses.
Results
Surrogate Variables
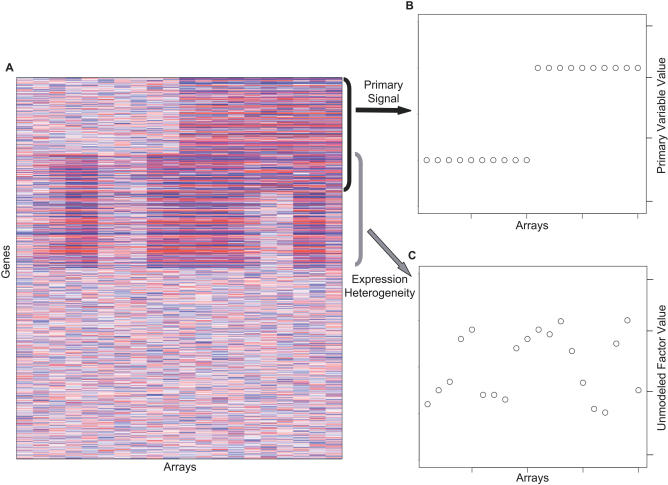
We have developed an approach called surrogate variable analysis that appropriately borrows information across genes to estimate the large-scale effects of all unmodeled factors directly from the expression data. Figure 2A shows a simulated example of EH. The primary variable distinguishes the first ten arrays from the last ten (Figure 2B); however, the unmodeled factor may have a variety of effects on expression (Figure 2C). The SVA approach flexibly captures signatures of EH, including highly irregular patterns not following any simple model, by estimating the signatures of EH in the expression data themselves rather than attempting to estimate specific unmodeled factors such as age or gender. After the surrogate variables are constructed, they are then incorporated into any subsequent analysis as covariates in the usual way. The SVA algorithm, described in mathematical detail in Materials and Methods, can conceptually be broken down into four basic steps: (Step 1) Remove the signal due to the primary variable(s) of interest to obtain a residual expression matrix. Apply a decomposition to the residual expression matrix to identify signatures of EH in terms of an orthogonal basis of singular vectors that completely reproduces these signatures. Use a statistical test to determine the singular vectors that represent significantly more variation than would be expected by chance. (Step 2) Identify the subset of genes driving each orthogonal signature of EH through a significance analysis of associations between the genes and the EH signatures on the residual expression matrix. (Step 3) For each subset of genes, build a surrogate variable based on the full EH signature of that subset in the original expression data. (Step 4) Include all significant surrogate variables as covariates in subsequent regression analyses, allowing for gene-specific coefficients for each surrogate variable.
Figure 2. Example of Expression Heterogeneity.
(A) A heatmap of a simulated microarray study consisting of 1,000 genes measured on 20 arrays.
(B) Genes 1–300 in this simulated study are differentially expressed between two hypothetical treatment groups; here the two groups are shown as an indicator variable for each array.
(C) Genes 201–500 in each simulated study are affected by an independent factor that causes EH. This factor is distinct from, but possibly correlated with, the group variable. Here, the factor is shown as a quantitative variable, but it could also be an indicator variable or some linear or nonlinear function of the covariates.
The four-step procedure is necessary both to ensure that the surrogate variables indeed estimate EH and not the signal from the primary variable (Step 1), to ensure an accurate estimate of each surrogate variable by identifying the specific subset of genes driving each EH signature (Step 2), to allow for correlation between the primary variable and the surrogate variables by building the surrogate variables on the original expression data (Step 3), and to take into account the fact that a surrogate variable may have a different effect on each gene (Step 4). The third and fourth steps are particularly important for maintaining unbiased significance with SVA, as demonstrated below.
Definition of a Correct Procedure
The overall goal of SVA is to provide a more accurate and reproducible parsing of signal and noise in the analysis of an expression study when EH is present. One way in which signal is commonly quantified is through a significance analysis [23]. The most basic definition of a significance analysis being performed “correctly” is if the null distribution is calculated properly [24]. A straightforward means for determining whether this is true is to assess whether the p-values corresponding to true null hypotheses are Uniformly distributed between zero and one. Indeed, p-values are specifically defined so that those corresponding to true null hypotheses have a Uniform(0,1) distribution if and only if the null distribution has been correctly calculated [25]. Throughout this paper, we examine the distribution of p-values from null genes to determine whether various procedures are able to recover the correct null distribution in the presence of EH. To assess statistically any deviations from the Uniform distribution for the null p-values, we apply a nested Kolmogorov-Smirnov test that is robust to chance fluctuations that may be present in a single simulated dataset (see Text S1).
Simulated Examples
We performed a simulation study to investigate the properties of SVA with respect to large-scale significance testing. Specifically, we show that the SVA algorithm (a) accurately estimates signatures of expression heterogeneity, (b) corrects the null distribution of p-values from multiple hypothesis tests, (c) improves estimation of the false discovery rate (FDR) [23,26], and (d) is robust to confounding between the primary variable and surrogate variables. The primary variable for our simulation was a binary variable indicating two disease classes. We simulated 1,000 expression studies, drawn from the same hypothetical population. For each study, we simulated expression for 1,000 genes on 20 arrays divided between the two disease states. The first 300 genes were simulated to be differentially expressed between disease states and genes 200–500 were affected by an independent unobserved factor to simulate a randomized study (Materials and Methods).
Surrogate variables accurately estimated.
We first assessed the accuracy of the surrogate variables estimated from SVA. In 99.5% of the simulated studies, a permutation procedure [27] correctly identified one significant surrogate variable. Since there is only one unmodeled factor that was simulated in this study, we assessed the accuracy of the surrogate variable estimation by correlation. (If there is more than one surrogate variable or more than one unmodeled factor, then one must assess the accuracy by using some sort of multiple regression and calculating an R2 value.) The average correlation between the estimated surrogate variable and the true unmodeled factor over all 1,000 experiments was 0.95 with a standard deviation of 0.05. Each surrogate variable is a weighted average of the expression measurements over a subset of genes. We chose a liberal adaptive cutoff for determining the number of genes affected by each orthogonal EH signal to avoid overfitting. The SVA algorithm correctly identifies the genes affected by the unmodeled factor. On average, 30.5% of the truly affected genes were identified as affected, whereas only 9.9% of the truly unaffected genes were identified as affected.
Correct p-value distribution.
It is well known that in a significance analysis, p-values corresponding to null genes should be Uniformly distributed (i.e., “flat”) [28]. Statistics has classically dealt with effects from unmodeled factors by performing randomized studies. In our simulations, the unmodeled factor was independently realized, which is equivalent to randomizing the unmodeled factor with respect to the primary variable. Because of this, the p-values corresponding to any single null gene over many simulated datasets follow the Uniform distribution. However, for any given experiment, a single randomization is applied to all genes. Therefore, the thousands of p-values resulting from a single microarray study are not the same as thousands of p-values resulting from independent randomizations of an unmodeled factor. The dependence across genes induced by EH can result in major fluctuations and bias in the p-values for the null genes for any single expression study, even in a well designed, randomized study. This bias generally takes the form of a global deviation of the null p-values from the Uniform distribution. Specifically, if the unmodeled factor is correlated with the primary variable, the null p-values will be too small, biased towards zero. If the unmodeled factor is uncorrelated with the primary variable, the null p-values will be too big, biased towards one.
These noteworthy fluctuations and biases in the null p-values can be seen in nine representative datasets from our simulation study in Figure S2 and across all 1,000 simulated datasets in Figure 1B. The bias results in incorrect assessment of significance, regardless of the particular significance measure chosen [24]. By applying the SVA algorithm to adjust the significance analysis, the p-values from the null genes for any single experiment are now corrected toward the Uniform distribution. This can be seen when SVA is applied to these same nine datasets in Figure S3 and across all 1,000 simulated datasets in Figure 1B. Figure 1B shows that the null p-values consistently follow the Uniform distribution when SVA is applied, but they consistently do not follow the Uniform in a typical unadjusted analysis. To confirm that SVA is robust to the distribution of the gene specific error, we ran a second independent simulation study where the residuals were drawn from a published microarray study (Materials and Methods). Figure S4 shows that behavior of the null p-values is corrected by SVA, which is qualitatively the same as in the case of purely simulated data.
It should also be noted that p-values corresponding to differentially expressed genes will be similarly affected; the loss in power can be seen in Figure 1C. Although, note that power versus FDR is calculated in Figure 1C when we know the correct answers, which clearly will not be reflected in actual studies where an unadjusted analysis produces an incorrect set of null p-values. Therefore, the application of SVA can result in empirical increases or decreases in power, depending on whether the null p-values are spuriously pushed towards zero or one, even though SVA tends to only provide increases in the true power.
Gene ranking more accurate and stable.
Perhaps most importantly, SVA also results in a more powerful and reproducible ranking of genes for differential expression. This can be seen in Figures 1A and S5; SVA-adjusted analyses provide gene rankings comparable to the scenario where there is no heterogeneity, whereas an unadjusted analysis allows for incorrect and highly variable gene rankings. This is arguably the most important feature of SVA, since an accurate and reproducible gene ranking is key for making biological inference when only a subset of genes will be selected for future study. In other words, these results suggest that SVA would yield results reproducible on the level that we would expect given that the primary variable is the only source of signal.
Improved FDR estimation.
There has been much recent interest in the effect of expression dependence across genes on estimates of multiple testing significance measures. Large-scale dependence has been shown to be particularly problematic for estimating FDR, as dependence across genes increases the variance of most standard FDR estimators [1,2,29–35]. EH represents large-scale dependence across genes that may significantly affect estimates of the FDR and related measures. To evaluate the potential impact of SVA in this situation, we performed a simulation study as described above. However, in this case, to create large-scale dependence, we let genes 201–1,000 be affected by the unmodeled factor. SVA reduces the variability in both the estimate of the proportion of null hypotheses and the q-values for each study (Figure S6). Furthermore, the behavior of the SVA-adjusted FDR estimates is almost identical to the behavior under the scenario with no EH.
Robustness to confounding in observational studies.
To assess the accuracy of the SVA algorithm in the case where the primary variable and unmodeled factors are heavily correlated, we performed a second simulation study. The set-up for the second simulation study was identical to that for the original study above, except in this case the unmodeled factor was simulated such that the average correlation with the primary variable was 0.50 with a standard deviation of 0.16. Under this model, the unobserved factor is both correlated with the primary variable and affects an overlapping set of genes. This is representative of the potential confounding present in observational microarray studies (see Disease Class below) and that which happens by chance in a non-negligible subset of randomized studies. Even in this set-up, the permutation hypothesis test correctly identified a single surrogate variable in 94.5% of the simulated datasets. Further, the average correlation between the estimated surrogate variable and the true unmodeled factor over 1,000 datasets was 0.94 with a standard deviation of 0.22. Thus, SVA accurately estimates the unobserved factor even when there is strong dependence between the primary and unobserved factors, with a subset of genes affected by both. SVA also provided a correct Uniform distribution of null p-values as in the above randomized study scenario.
Proof of Concept: Genetics of Gene Expression in Yeast
Several recent studies have carried out the genetic dissection of expression variation at the genome-wide level [10,11,15]. Brem et al. [10, 21] measured expression genome wide in 112 segregants of a cross between two isogenic strains of yeast. They also obtained genotypes for each segregant at markers covering 99% of the genome (Materials and Methods). It was shown that many gene expression traits are cis-linking, i.e., the quantitative trait locus (QTL) linkage peak coincided with the physical location of the open reading frame for the expression trait [36]. At the same time, it was also shown that a number of gene expression traits are trans-linking, with linkage peaks at loci distant from the physical location of their open reading frames. In particular, several “pivotal” loci each appear to influence the expression of hundreds or even thousands of gene expression traits. Similar highly influential loci have been observed in other organisms [11,15]. These pivotal loci act as a major source of EH, regardless of whether genotypes have been measured in an expression study.
As proof of concept, the Brem et al. [10,21] dataset was used to show that well-defined EH exists in actual studies and that SVA can properly capture and incorporate this EH structure into the statistical analysis of measured variables of interest. First, we analyzed the full dataset to identify the expression traits under the influence of these pivotal trans-acting loci, as well as the patterns of EH induced by these loci. Then we applied SVA to only the expression data, ignoring the genotype data to identify relevant surrogate variables capturing EH. Linkage analysis was performed again including the surrogate variables as covariates, showing that the effects from the pivotal loci are now negligible. In other words, SVA was able to capture and remove the effects of these few pivotal loci without the need for genotypes.
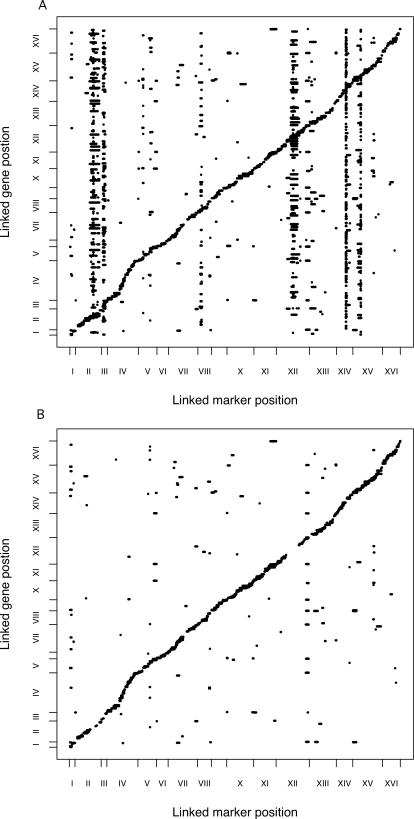
A number of expression traits have significant trans-linking eQTL mapping to pivotal loci on Chromosomes II, III, VIII, XII, XIV, and XV (Figure 3A). In the SVA-adjusted analysis, the majority of the trans-linkages to the pivotal loci have been eliminated (Figure 3B). The pervasive trans-linkage signal mapping to the pivotal loci can be viewed as global expression heterogeneity. The reduction in trans-linkage to these loci in the SVA-adjusted significance analysis indicates that SVA effectively captures genetic EH.
Figure 3. SVA Captures EH Due to Genotype.
(A) A plot of significant linkage peaks (p-value < 1e−7) for expression QTL in the Brem et al. [10,21] study by marker location (x-axis) and expression trait location (y-axis).
(B) Significant linkage peaks (p-value < 1e−7) after adjusting for surrogate variables. Large trans-linkage peaks on Chromosomes II, III, VII, XII, XIV, and XV have been eliminated without reducing cis-linkage peaks.
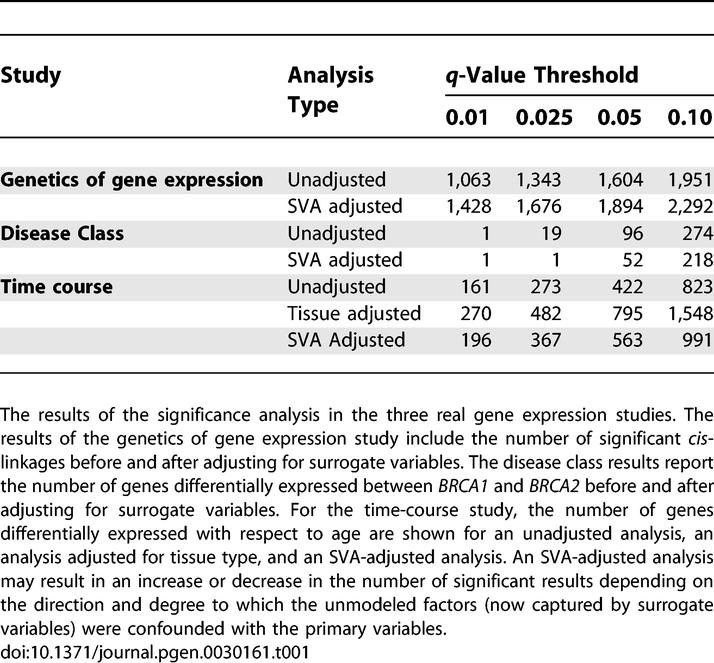
Pivotal trans-linkage signals indicate large-scale effects of a few loci. However, subtle and potentially more interesting cis-linkage may be lost in the presence of substantial genetic heterogeneity. To assess the impact of SVA on power to detect cis-linkage, we calculated linkage p-values only for markers located within three centimorgans of the open reading frame of each trait. On chromosomes without a pivotal locus (Chromosomes I, IV, V, VI, VII, IX, X, XI, and XIII) the SVA-adjusted analysis finds substantially more cis-linkage signal. At an FDR cutoff of 0.05, the adjusted analysis finds 1,894 significant cis-linkages, compared with 1,604 for the unadjusted analysis. This increase is consistent across a range of FDR cutoffs (Table 1) and illustrates the potential increase in power obtained from applying SVA.
Table 1.
Significance Results
SVA Applied to Human Expression Studies
We applied the SVA approach to two human studies [7,22], representing the two common human study designs: disease state and timecourse.
Disease class.
Hedenfalk et al. [22] measured expression in seven BRCA1 and eight BRCA2 mutation-positive tumor samples (Materials and Methods). The goal of the study was to identify genes that showed differential expression across breast cancer tumor subtypes defined by these germline mutations.
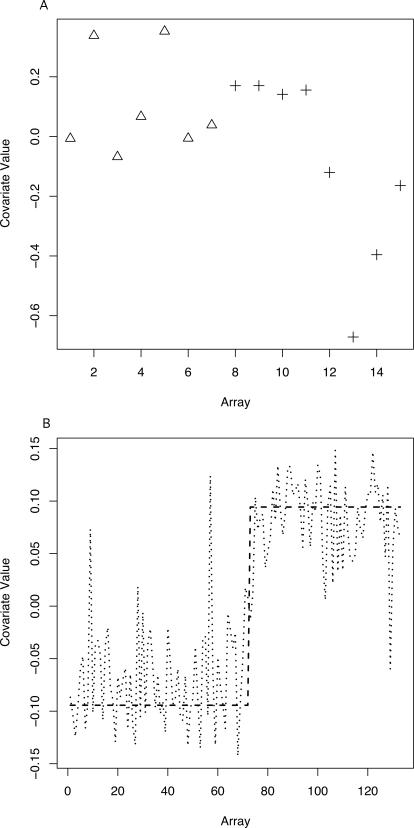
Hierarchical clustering [37] of the data reveals notable substructure within the BRCA2 samples [38]. We applied SVA and identified a single surrogate variable that appears to capture this trend (Figures 4A and S7A). We included this surrogate variable in a significance analysis comparing BRCA1 and BRCA2 tumors (Materials and Methods). The adjusted analysis finds fewer significant genes at standard FDR cutoffs (Table 1). This can be understood in the context of substructure within the BRCA2 group. Many of the genes declared differentially expressed at the most extreme levels of significance are highly associated with the top surrogate variable. Thus, differential expression for a number of genes is driven primarily by expression heterogeneity. Adjusting for the top surrogate variable eliminates spurious differential expression due to EH. As an example, eukaryotic translation initiation factor 2 (EIF2S2) is declared differentially expressed with a q-value of 0.09 in the unadjusted analysis. However, the first four BRCA2 samples show nearly identical expression values to the BRCA1 samples for this gene (Figure S7B). Thus, it is unlikely that differential expression is being driven by the difference in BRCA genotypes, but rather by some other confounding factor due to the observational nature and small sample size of the study.
Figure 4. Surrogate Variables from Human Studies.
(A) A plot of the top surrogate variable estimated from the breast cancer data [22]. The BRCA1 group is relatively homogeneous (triangles), but the BRCA2 group shows substantial heterogeneity (pluses).
(B) A plot of tissue type versus array for the Rodwell et al. [7] study (dotted line) and the top surrogate variable estimated from the expression data when tissue was ignored (dashed line). There is strong correlation between the top surrogate variable and the tissue type variable.
As shown above, SVA also increases the accuracy and stability of the ordering of the significant gene lists (see Simulated Examples). Since it is standard practice to examine only the most significant genes for further study, an SVA-adjusted analysis may result in completely distinct biological conclusions. For example, Figure S8 shows a substantial reordering of genes for significance when applying SVA, including a number of highly significant genes in an adjusted analysis that moved substantially down in ranking when SVA was applied. These genes may represent spurious signal due to the confounding shown earlier that would reduce the quality of the gene list.
Time-course sampling.
Rodwell et al. [7] measured genome-wide expression in kidney tissue samples from 133 patients (Materials and Methods). The goal of the study was to identify genes whose expression changed with age. We applied a recently developed procedure for time-course significance analysis to identify differential expression with respect to age [8]. In these data, tissue type had a strong impact on the expression of thousands of genes. We first performed a time-course differential expression analysis with tissue type included as a covariate. We also performed a second differential expression analysis ignoring tissue type.
We then applied SVA to the expression data ignoring the tissue information. The top surrogate variable identified by SVA had a correlation of 0.86 with tissue type (Figure 4B). The SVA algorithm identified 84% of the genes as likely to be associated with the top surrogate variable, indicating pervasive signal due to tissue type, as can be directly verified. To determine if this surrogate variable captured the overall effect of tissue type, we performed a third differential expression analysis ignoring tissue type and including the top surrogate variable as a covariate.
At standard q-value cutoffs, the results of the analysis adjusted for the top surrogate variable appeared to be very similar to the results when the true tissue type was included (Table 1). At a standard q-value cutoff of 0.05, 100% of the 422 genes declared significant by the unadjusted analysis were declared significant by the tissue-adjusted analysis. At the same cutoff, 96% of the 538 genes declared significant in the SVA-adjusted analysis were also declared significant in the tissue-adjusted analysis. That is, 116 genes were significant in the SVA-adjusted analysis that were also significant in the tissue-adjusted analysis, but were not significant in the unadjusted analysis. These genes represent an increase in power to detect differential expression after adjusting for a surrogate variable in place of an unmodeled confounding factor.
Comparison with Existing Methods
Regression on eigenvectors.
There are several well-established statistical approaches for partitioning of sources of variation among multiple variables into components [39]. The classical singular value decomposition (or principal components) approach has been successfully applied in several areas of genomics. For example, Alter et al. applied the singular value decomposition to identify significant trends in gene expression studies [40]. They showed that the right singular vectors, or “eigengenes,” represent trends that account for a large proportion of the variation in the expression matrix. Recently, Price et al. [41] also performed this singular value decomposition of whole-genome SNP genotypes (coded as 0, 1, or 2) in order to account for systematic sources of variation due to population substructure. Both of these methods extract and utilize patterns of variation from the entire matrix of genomic data without supervision from primary variables.
When performing a significance analysis of an expression study with respect to primary variables, one cannot employ this classical approach. As opposed to association studies, where population structure has genome-wide effects at a signal relatively much stronger than the primary variable, the signal structure in expression studies tends to be much more complex. There can be multiple levels of signal from multiple sources that each affect certain subsets of genes, making it important to supervise the decomposition with respect to known primary variables and these subsets of genes.
To demonstrate these issues, we considered two straightforward significance analysis applications of the well-established singular value decomposition approach previously utilized in genomics [40,41]. The first application identifies significant eigengenes by the same permutation-based algorithm as in our SVA approach. The eigengene with the highest absolute correlation with the primary variable is removed and the remaining significant eigengenes are included as covariates in the significance analysis. The second algorithm identifies significant eigengenes in the residuals of the regression of gene expression on the primary variable, again using a permutation-based algorithm. All significant eigengenes identified in the residuals are included in the significance analysis. Both algorithms do not produce consistently accurate results (Figures S9 and S10), and sometimes their adjustments produce more bias than making no adjustment at all. The eigengenes calculated from the entire expression matrix capture the signal due to both the unmodeled factor and the primary variable, which results in biased estimation of the unmodeled factor. The eigengenes calculated from the residuals do not take into account possible overlapping signal between the primary variable and unmodeled factors, often resulting in over-fitting.
SVA is a new methodological development aimed at overcoming the issues not addressed by existing methods. Rather than decomposing the entire expression matrix (or genotype matrix), SVA performs what could be called a “supervised factor analysis” of the expression data (Materials and Methods). Specifically, SVA decomposes the expression variation with respect to the primary variables already included in the model. Our multi-step approach for estimating surrogate variables uses the eigengenes from carefully defined subsets of genes in the original expression matrix that correspond to patterns observed in a residual expression matrix where the main effects of the primary variables have been removed. This allows us to decompose the variation in such a way that distinct sets of genes (but possibly overlapping) drive each surrogate variable, where the surrogate variables may be correlated with the primary variables. It also does not require any assumptions about the relative strength of signal due to each source of variation.
Multiple testing dependence.
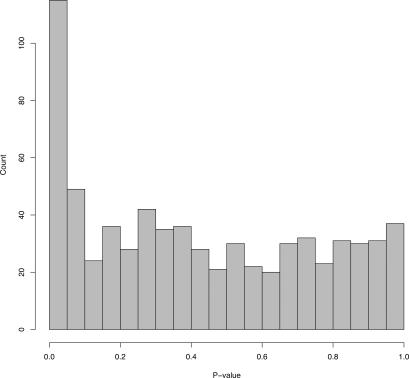
It is clear that EH induces widespread dependence in expression variation across genes. EH is therefore related to the issue of multiple testing dependence, which has been recognized as an important problem [1,2,30]. A number of methods have been proposed for adjusting for dependence in multiple tests that make adjustments directly once the tests are summarized as p-values or test-statistics, rather than the original dataset [31–35]. It does not appear that these multiple testing procedures can solve the problem of EH at the level of generality of SVA. Figure 5 shows a histogram composed of all null p-values affected by EH from the simulation study. Without the presence of EH, these null p-values would be Uniformly distributed between zero and one. However, it is also possible to produce a set of p-values from an experiment unaffected by EH, where a subset of tests are true alternatives and have p-values pushed towards zero so that they are indistinguishable from Figure 5. In other words, by only observing the set of p-values in Figure 5, it is not possible to know whether they are all null and affected by EH, or whether they are unaffected by EH and a subset are true alternatives.
Figure 5. Null p-Values under Heterogeneity.
A histogram of the null p-values from a single simulated experiment affected by heterogeneity. The distribution of these p-values appears identical to a complete set of p-values from an experiment that is not subject to heterogeneity. Therefore, it is not possible to identify and account for heterogeneity by analyzing one-dimensional p-values or test-statistics (see also Text S1).
If the original data are ignored and an adjustment for EH is applied to the p-values, then the only unbiased adjustment is to make all p-values larger so that the histogram in Figure 5 is transformed to a flat, Uniformly distributed histogram. Therefore, if one adjusts for EH based only on p-values, then all p-value histograms that look like Figure 5 should be made flat. By producing datasets with stronger EH, it is possible to produce histograms where the p-values are pushed even more strongly towards zero because of the stronger dependence. This argument shows that any method that adjusts for EH in general at the level of p-values must make all p-value histograms Uniformly distributed. The same argument holds for test-statistics, where they would have to be transformed to be distributed as their “theoretical null” distribution (Figure S11). Therefore, it does not appear that one can generally adjust for EH based only on p-values or test-statistics, especially when considering examples such as that in Figure 5. This point can be further supported with a more theoretical argument (Text S1). Additionally, methods that adjust for what is typically defined as multiple testing dependence do not usually take into account the fact that the sources of dependence may have signal that overlaps with the primary variables of interest, whereas SVA does. It appears that the framework presented here may be a generalization of the multiple testing problem, but this issue requires further investigation.
Discussion
Expression heterogeneity due to technical, genetic, environmental, or demographic variables is common in gene expression studies. Here we have introduced a new method, SVA, for identifying, estimating, and incorporating sources of EH in an expression analysis. SVA uses the expression data itself to identify groups of genes affected by each unobserved factor and estimates the factor based on the expression of those genes. Simulations show that SVA accurately detects expression heterogeneity and improves significance analyses. We performed SVA on experiments involving recombinant inbred lines, individuals of varying disease state, and expression measured over time to illustrate the broad range of studies on which SVA can be applied. One advantage of the SVA approach is the ability to disentangle correlated and overlapping differential expression signals. This approach may be particularly useful in clinical studies, where a large number of clinical variables may have a complicated joint impact on expression. We have implemented SVA in an open source package available for downloading at http://www.genomine.org/sva/.
Materials and Methods
Expression data.
Three publicly available datasets were employed to represent a broad range of gene expression studies performed in practice. The first dataset consists of gene expression measurements for 6,216 genes in 112 segregants of a cross between two isogenic strains of yeast, as well as genotypes across 3,312 markers [10,21]. The second dataset consists of gene expression for 3,226 genes in seven BRCA1 and eight BRCA2 mutation–positive tumor samples [22]; several genes with apparent outliers were removed as described [23] for a total of 3,170 genes. The third dataset consists of gene expression measurements in kidney samples from normal kidney tissue obtained at nephrectomy from 133 patients [7]; the 34,061 genes analyzed in [8] were also analyzed here. Seventy-four of the tissue samples were obtained from the cortex and 59 from the medulla. Details of the protocol for each study appear in the corresponding references. All expression data were analyzed on the log scale.
Linkage analysis of yeast cross.
The SVA algorithm identified 14 significant surrogate variables from the expression data. We performed both an unadjusted and an SVA-adjusted linkage analysis for each expression trait. In the unadjusted analysis, linkage p-values were calculated from an F-test comparing an additive genetic model to the null model of no genetic association [42]. SVA-adjusted p-values were calculated from an F-test comparing the full model of genetic association and the null model of no association, both models including all significant surrogate variables as additive terms.
Simulation details.
For each study, we simulated expression for 1,000 genes on 20 arrays divided between the two disease states. For simplicity, the expression measurements for each gene were drawn from a normal distribution with mean zero and variance one. We simulated expression heterogeneity with a dichotomous unmodeled factor independent of the disease state. The mean differences between disease states and states of the unmodeled factor were drawn from two independent normal distributions. For the real data example, we calculated the residuals from the regression of BRCA tumor type on expression for the Hedenfalk data [22]. Then, for each simulated study, we independently permuted each row of the expression data to create a matrix of residuals. To this matrix, we added signal, as in the case of the purely simulated data. The simulation studies were based on data generated using the R programming language [43]. All differential expression analyses were performed by a t-test based on standard linear regression. The genes were ranked for relative significance by the absolute values of their t-statistics.
Analysis of the human studies.
Differential expression was calculated using a t-test based on standard linear regression for the disease class data. The method of Storey et al. [8] was applied for the time-course data. q-Values were estimated using previously described methodology [23].
Statistical model for SVA.
Let Xmxn = (x1,..,xm)T be the normalized m × n expression matrix with n arrays for m genes, where xi = (xi1,..,xin)T is the vector of normalized expression for gene i. Let y = (y1,..,yn)T be a vector of length n representing the primary variable of interest.
Without loss of generality model xij = μi + fi(yj) + eij, where μi is the baseline level of expression, fi(yj) = E(xij | yj) − μi gives the relationship between measured variable of interest and gene i, and eij is random noise with mean zero. As a simple example, for a dichotomous outcomes yj ∈ (−1,1} we would employ the linear model xij = μi + βi yj +eij and estimate μi and βi by least squares. We could then perform a standard test of whether βi = 0 or not for each gene. This hypothesis test is exactly equivalent to performing a test of differential expression between the two classes.
Suppose in a microarray study there are L biologically meaningful unmodeled factors, such as age, environmental exposure, genotype, etc. Let gℓ = (gℓ1,...,gℓn) be an arbitrarily complicated function of the ℓth factor across all n arrays, for ℓ=1,2,...,L. Therefore, we can now model the expression for gene i on array j as xij = μi + fi (yj)+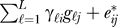 , where γℓi is a gene-specific coefficient for the ℓth unmodeled factor. If unmodeled factor ℓ does not influence the expression of gene i, then γℓi = 0. The fact that we employ an additive model is actually quite general: it has been shown that even complicated nonlinear functions of factors can be represented in an additive fashion for a reasonable choice of a nonlinear basis [44]; we simply define the gℓ to be as nonlinear as necessary and make L as large as necessary to best fit the additive effect. Since there are n arrays, each gene's expression can be modeled by at most n linearly independent factors, and hence any dependence structure between genes can be represented using L ≤ n vectors in this additive fashion.
, where γℓi is a gene-specific coefficient for the ℓth unmodeled factor. If unmodeled factor ℓ does not influence the expression of gene i, then γℓi = 0. The fact that we employ an additive model is actually quite general: it has been shown that even complicated nonlinear functions of factors can be represented in an additive fashion for a reasonable choice of a nonlinear basis [44]; we simply define the gℓ to be as nonlinear as necessary and make L as large as necessary to best fit the additive effect. Since there are n arrays, each gene's expression can be modeled by at most n linearly independent factors, and hence any dependence structure between genes can be represented using L ≤ n vectors in this additive fashion.
Due to this formulation, the inter-gene dependent eij have now been replaced with 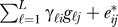 , where
, where 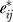 is the true gene-specific noise, now sufficiently independent across genes. In other words, we have broken the error eij into two terms, one that represents dependent variation across genes due to unmodeled factors,
is the true gene-specific noise, now sufficiently independent across genes. In other words, we have broken the error eij into two terms, one that represents dependent variation across genes due to unmodeled factors, 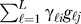 , and one that represents gene-specific independent fluctuations in expression
, and one that represents gene-specific independent fluctuations in expression 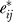 .
.
It is not possible in general to directly estimate the unmodeled gℓ, and SVA does not attempt to do so. The key observation is to note that for L vectors in n space, it is possible to identify an orthogonal set of vectors hk , k = 1,...,K (K≤L) that spans the same linear space as the gℓ In other words, for any set of vectors gℓ and coefficients γℓi, it is possible to identify mutually orthogonal vectors hk and coefficients λki such that 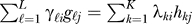 and
and
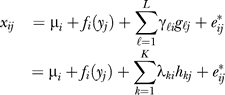 |
Therefore, we do not need to estimate the specific variables gℓ. We only need to estimate the linear combination 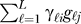 , so we can choose a set of vectors that spans the same space but is statistically tractable. Here we choose the set of K orthogonal vectors (denoted by the hk) to be those that are the right non-zero singular vectors provided by the singular value decomposition of the m × n matrix with (i, j) entry
, so we can choose a set of vectors that spans the same space but is statistically tractable. Here we choose the set of K orthogonal vectors (denoted by the hk) to be those that are the right non-zero singular vectors provided by the singular value decomposition of the m × n matrix with (i, j) entry 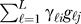 . This justifies the use of the singular value decomposition to identify orthogonal signatures of expression heterogeneity for surrogate variable estimates. We call these h1,h2,...,hK the “surrogate variables.”
. This justifies the use of the singular value decomposition to identify orthogonal signatures of expression heterogeneity for surrogate variable estimates. We call these h1,h2,...,hK the “surrogate variables.”
An intuitive question that arises from an inspection of this formulation is about the model assumptions of the gℓj. Whereas the term fi (yj) is a model of the measured variable, yj, it is not generally possible to analogously formulate gℓj as a function of a well-defined, measured variable. Since we estimate the outcomes 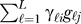 directly from the expression data (as
directly from the expression data (as 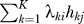 ), it is not necessary to determine a model of the gℓj in terms of a biologically meaningful variable. Thus, we can bypass the need to know what the most relevant model of a measured variable is for gℓj for the purposes of estimating the EH.
), it is not necessary to determine a model of the gℓj in terms of a biologically meaningful variable. Thus, we can bypass the need to know what the most relevant model of a measured variable is for gℓj for the purposes of estimating the EH.
SVA algorithm.
The goal of the SVA algorithm is therefore to identify and estimate the surrogate variables, hk , = (hk1,...,hkn)T, based on certain consistent patterns of expression variation. Methods for empirically identifying [37] and estimating [40] expression trends or clusters have previously been developed. However, care must be taken when estimating expression trends for use in subsequent analyses of measured variables of interest. Specifically, surrogate variables must represent signal due to sources other than the primary variable and allow for potential overlap with the primary variable. The SVA algorithm is designed to estimate surrogate variables that meet both requirements. We assume that n < m and, for simplicity, that there is only a single primary variable; the extension to multiple primary variables simply requires one to include all of them in the model fit occurring in each Step 1 below.
The algorithm is decomposed into two parts: detection of unmodeled factors and construction of surrogate variables. The basic form of the first algorithm has been proposed previously [27]. The second algorithm has been proposed and justified in this manuscript
Algorithm to detect unmodeled factors.
1. Form estimates 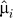 and
and 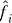 by fitting the model xij = μi + fi (yj) + eij, and calculate the residuals rij = xij −
by fitting the model xij = μi + fi (yj) + eij, and calculate the residuals rij = xij −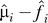 (yj) to remove the effect of the primary variable on expression. Form the m × n residual matrix R, where the (i, j) element of R is rij.
(yj) to remove the effect of the primary variable on expression. Form the m × n residual matrix R, where the (i, j) element of R is rij.
2. Calculate the singular value decomposition of the residual expression matrix R = UDVT.
3. Let dℓ be the ℓth eigenvalue, which is the ℓth diagonal element of D, for ℓ=1,...,n. If df is the degrees of freedom of the model fit μ̂i + f̂i (yj), then by construction the last df eigenvalues are exactly zero and we remove them from consideration. For eigengene k=1,..., n-df set the observed statistic to be
which is the variance explained by the kth eigengene.
4. Form a matrix R* by permuting each row of R independently to remove any structure in the matrix. Denote the (i, j) entry of R* by 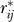 .
.
5. Fit the model 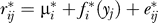 and calculate the residuals
and calculate the residuals 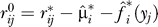 to form the m × n model-subtracted null matrix R0.
to form the m × n model-subtracted null matrix R0.
6. Calculate the singular value decomposition of the centered and permuted expression matrix R0 = U0D0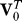 .
.
7. For eigengene k form a null statistic
as above, where d0ℓ is the ℓth diagonal element of D0.
8. Repeat steps 4−7 a total of B times to obtain null statistics 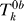 for b = 1,...,B and k = 1,...,n-df.
for b = 1,...,B and k = 1,...,n-df.
9. Compute the p-value for eigengene k as:
Since eigengene k should be significant whenever eigengene k′ is (where k′>k), we conservatively force monotonicity among the p-values. Thus, set pk = max (pk−1, pk) for k = 2,...,n-df.
10. For a user-chosen significance level 0≤α≤1, call eigengene k a significant signature of residual EH if pk ≤ α.
Algorithm to construct surrogate variables.
1. Form estimates 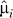 and
and 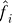 by fitting the model xij.= μi + fi(yj) + eij, and calculate the residuals rij = xij −
by fitting the model xij.= μi + fi(yj) + eij, and calculate the residuals rij = xij −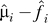 (yj) to remove the effect of the primary variable on expression. Form the m × n residual matrix R, where the (i, j) element of R is rij.
(yj) to remove the effect of the primary variable on expression. Form the m × n residual matrix R, where the (i, j) element of R is rij.
2. Calculate the singular value decomposition of the residual expression matrix R = UDVT. Let ek = (ek1,...,ekn)T be the kth column of V (for k=1,...,n). These ek are the residual eigengenes and represent orthogonal residual EH signals independent of the signal due to the primary variable.
3. Set 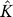 to the number of significant eigengenes found by the above algorithm. Note that “significant” means that the eigengene represents a greater proportion of variation than expected by chance.
to the number of significant eigengenes found by the above algorithm. Note that “significant” means that the eigengene represents a greater proportion of variation than expected by chance.
For each significant eigengene
ek k=1,...,
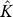 .
.
4. Regress ek on the xi (i = 1,...,m) and calculate a p-value testing for an association between the residual eigengene and each gene's expression. This p-value measures the strength of association between the residual eigengene ek and the expression for gene i.
5. Let π0 be the proportion of genes with expression not truly associated with ek; form an estimate 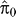 , as described previously [23] and estimate the number of genes associated with the residual eigengene by
, as described previously [23] and estimate the number of genes associated with the residual eigengene by 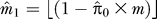 . Let
. Let  be the indices of the genes with
be the indices of the genes with 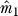 smallest p-values from this test.
smallest p-values from this test.
6. Form the 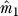 × n reduced expression matrix
× n reduced expression matrix 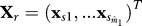 . Since
. Since 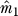 is an estimate of the number of genes associated with residual eigengene k, the reduced expression matrix represents the expression of those genes estimated to contain the EH signature represented by some hk as described above. As was done for R, calculate the eigengenes of Xr, and denote these by
is an estimate of the number of genes associated with residual eigengene k, the reduced expression matrix represents the expression of those genes estimated to contain the EH signature represented by some hk as described above. As was done for R, calculate the eigengenes of Xr, and denote these by 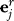 for j=1,...,n.
for j=1,...,n.
7. Let j* = argmax1≤j≤n
cor
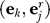 and set
and set 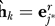 . In other words, set the estimate of the surrogate variable to be the eigengene of the reduced matrix most correlated with the corresponding residual eigengene. Since the reduced matrix is enriched for genes associated with this residual eigengene, this is a principled choice for the estimated surrogate variable that allows for correlation with the primary variable.
. In other words, set the estimate of the surrogate variable to be the eigengene of the reduced matrix most correlated with the corresponding residual eigengene. Since the reduced matrix is enriched for genes associated with this residual eigengene, this is a principled choice for the estimated surrogate variable that allows for correlation with the primary variable.
8. In any subsequent analysis, employ the model xij = μi + fi(yj) + 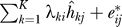 , which serves as an estimate of the ideal model xij = μi + fi(yj) +
, which serves as an estimate of the ideal model xij = μi + fi(yj) + 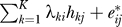 .
.
The singular value decomposition is employed in these SVA algorithms. It may be possible to utilize other decomposition methods, but since the singular value decomposition provides uncorrelated variables that decompose the data in an additive linear fashion with the goal of minimizing the sum of squares, we found this to be the most appropriate decomposition. If the primary variables are modeled for data that are not continuous, then it may make sense to decompose the variation with respect to whatever model-fitting criteria will be employed
Software.
SVA has been made freely available as an R package at http://www.genomine.org/sva/.
Supporting Information
Heatmaps of hierarchically clustered gene expression data for a random subset of 1,000 genes from three studies are shown. (A) Hedenfalk et al. [22] compared gene expression across tumor subtypes defined by germline BRCA mutations (yellow divides BRCA tumor subtypes), (B) Brem et al. [10,21] measured expression in naturally recombining yeast populations, and (C) Rodwell et al. [7] measured gene expression in kidney samples for patients ranging in age from 27–92 y.
(849 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression data. The null p-values should be Uniformly distributed, or “flat,” for each experiment. However, across independently simulated datasets, the null p-values range from being conservatively biased to anticonservatively biased depending on the configuration of the unmeasured or unmodeled factor.
(17 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, adjusted by SVA. The p-values for the null genes in each simulated experiment are Uniformly distributed. None of these deviates from the Uniform according to a Kolmogorov-Smirnov test.
(18 KB PDF)
For each simulated dataset based on the permuted residuals from the Hedenfalk et al. study, a nested Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution. A quantile–quantile plot of the one thousand Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the unadjusted analysis (dashed line). The grey line represents the expected quantiles. It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the unadjusted analysis does not, due to EH.
(62 KB PDF)
A plot of the true rank (according to signal-to-noise ratio) versus the significance test–based average rank (black) plus or minus one standard deviation (red) for each differentially expressed gene in simulated studies (A) affected by EH with an unadjusted analysis, (B) affected by EH with an SVA-adjusted analysis, and (C) unaffected by EH.
(439 KB PDF)
(A) A histogram of the estimates of the proportion of true nulls π0 for studies affected by EH. (B) A histogram of the estimates of the proportion of true nulls π0 for studies affected by EH, after adjusting for SVA. (C) A histogram of the estimates of the proportion of true nulls π0 for studies without EH. (D) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies affected by EH. (E) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies affected by EH, adjusted by SVA. (F) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies without EH.
(265 KB PDF)
(A) A plot of the top surrogate variable from the breast cancer data of Hedenfalk et al. [22]; triangles are BRCA1, pluses are BRCA2. (B) A plot of the expression for eukaryotic translation initiation factor 2, EIF2S2, which follows a similar pattern to the top surrogate variable.
(87 KB PDF)
A plot of the p-value rankings for the SVA-adjusted versus unadjusted significance analysis of the breast cancer data [22], showing substantial differences in the rankings obtained from the two analyses. The red line represents equality of ranking between the two procedures.
(181 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, after adjustment by the first regression on standard eigengenes algorithm.
(16 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, after adjustment by the second regression on standard eigengenes algorithm.
(16 KB PDF)
For 1,000 simulated datasets based on the Normal residuals, a nested Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution. A quantile–quantile plot of the one thousand Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the “Empirical Null” technique [31,32]. The grey line represents the expected quantiles. It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the “Empirical Null” adjusted null p-values do not.
(62 KB PDF)
(50 KB PDF)
Acknowledgments
We thank the investigators of the Inflammation and the Host Response to Injury Consortium (http://www.gluegrant.org/) for their helpful feedback and support on this work. We also thank Gary Churchill for several helpful conversations related to this work.
Abbreviations
- EH
expression heterogeneity
- FDR
false discovery rate
- QTL
quantitative trait locus
- SVA
surrogate variable analysis
Footnotes
A previous version of this article appeared as an Early Online Release on August 1, 2007 (doi:10.1371/journal.pgen.0030161.eor).
Author contributions. JTL and JDS conceived and designed the experiments, performed the experiments, contributed reagents/materials/analysis tools, and wrote the paper. JTL analyzed the data.
Funding. This research was supported in part by NIH grants U54 GM2119 (PI: Ronald Tompkins) and R01 HG002913.
Competing interests. The authors have declared that no competing interests exist.
References
- Qiu X, Xiao Y, Gordon A, Yakovlev A. Assessing stability of gene selection in microarray data analysis. BMC Bioinformatics. 2006;7:50. doi: 10.1186/1471-2105-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanov L, Yakovlev A. Treating expression levels of different genes as a sample in microarray data analysis: is it worth a risk? Stat Appl Genet Mol Biol. 2006;5:art9. doi: 10.2202/1544-6115.1185. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, et al. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci U S A. 2000;97:8409–8414. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:2191–2201. doi: 10.1371/journal.pbio.0020427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Xiao W, T LJ, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Tseng G, Oh M, Rohlin L, Liao J, Wong W. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 2001;29:2540–2557. doi: 10.1093/nar/29.12.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dudoit S, Luu P, Lin D, Peng V, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui X, Klebanov L, Yakovlev A. Correlation between gene expression levels and limitations of the empirical bayes methodology for finding differentially expressed genes. Stat Appl Genet Mol Biol. 2005;4:art34. doi: 10.2202/1544-6115.1157. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber T, Devlin JL, Ewens KG, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37:31–37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- Nguyen DM, Sam K, Tsimelzon A, Li X, Wong H, et al. Molecular heterogeneity of inflammatory breast cancer: A hyperproliferative phenotype. Clin Cancer Res. 2006;12:5047–5054. doi: 10.1158/1078-0432.CCR-05-2248. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, et al. Flourescent cdna microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress response. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Dabney AR, Storey JD. A new approach to intensity-dependent normalization of two-channel microarrays. Biostatistics. 2007;8:128–139. doi: 10.1093/biostatistics/kxj038. [DOI] [PubMed] [Google Scholar]
- Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, et al. Gene-expression profiles in hereditary breast cancer. New Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney AR, Storey JD. A reanalysis of a published Affymetrix genechip control dataset. Genome Biol. 2006;7:401. doi: 10.1186/gb-2006-7-3-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JA. Mathematical statistics and data analysis. 2nd edition. Belmont (California): Duxbury Press; 1995. [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Royal Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
- Buja A, Eyuboglu N. Remarks on parallel analysis. Multivariate Behav Res. 1992;27:509–540. doi: 10.1207/s15327906mbr2704_2. [DOI] [PubMed] [Google Scholar]
- Lehman EL, Romano JP. Testing statistical hypotheses. New York: Springer-Verlag; 2005. [Google Scholar]
- Owen AB. Variance of the number of false discoveries. J Royal Stat Soc Ser B. 2005;67:411–426. [Google Scholar]
- Qiu X, Yakovlev A. Some comments on instability of false discovery rate estimation. J Bioinform Comput Biol. 2006;4:1057–1068. doi: 10.1142/s0219720006002338. [DOI] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J Am Stat Assoc. 2004;99:96–104. [Google Scholar]
- Efron B. Correlation and large-scale simultaneous significance testing. J Am Stat Assoc. 2007;102:93–103. [Google Scholar]
- Cai GQ, Sarkar SK. Modified simes' critical values under positive dependence. J Stat Plan Inference. 2006;136:4129–4146. [Google Scholar]
- Benjamini Y, Yekultieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Pawitan Y, Calza S, Ploner A. Estimation of false discovery proportion under general dependence. Bioinformatics. 2006;22:3025–3031. doi: 10.1093/bioinformatics/btl527. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns Proc. Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Ringer M, Ben-Dor A, Yakhini Z, Chen Y, et al. Molecular classification of familial non-brca1/brca2 breast cancer. Proc Natl Acad Sci U S A. 2003;100:2532–2537. doi: 10.1073/pnas.0533805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate analysis. London: Academic Press; 1980. [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, SN A, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Storey JD, Akey JM, Kruglyak L. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3:1380–1390.:e267. doi: 10.1371/journal.pbio.0030267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2004. [Google Scholar]
- Hastie T, Tibshirani R. Generalized additive models. New York: Chapman & Hall; 1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heatmaps of hierarchically clustered gene expression data for a random subset of 1,000 genes from three studies are shown. (A) Hedenfalk et al. [22] compared gene expression across tumor subtypes defined by germline BRCA mutations (yellow divides BRCA tumor subtypes), (B) Brem et al. [10,21] measured expression in naturally recombining yeast populations, and (C) Rodwell et al. [7] measured gene expression in kidney samples for patients ranging in age from 27–92 y.
(849 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression data. The null p-values should be Uniformly distributed, or “flat,” for each experiment. However, across independently simulated datasets, the null p-values range from being conservatively biased to anticonservatively biased depending on the configuration of the unmeasured or unmodeled factor.
(17 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, adjusted by SVA. The p-values for the null genes in each simulated experiment are Uniformly distributed. None of these deviates from the Uniform according to a Kolmogorov-Smirnov test.
(18 KB PDF)
For each simulated dataset based on the permuted residuals from the Hedenfalk et al. study, a nested Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution. A quantile–quantile plot of the one thousand Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the unadjusted analysis (dashed line). The grey line represents the expected quantiles. It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the unadjusted analysis does not, due to EH.
(62 KB PDF)
A plot of the true rank (according to signal-to-noise ratio) versus the significance test–based average rank (black) plus or minus one standard deviation (red) for each differentially expressed gene in simulated studies (A) affected by EH with an unadjusted analysis, (B) affected by EH with an SVA-adjusted analysis, and (C) unaffected by EH.
(439 KB PDF)
(A) A histogram of the estimates of the proportion of true nulls π0 for studies affected by EH. (B) A histogram of the estimates of the proportion of true nulls π0 for studies affected by EH, after adjusting for SVA. (C) A histogram of the estimates of the proportion of true nulls π0 for studies without EH. (D) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies affected by EH. (E) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies affected by EH, adjusted by SVA. (F) A plot of observed FDR versus true FDR (grey) and average observed FDR versus true FDR (red) for simulated studies without EH.
(265 KB PDF)
(A) A plot of the top surrogate variable from the breast cancer data of Hedenfalk et al. [22]; triangles are BRCA1, pluses are BRCA2. (B) A plot of the expression for eukaryotic translation initiation factor 2, EIF2S2, which follows a similar pattern to the top surrogate variable.
(87 KB PDF)
A plot of the p-value rankings for the SVA-adjusted versus unadjusted significance analysis of the breast cancer data [22], showing substantial differences in the rankings obtained from the two analyses. The red line represents equality of ranking between the two procedures.
(181 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, after adjustment by the first regression on standard eigengenes algorithm.
(16 KB PDF)
Histograms of the null p-values for nine independent realizations of the simulated gene expression experiment, after adjustment by the second regression on standard eigengenes algorithm.
(16 KB PDF)
For 1,000 simulated datasets based on the Normal residuals, a nested Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution. A quantile–quantile plot of the one thousand Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the “Empirical Null” technique [31,32]. The grey line represents the expected quantiles. It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the “Empirical Null” adjusted null p-values do not.
(62 KB PDF)
(50 KB PDF)