Abstract
Cancer is a disease of extreme heterogeneity. Microarray analysis has identified thousands of genes that are transcriptionally up- or down-regulated in tumor samples; molecularly characterized lesions that play a causative role in tumorigenesis constitute more than 1 percent of the human genome. Such a large number of “cancer genes” stirs the debate of whether it is relevant to continue classifying cancer as a single condition. Yet, a discrete set of cellular processes has been found to underlie such complexity. Their deregulation has been proposed to act as a common denominator that enables tumors to evade cellular barriers to proliferation and metastasis. Efforts have been made to identify and model the mechanistic origins of cancer. Two such models are discussed here: the multistage model of cancer and the cancer platform model. The former suggests cancer arises by the sequential acquisition of mutations leading to the progressive erosion of normal cellular control mechanisms. In contrast, the latter reduces cancer initiation to two interdependent conditions: sustained proliferation with the concomitant inhibition of cell death. This review proposes that a third condition — cellular differentiation — should be added to the cancer platform model. Differentiation can act as a fail-safe mechanism against unrestrained cellular growth — much like cell death. Clinical implications of the different models are also analyzed.
The Complexity of the Cancer Phenotype
Cancer is an extremely heterogeneous disease; tumors in different tissues display strikingly different behaviors. For example, tumors of the pancreas tend to be highly aggressive, while prostate tumors are more frequently organ-confined. Tumors that arise in the same tissue can even exhibit an array of cellular pathologies, ranging from benign hyperplasias to highly invasive malignancies [1-3]. Cancer is also a complex disease involving the deregulation of multiple signal transduction pathways. Since the discovery of the first tumor-promoting gene, hundreds of genes have been shown to play a role in tumor initiation and progression, and research continues to uncover many more. Microarray analysis has identified thousands of genes that are transcriptionally upregulated or downregulated in cancer samples [4-6]. It remains unclear which transcriptionally deregulated genes in an individual tumor play a causative role in tumor initiation and maintenance and which ones represent bystanders with no selective advantage. Regardless of the role specific genes may play in cancer progression, these studies underscore the fact that by the time a tumor is histologically identified, it has accumulated a large number of molecular lesions. In addition, a recent literature survey of all published cancer genes identified 291 genes for which there are molecularly characterized mutations and evidence of a causative role in tumorigenesis. These genes represent more than 1 percent of the human genome [7]. Yet this number is a conservative estimate, since the study did not consider epigenetic regulation of gene expression. The large number of mutations found in tumor samples raises the question of whether it is biologically meaningful to classify cancer as a single disease entity. Is there a common thread that underlies most, if not all, human malignancies? Are there biological rules that govern cancer initiation and progression?
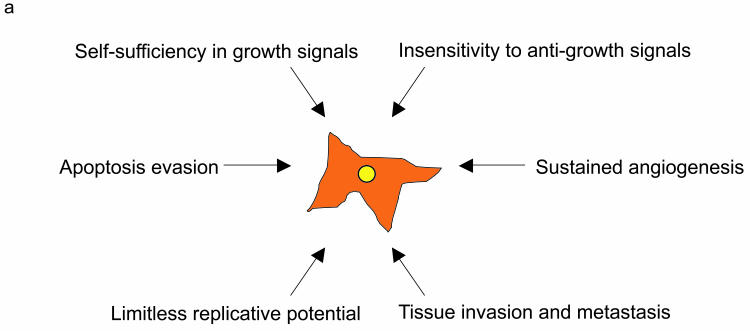
Despite the heterogeneity observed in cancer, most tumors share certain characteristics: self-sufficiency in growth signals, insensitivity to anti-growth signals, evasion of apoptosis, acquisition of a limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis. These common traits, which have been termed “the hallmarks of cancer,” allow tumors to breach cellular barriers against expansion and metastasis [8]. The similarity in the cellular processes subverted in all cancer cells, regardless of their tissue of origin, likely indicates common tumor-initiation mechanisms for the complex pathologies observed in clinical tumor samples.
The Multi-Stage Model of Cancer
Cellular transformation — the process by which a normal cell is “transformed” into a malignant one — is thought to take place through the accumulation of mutations, as well as epigenetic changes, that activate oncogenes or downregulate tumor-suppressor genes and lead to uncontrolled clonal expansion. Oncogenes originally were identified as the transforming agents of tumor viruses. It was later found that oncogenes were mutated versions of normal cellular genes, or proto-oncogenes, which had been incorporated into the viral genome by recombination. Mutations or epigenetic events, leading to the deregulated activity or increased expression of cellular oncogenes, are found in most cancers. Oncogene activation is implicated in the positive control of cellular growth, and mutations in oncogenes are generally dominant. In contrast, tumor suppressor genes function as negative regulators of cellular growth. Mutations in tumor-suppressors usually inactivate gene function and are generally recessive. Thus, inactivation of both copies of a tumor-suppressor gene is usually necessary for tumor development [9]. Efforts to define the mechanistic origins of cancer have focused on identifying such genes, as well as the pathways they regulate.
Several lines of evidence indicate that mutations in a single oncogene or tumor-suppressor are insufficient to give rise to cancer. First, most cancers develop late in life and the incidence of disease increases dramatically with age. Statistical analysis of epidemiological data shows four to five rate-limiting steps as necessary for cancer to occur, implying that a cell needs to accumulate four to five sequential genetic lesions in key regulatory pathways in order to become malignant [10]. Second, in vitro experiments using cell lines, as well as in vivo models of cancer, confirm the multiple-hit hypothesis for the majority of cancers, with retinoblastoma and certain types of leukemia being exceptions to the rule.
Initial research using transforming retroviruses, which contained activated versions of normal growth-controlling genes, indicated that alterations in a single gene could lead to transformation of rodent cells in culture [11-14]. However, the cells used in these initial cancer studies were immortal and could therefore proliferate indefinitely. In addition, these cell lines most likely had acquired a series of other genetic alterations in culture. When these experiments were repeated with primary cell lines, it was found that activation of at least one pair of oncogenes was required for transformation. Research by Land et al. [15] and Sinn et al. [16], along with experiments carried out with other sets of oncogenes, confirmed that at least two cooperating mutations are required for cancer.
Clinically, tumors are histologically classified as presenting with different “grades,” which correspond to a set of physiological markers (such as loss of differentiation, abnormal ploidy, and morphology) and correlate with patient outcome. Higher grade tumors have a more negative prognosis, while low-grade tumors are often considered early lesions and may progress to more invasive, high-grade disease. These observations have led to the hypothesis that cancer progression can be dissected into a small number of crucial steps whose sequential deregulation is critical for the clinical progression from low- to high-grade cancer. Several pathways with key functions in normal cell biology are deregulated in tumors:
Cell cyle entry
Normal cells tightly regulate cell cycle progression via a number of crucial proteins that work at the cell-cycle checkpoints to integrate information from both the external and internal cellular environments. One such protein is the tumor-suppressor protein Rb. In its hypo-phosphorylated state, Rb binds to transcription factor E2F and prevents it from activating cellular genes involved in DNA replication. When Rb becomes hyper-phosphorylated, transcription factor E2F is released, allowing for the transcription of genes essential for DNA synthesis [17-19]. Alterations at any level of the cell cycle control hierarchy leading to the disruption of the normal function of Rb can be found in most cancers.
Cell growth arrest, apoptosis, and senescence
Cell growth arrest, death, and senescence are essential mechanisms that not only regulate normal development but also prevent the accumulation of harmful mutations. Proteins that stimulate these processes are likely downregulated in cancer cells due to selective pressure to proliferate incessantly. The expression of several pro-apoptotic proteins, such as p53, has been found to be either reduced or eliminated in cancer cells [20].
In addition, cellular senescence, which regulates normal cellular lifespan, is also disrupted in cancer. Telomere length is thought to play a crucial role in modulating genomic stability and cellular lifespan. Telomeres are repetitive sequences at the ends of chromosomes that prevent them from being recognized as products of DNA fragmentation. Telomere length is shortened with each DNA replication until it reaches a critical length below which the cell can no longer divide and becomes senescent [21]. However, when cultured in vitro, a subset of tumor cells was found to have acquired an unlimited replicative lifespan. These cells maintain stable telomere lengths through the upregulation of the enzyme telomerase, which extends telomeric DNA [22-24].
Growth factor signaling
Growth factors provide environmental cues to regulate cell growth and proliferation. Growth-factor independence is one of the hallmarks of cancer. Constitutive activation or elevated expression of membrane receptors, such as EGFR and PDGFR, as well as non-receptor proteins that relay growth-factor signaling, such as ras or myc, is present in the majority of cancers [25-29].
Invasion and metastasis
Cancer cells have the ability not only to grow in the tissue of origin, but also propagate and colonize distant sites. The processes of invasion and metastasis have been compared with the normal physiological processes of embryonic cell migration and wound healing. Several of the molecules responsible for these physiological processes are deregulated in invasive malignancies. Cancer cell migration is regulated by integrins, matrix-degrading enzymes, and cell-to-cell adhesion and communication molecules. In particular, the adhesion molecule E-cadherin has emerged as a key regulator of metastasis initiation. Loss of E-cadherin expression in the primary tumor weakens epithelial cell-cell contacts and is thought to allow the release of invasive cells [30].
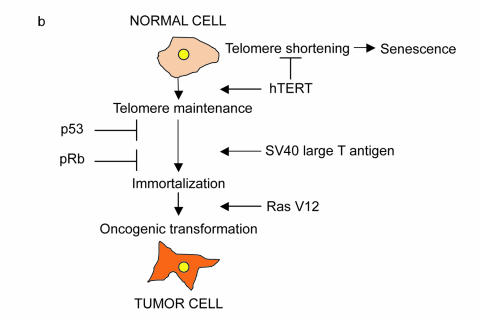
Experimental evidence supports that the pathways described above play a key role in tumorigenesis. Most notably, in vitro models with human cells have been able to reconstitute cancer progression using defined molecular alterations. It was found that at least four pathways must be altered in order for tumor progression to occur: maintenance of telomere length (achieved by expressing human telomerase), deregulated cell-cycle entry (inactivation of Rb), deregulated cell growth arrest and apoptosis (inactivation of p53), and growth-factor independence (by oncogenic ras overexpression) [31] [Figure 1a and Figure 1b]. It remains to be explored how different oncogenes and tumor-suppressors found in tumor samples contribute to these cancer pathways and how they interact with each other to reinforce their tumorigenic potential.
Figure 1a.
Schematic representation of the “Hallmarks of Cancer” proposed by Hanahan and Weinberg. Cancer arises by the step-wise accumulation of mutations in key signal transduction pathways that lead to the acquisition of a common set of capabilities. While all cancers share a common set of properties, the particular combination of mutations that allow unrestrained growth will be tumor-specific.
Figure 1b.
Genetic pathways required for the transformation of human cells. Deregulation of four genetic pathways is sufficient to convert normal human cells to cancer cells in vitro. Expression of the SV40 protein large T antigen antagonizes Rb and p53 function, thus allowing unrestrained cell cycle entry and resistance to apoptosis. Overexpression of telomerase maintains telomere length while activated Ras (RasV12) confers cells with growth-factor independence.
The Cancer Platform Model
The multi-stage model of cancer postulates that a series of sequential events that progressively bypass cellular growth-control mechanisms are needed for tumorigenesis. In contrast, the cancer platform model posits that the principles governing cancer initiation can be further reduced to two interdependent conditions — stimulation of proliferation with a simultaneous block of cell death within a single cell. In this model, proliferation and death pathways are linked not only in tumor initiation, but also in normal development.
Early experiments with transforming retroviruses showed overexpression or activation of oncogenes frequently led to growth arrest or apoptosis. To date, most oncogenes have been found to either sensitize cells to cell death, directly cause cell death, or promote growth arrest. For example, activation of ras in rat embryo fibroblasts leads to growth arrest, while activation of src and myc promotes cell death [32-34]. These observations suggest that uncontrolled cellular proliferation could trigger these pathways as a means to control unrestrained growth. However, proliferation and death also are part of normal development, and a high-proliferative rate is required for certain developmental periods. Therefore, a cell would have to be able to distinguish between a normal high proliferative rate and the abnormal cell proliferation characteristic of cancer cells in order to appropriately activate cell death pathways in response to excessive cell growth.
During development, environmental cues — most importantly, growth factor and nutrient availability — determine whether a cell is able to proliferate. Thus, it was proposed that the activation of pathways leading to cellular growth stimulates both proliferation and death; only when trophic environmental factors that support growth by blocking cell death are present is a cell able to proliferate. The ability to stimulate both proliferation and death has been recognized for several oncogenes, including c-myc, ras, e2f, v-jun, CDKs and cyclins. Furthermore, it was found that in fibroblasts, myc-induced apoptosis could be inhibited by serum or IGF and that E2F induced high rates of cell death in the absence of serum. These results suggest environmental signals are indeed crucial for oncogenic stimulation of proliferation [35,36]. The interdependence between proliferation and death could represent an evolutionary response against cancer progression.
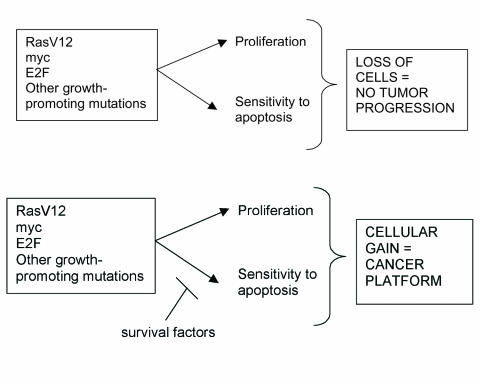
According to the cancer platform model, cancer is a rare occurrence due to the low statistical probability of one cell gaining two mutations simultaneously in a single, rate-limiting step of cancer initiation. Once a “cancer platform” of uncontrolled cellular expansion is established, the interaction of this expanding cellular mass with its environment will give rise to subsequent mutations that allow for other traits of malignancies to develop: angiogenesis, immune evasion, invasion, and metastasis. In addition, since apoptosis is frequently triggered in DNA-damage response pathways to eliminate unwanted cells, blocking apoptosis can lead to an increase in mutation rates. Finally, several of the characteristics of cancer cells may reflect intrinsic properties of proliferating cells and expanding tissues, rather than the accumulation of new mutations, as well as properties of the cells and/or tissue of origin. [37,38]. For example, in comparison to more differentiated cells, oncogenic mutations in a stem cell may give rise to more malignant, undifferentiated cancers with self-renewal capacity [Figure 2].
Figure 2.
Signals that induce cell proliferation simultaneously engage the apoptosis machinery unless pro-survival factors are present. Oncogene activation and tumor-suppressor loss stimulate proliferative and apoptotic pathways leading to a net loss of cells. Cancer will arise only when a cell sustains mutations that simultaneously promote proliferation while blocking cell death (or providing survival factors) thus providing a cancer platform.
Both models discussed above propose that the complexity of the cancer phenotype can be reduced to a small set of common pathways that must be deregulated for cancer progression. The cancer platform model identifies two processes of key importance in tumorigenesis: cellular proliferation and death. This has important therapeutic significance since it implies that cancer cells could be eliminated by targeting either the oncogenic lesion that confers proliferative advantage to the tumor cells or the apoptotic pathways deregulated in these cells. Understanding the mechanisms of oncogenic stimulation of proliferation and death is important to dissect specific cancer-initiation pathways and to develop therapeutics.
Genetic Pathways in Cancer
Even though the same processes seem to be deregulated in all cancer cells, tumors arising in different cells or tissues may preferentially deregulate specific pathways that contribute to these processes. In addition, specific oncogenes or tumor-suppressors may be more frequently mutated or exhibit altered expression in some tissues. It is possible there is a genetic signature in different types of cancer determined both by the tissue and cells of origin and by the oncogenic and tumor-suppressive lesions it has undergone. Moreover, if deregulation of an oncogene activates a specific cell death pathway, it is likely that tumors in which this oncogene is deregulated also successfully have blocked that pathway. While mutations occur at random, once the first (or first two) lesions have been selected for and fixed in a clonal population, the new mutations the tumor acquires could be influenced by external environmental selection pressures as well as internal selection pressures of the mutations already selected. Thus, tumor initiating mutations may predict what types of mutations may occur later in the life of a tumor.
Colon cancer is one of the few malignancies for which a genetic pathway has been defined. Colorectal cancers follow a defined histological pattern of development from adenomas to carcinomas; each of these histological changes is accompanied by mutations in specific genes in a large percentage of tumors [39]. More recently, microarray analysis has been used to generate expression-based classifications of different tumor types. Most tumors show characteristic expression signatures recognizable both for individual tumors and for tumor families with shared characteristics. Further, molecular classification of tumors has revealed different tumors show similarities that can be ascribed to the tissue or cell-type of origin. In addition, tumors’ molecular signatures can be grouped to predict clinical outcome. For example, analysis of histologically indistinguishable breast cancer samples identified four subgroups: ER+-luminal like, HER2+, normal breast, and basal-like; of these four, the last was a predictor of poor outcome. These findings suggest there are subsets of mutations that correlate with specific types of cancer, as well as subsets of genes that correlate with the degree of malignancy of specific tumors [6,40].
The concept that there is a genetic signature to cancer is compatible with all of the models discussed so far. In principle, it would be possible to describe pathways for tumors in different tissues and with different cellular origins, which could predict outcome and help design specific therapies. The existence of genetic pathways may imply that late-stage tumors are still dependent on the original lesions for survival. Alternatively, new mutations may not be influenced by earlier ones. Once a specific process is thwarted, as in the bypassing of barriers against uncontrolled proliferation, new mutations are selected independently of the original mutations. If the first approach is correct, understanding tumor-initiating events in the context of different molecular lesions will be crucial to develop effective cancer therapies.
Oncogenes as Therapeutic Agents
If tumors remain dependent on their initial transforming oncogenic mutations for growth and survival, oncogene inactivation could lead to tumor regression, even in malignant cancers. This hypothesis has been tested using inducible mouse models of cancer [41]. In particular, several studies evaluating the overexpression of the myc oncogene in lymphoid and epidermal tissues showed that the inactivation of myc led to sustained tumor regression with concomitant promotion of either differentiation or apoptosis [42-45]. However, in other models, a fraction of tumor cells were found to be refractory to myc inactivation; these cells presumably had acquired new mutations that allowed myc-independent growth [46-49], suggesting that while mutations that give rise to tumors are often interdependent, new lesions also can arise independently of pre-existing ones, often replacing their function. Therefore, targeting tumor-initiating mutations may not eliminate all tumor cells.
Metastasis represents the main cause of treatment failure for cancer patients, since even complete resection of the primary tumor can leave behind undiscovered micrometastases. The traditional model of metastatic progression postulates that only a small subset of cells from the primary tumor have acquired the requisite mutations to metastasize to distant sites, where new mutations are accumulated as a response to the different selective pressures of a novel environment [50]. However, recent data suggest that most cells in primary tumors with metastatic potential already contain the lesions necessary for metastasis and, possibly, for survival in a foreign environment. Microarray analysis compared patterns of gene expression in lymph node-negative breast cancer patients with their known five-year survival and recurrence rates. Seventy genes were identified that could predict clinical outcome with a combined 83 percent accuracy [51]. In addition, it was found that solid tumors of different origin shared the same metastatic signature, implying there is a common set of molecules regulating metastasis in a variety of primary tumors [52,53]. If this model is correct, it follows that mutations involved in tumor initiation also may be predictive of clinical outcome. The ability to identify and understand the molecular signatures of metastatic and non-metastatic primary tumors would provide new prognostic markers. In addition, if mutations that confer metastatic potential are present in the primary tumor, and if metastatic lesions remain dependent on the original oncogenic mutations for their survival, targeting these genes also may be an effective therapy against metastatic spread. Delineation of the genetic pathways involved in specific tumors will be crucial for identifying these initial oncogenic mutations.
Outlook: Differentiation as an Element of Tumor Initiation
Different models of cancer initiation have focused on deregulation of proliferation and cell death as the main engines of cancer progression. However, impaired differentiation is a characteristic of most cancers, as a decrease in the degree of differentiation correlates with highly malignant lesions. Several oncogenes have been shown to regulate cell-fate decisions. Thus, depending on the cellular context, oncogenes can promote not only proliferation and death, but also differentiation, which can act as a failsafe mechanism against unrestrained growth [54]. Expression of c-myc in bone marrow cells leads to a loss of cell-renewal activity in hematopoietic stem cells leading to differentiation [55]; ras and src are highly expressed in developing neurons and their overexpression leads to neurite outgrowth in PC12 cells [56-59]. In addition, oncogene activation does not always lead to cell death and may even protect against it. In these cases, terminal differentiation could be an effective mechanism to thwart tumor progression [60].
The dual cancer platform may not be sufficient for cancer progression in all contexts, and a third axis may be needed: cellular differentiation. In this expanded model, only when oncogene-induced differentiation effectively is blocked by additional mutations or when the cellular environment fosters the proliferating function of the oncogene will tumors arise. Promoting proliferation while simultaneously preventing differentiation thus may constitute in specific situations a sufficient platform for cancer expansion. In others, the simultaneous blockade of apoptosis and differentiation, together with the promotion of proliferation, may be needed to establish a cancer platform. Mutations that block cellular differentiation likely will have oncogenic capabilities in the context of molecular lesions that deregulate proliferation and prevent cell death. Identification of genes responsible for cell-fate determination may thus provide new insights into mechanisms of cancer initiation as well as provide novel targets for cancer therapies.
References
- Screening for prostate cancer: recommendation and rationale2. Ann Intern Med. 2002;137:915–916. doi: 10.7326/0003-4819-137-11-200212030-00013. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- Troyer DA, Mubiru J, Leach RJ, et al. Promise and challenge: Markers of prostate cancer detection, diagnosis and prognosis. Dis Markers. 2004;20:117–128. doi: 10.1155/2004/509276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Cooper GM. Oncogenes. 2nd ed. Boston: Jones and Bartlett Publishers; 1995. [Google Scholar]
- Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop M. Viral Oncogenes. Cell. 1985;42:23–38. doi: 10.1016/s0092-8674(85)80098-2. [DOI] [PubMed] [Google Scholar]
- Krontiris TG, Cooper GM. Transforming activity of human tumor DNAs. Proc Natl Acad Sci USA. 1981;78:1181–1184. doi: 10.1073/pnas.78.2.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C, Padhy LC, Murray M, et al. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- Shih C, Shilo BZ, Goldfarb MP, et al. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc Natl Acad Sci USA. 1979;76:5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Sinn E, Muller W, Pattengale P, Tepler I, et al. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Slee EA, O'Connor DJ, Lu X. To die or not to die: how does p53 decide? Oncogene. 2004;23:2809–2818. doi: 10.1038/sj.onc.1207516. [DOI] [PubMed] [Google Scholar]
- Masutomi K, Hahn WC. Telomerase and tumorigenesis. Cancer Lett. 2003;194:163–172. doi: 10.1016/s0304-3835(02)00703-6. [DOI] [PubMed] [Google Scholar]
- Hiyama E, Hiyama K. Telomerase as tumor marker. Cancer Lett. 2003;194:221–233. doi: 10.1016/s0304-3835(02)00709-7. [DOI] [PubMed] [Google Scholar]
- Kim NW. Clinical implications of telomerase in cancer. Eur J Cancer. 1997;33:781–786. doi: 10.1016/S0959-8049(97)00057-9. [DOI] [PubMed] [Google Scholar]
- Shay JW, Gazdar AF. Telomerase in the early detection of cancer. J Clin Pathol. 1997;50:106–109. doi: 10.1136/jcp.50.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn NH, Moses HL, Stanbridge EJ, editors. Growth factors, tumor promoters, and cancer genes. Triton Biosciences-UCLA Symposium; 1986 April 6-13; Steamboat Springs, Colorado. New York: Liss; 1986. [Google Scholar]
- Feramisco J, Ozanne B, Stiles CD, editors. Growth Factors and Transformation. Vol. 3, Cancer Cells. New York: Cold Spring Harbor Laboratories; 1985. [Google Scholar]
- Guroff G. Oncogenes, genes, and growth factors. New York: Wiley; 1987. [DOI] [PubMed] [Google Scholar]
- Kudlow JE. Biology of growth factors : molecular biology, oncogenes, signal transduction, and clinical implications. New York: Plenum Press; 1988. [Google Scholar]
- Parker PJ, Katan M. Molecular biology of oncogenes and cell control mechanisms. New York: E Horwood; 1990. [Google Scholar]
- Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2005;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, et al. Creation of human tumour cells with defined genetic elements. Nature. 1990;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Hirakawa T, Ruley HE. Rescue of cells from ras oncogene-induced growth arrest by a second, complementing, oncogene. Proc Natl Acad Sci USA. 1988;85:1519–1523. doi: 10.1073/pnas.85.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavoloni N, Inoue H, Sabe H, et al. v-src transformation of rat embryo fibroblasts. Inefficient conversion to anchorage-independent growth involves heterogeneity of primary cultures. J Cell Biol. 1994;126:475–483. doi: 10.1083/jcb.126.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Fanidi A, Evan GI. Oncogenes and cell death. Curr Opin Genet Dev. 1994;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Hueber AO, Evan GI. Traps to catch unwary oncogenes. Trends Genet. 1998;14:364–367. doi: 10.1016/s0168-9525(98)01520-0. [DOI] [PubMed] [Google Scholar]
- Evan G. Cancer — a matter of life and cell death. Int J Cancer. 1997;71:709–711. doi: 10.1002/(sici)1097-0215(19970529)71:5<709::aid-ijc2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW. Cancer revoked: oncogenes as therapeutic targets. Nat Rev Cancer. 2003;3:375–380. doi: 10.1038/nrc1070. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, et al. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1993;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Rudolph B, Hueber AO, Evan GI. Reversible activation of c-Myc in thymocytes enhances positive selection and induces proliferation and apoptosis in vitro. Oncogene. 2000;19:1891–1900. doi: 10.1038/sj.onc.1203508. [DOI] [PubMed] [Google Scholar]
- Beer S, Zetterberg A, Ihrie RA, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, et al. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Berns A. Oncogene addiction: sometimes a temporary slavery. Cancer Cell. 2004;6:535–538. doi: 10.1016/j.ccr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89–96. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Woelfle U, Cloos J, Sauter G, et al. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003;63:5679–5684. [PubMed] [Google Scholar]
- Kalia LV, Gingrich JR, Salter MW. Src in synaptic transmission and plasticity. Oncogene. 2004;23:8007–8016. doi: 10.1038/sj.onc.1208158. [DOI] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CA, Cox ME, Ward DC, et al. c-src and other proto-oncogenes implicated in neuronal differentiation. Mol Chem Neuropathol. 1989;10:1–14. doi: 10.1007/BF02969481. [DOI] [PubMed] [Google Scholar]
- Kremer NE, D'Arcangelo G, Thomas SM, et al. Signal transduction by nerve growth factor and fibroblast growth factor in PC12 cells requires a sequence of src and ras actions. J Cell Biol. 1991;115:809–819. doi: 10.1083/jcb.115.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Beau JM, Tedeschi B, Walter G. Increased expression of pp60c-src protein-tyrosine kinase during peripheral nerve regeneration. J Neurosci Res. 1991;28:299–309. doi: 10.1002/jnr.490280217. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P, Der CJ, Verma IM. ras-induced neuronal differentiation of PC12 cells: possible involvement of fos and jun. Mol Cell Biol. 1989;9:3174–3183. doi: 10.1128/mcb.9.8.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JA. Apoptosis and tumourigenesis. Curr Opin Genet Dev. 2002;12:67–72. doi: 10.1016/s0959-437x(01)00266-0. [DOI] [PubMed] [Google Scholar]