Abstract
Myosin VII (M7) and talin are ancient and ubiquitous actin-binding proteins with conserved roles in adhesion. Talin serves to link membrane receptors to the underlying actin cytoskeleton and forms a complex with M7 in Dictyostelium. The levels of talinA are tightly linked to M7 levels in Dictyostelium. Cells lacking M7 exhibit an 80% decrease in steady-state levels of talinA, whereas increased levels of M7 result in concomitant increases in total talinA. In contrast, changes in talinA levels do not affect M7 levels. Immunoprecipitation reveals that talinA and M7 are associated with each other in membrane fractions. Fluorescence recovery after photobleaching experiments on green fluorescent protein (GFP)-M7 cells expressing different levels of the M7 and talinA show that changes in the overall amounts of these two proteins influences the dynamics of membrane-associated M7. The recovery of GFP-M7 on the membrane is faster in cells lacking talinA and limited in the presence of excess amounts of talinA and M7. These results establish that M7 stabilizes talinA in the cytosol and, in return, talinA regulates the residence time of M7 at the plasma membrane, suggesting that these two proteins are both part of the same dynamic adhesion complex on the plasma membrane.
INTRODUCTION
Efficient cellular migration is critically dependent on timely regulation of cell–cell and cell–substrate contacts. The two general modes of movement used by eukaryotic cells can be defined by the nature of the adhesive contacts that cells make with the substrata. The widely studied fibroblastic mode of migration is characterized by relatively high-affinity focal contacts between the cell and substrata, whereas the amoeboid mode of migration made by lymphocytes and Dictyostelium uses lower affinity, broader contacts that permit rapid movement. The two modes of adhesion employ the same general molecular machinery: a transmembrane adhesion receptor(s) linked to the cortical actin cytoskeleton via one or more adaptor proteins, suggesting that the differences may arise from variations in the strength or duration of the adhesive complex. Talin plays a key role in both fibroblastic and amoeboid motility. Detailed studies in mammalian cells have established its role in activation of integrin binding to its ligand and tension sensing in focal contacts (Nayal et al., 2004) and it is essential for cell–substrate adhesion in amoeboid cells (Niewöhner et al., 1997).
Talin is present in a wide range of organisms, from amoebozoa to humans (Senetar and McCann, 2005), consistent with this protein having an important and highly conserved role in cellular adhesion. It is a large protein (∼230 kDa) comprised of an N-terminal FERM domain, a long central rod and a C-terminal I/LWEQ actin binding domain. FERM domains can be considered a signature module for proteins that have a role in cellular adhesion, including cytoskeletal linker proteins such as ezrin and moesin and regulatory proteins such as focal adhesion kinase. The talin FERM domain binds to NPxY motifs on the cytoplasmic tails of transmembrane adhesion receptors such as integrins, but can also bind directly to membrane phospholipids. In addition to actin and membrane receptors, talin interacts with vinculin and PIPKI gamma in mammalian cells (Nayal et al., 2004). Talin can dimerize and effectively cross-link actin filaments into networks and bundles, suggesting additional roles in the general organization of cortical actin (Goldmann et al., 1997; Zhang et al., 1996). Consistent with its widespread distribution and ability to bind to both adhesion receptors and the actin cytoskeleton, talin is found in a range of different adhesion structures in addition to focal and sites of amoeboid cell contact with surfaces. It is present in podosomes, invasive structures found in transformed cells and the immunological synapse where it has been shown to be important for stabilizing the LFA-1–dependent (lymphocyte function–associated antigen) interaction between T-cells and antigen-presenting cells (Marchisio et al., 1988; Monks et al., 1998; Simonson et al., 2006).
The role of talin in the establishment of and signaling from focal contacts is well-known but is role in cell–substrate contact in amoeboid cells remains poorly understood. The ability of amoeboid cells to move relatively fast is the result of their making low-affinity, transient contacts with surfaces, suggesting either that talin association with integrins is not accompanied by activation of these receptors or that talin may interact with a different class of receptors altogether. Studies of talin function in lower eukaryotes reveals its essential role in the adhesion of amoeboid cells. For example, the social amoeba Dictyostelium discoideum expresses two talin homologues, talinA and talinB, both of which contribute to adhesion (Niewöhner et al., 1997; Tsujioka et al., 2004). TalinA, in particular, has been shown directly to have a critical role in both cell–cell and cell–substrate adhesion. Mutations in talinA result in defects in cellular adhesion, phagocytosis, and cytokinesis. This is similar to what has been found when talin function is disrupted in the higher eukaryotes (Nuckolls et al., 1992; Niewöhner et al., 1997; Priddle et al., 1998; Brown et al., 2002), consistent with a fundamental conservation of talin function in mediating cellular adhesion.
Myosin VII (M7) is the major talinA-binding partner in Dictyostelium (Tuxworth et al., 2005). The class VII myosins are also a family of highly conserved cytoskeletal proteins with roles in adhesion in different organisms (Tuxworth et al., 2001; El-Amraoui and Petit, 2005; Richards and Cavalier-Smith, 2005). Dictyostelium mutants lacking M7 exhibit defects in substrate adhesion and phagocytosis (Titus, 1999; Tuxworth et al., 2001), phenotypes similar to those of the talinA null mutant. Both proteins are found at the leading edge of migrating cells and in filopodia but they localize to the plasma membrane independently of one another (Kreitmeier et al., 1995; Tuxworth et al., 2001, 2005). M7 and talinA appear to be found in an exclusive complex with each other in the cytosol, suggesting that they operate as a complex to generate optimal adhesion during migration upon recruitment to the membrane. The relationship between these two conserved adhesion proteins was further investigated to better understand their potentially shared and individual contributions to adhesion.
MATERIALS AND METHODS
Strains, Cell Growth, and Maintenance
All Dictyostelium strains were maintained using standard methods (Sussman, 1987). Cells were grown on tissue culture plates in HL5 growth medium supplemented with 10,000 U/ml penicillin G (Fisher Scientific, Pittsburgh, PA) and 10 μg/ml streptomycin sulfate (Sigma Chemical Co., St. Louis, MO). The M7 null strain HTD17–1 (in Ax3; Tuxworth et al., 2001), and talinA null strain HG1666 (in Ax2; Niewöhner et al., 1997) were periodically selected in HL5 supplemented with 10 μg/ml Blasticidin S (ICN Biomedicals, Costa Mesa, CA). Transformants expressing green fluorescent protien (GFP)-M7 or GFP-M7 wild-type and mutant tails or tail fragments (see below) in the wild-type Ax2, M7, or talinA null mutant backgrounds were generated as previously described (Tuxworth et al., 2001, 2005) and maintained in HL5 supplemented with 10 μg/ml G418 (Fisher Chemical, Fairlawn, NJ).
GFP-M7 Expression Plasmids
The extrachromosomal GFP fusion expression plasmids for full-length M7 (pDTi112), full-length tail (pDTi35), ΔFERM1/ΔFERM2 tail (pDTi114), and Pro1 region (pDTi183) have been described previously (Tuxworth et al., 2005). GFP fusions of the tail lacking both MyTH4/FERM regions (ΔMFMF; nucleotides 2759–3795 and 4994–5968; amino acids 809-1154 plus 1602–1878), the coil-Pro1 region alone (cP; nucleotides 2759–3795; amino acids 809-1154), the SH3-Pro2 region alone (SP2; nucleotides 5192–5968; amino acids 1620–1878), combined Pro1-Pro2 regions (Pro1–2; nucleotides 3041–3795 and 4994–5968; amino acids 903-1154 plus 1554–1878) and combined Pro1-SH3 regions (PS; nucleotides 3041–3795 and 4994–5371; amino acids 903-1154 plus 1554–1679) of the M7 tail were generated by a combination of PCR (PCR), overlapping PCR, and TOPO-TA cloning (Invitrogen, Carlsbad, CA) using either Ax2 genomic DNA or available genomic clones as templates. All PCR-derived clones were verified by sequencing, and the inserts were ligated in-frame to pTX-GFP, an extrachromosomal expression plasmid that carries the Neo gene, conferring G418 resistance (Levi et al., 2000) resulting in pDTi179 (ΔMFMF), pDTi201 (cP), pDTi203 (SP2), pDTi204 (Pro1–2), and pDTi206 (PS) expression plasmids. Each GFP-expression plasmid was transformed into the appropriate strain and transformants selected for by growth in 10 μg/ml G418, screened for fluorescence, and then analyzed for expression of GFP fusions by Western blotting.
Quantitative Immunoblotting
A total of 3 × 106 cells of each strain were centrifuged at 2700 × g and resuspended in 0.1 ml ULSB (6 M urea, 4% SDS, 20% glycerol, 125 mM Tris, pH 7.5). Samples of 5, 7.5, and 10 μl were adjusted to 15 μl with ULSB and loaded on 6% SDS-PAGE gels. Electrophoresis, transfer to PVDF membrane (Millipore, Bedford, MA), and immunodetection was then performed. Immunodetection of the class I myosin myoB was used as a loading control. Raw data were plotted and linear regression analysis was performed; all R2 values for the analysis exceeded 0.9. Rabbit serum containing antibodies directed against the N-terminal portion of the M7 heavy-chain tail region (UMN87; Tuxworth et al., 2005) was used at 1:1000 for immunoblotting. A rabbit polyclonal antibody specific for the heavy chain of the class I myosin, myoB, was also used at 1:1000 (Novak et al., 1995). A mouse mAb specific for the N-terminus of Dictyostelium talinA, mAb 341 (Niewöhner et al., 1997) was initially a generous gift of Dr. Günther Gerisch (MPI, Martinsreid, Germany) and subsequently obtained from Developmental Studies Hybridoma Bank (Iowa City, IA). The antibody was used without dilution. All of the primary antibodies, with the exception of talinA, were diluted in 0.1% casein in phosphate-buffered saline at pH 7.4 (Bio-Rad, Hercules, CA). Washed blots were incubated with Alexa fluor 680– or 800–conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Molecular Probes, Eugene, OR) and detected and quantified with an Odyssey infared imaging system (LI-COR Biosciences, Lincoln, NE).
Kinetics of talinA Digestion
A total of 1 × 108 cells of wild-type strain Ax2 and M7 null strain HTD17 were washed twice and resuspended in 500 μl MES buffer (2 mM MgSO4, 0.2 mM CaCl2, 20 mM MES, pH 6.8). Cells were lysed on ice with 1% Triton X-100 (Anatrace, Maumee, OH) and then 50 μl removed and mixed with 50 μl ULSB at the indicated time points. These samples were heated at 100°C for 3 min, triplicate samples of increasing volumes were adjusted to 10 μl with additional ULSB and loaded on 6% SDS-PAGE gels as described above. Electrophoresis, transfer to PVDF membrane, and immunodetection was then performed. The data were plotted using Origin software (Rockware, Golden, CO).
RNA Isolation and cDNA Preparation
A total of 1 × 107 cells of wild-type strain Ax2 and M7 null strain HTD17 were resuspended in 1 ml of TRIZOL reagent (Invitrogen) and RNA isolated according to manufacturer's instructions. RNA samples were treated with DNAse I (Ambion, Austin, TX), quantified, and overall quality examined on a 1% agarose MOPS-formaldehyde gel. cDNA was prepared from 5 μg total RNA with Superscript III reverse transcriptase mix (Invitrogen) using a 20-base poly-dT primer. No RT reactions were performed to control for contaminating genomic DNA.
Quantitative PCR
Quantitative PCR was performed with the Roche LightCycler real-time PCR system (Roche Diagnostics, Indianapolis, IN) with Roche FastStart DNA Master mix (SYBR Green I). Optimal annealing temperatures to reduce secondary products were determined for each reaction by temperature gradient PCR, and melting temperature analysis was performed on real-time reactions to ensure that secondary products were not present. Relative amounts of sample RT product were standardized to H7, a control gene (Singleton et al., 1988). Annealing was performed for 5 s at 47°C, and extension time was 14 s. Primers for talA were talA3: 5′-CCATGGTTGCTGCAACAATCGTAGATGC-3′ (nucleotides 7092–7114) and talA4: 5′-CTCGAGTTAATTTTTATTATAATTTTGTTTTCTTG-3′ (nucleotides 7648–7676); primers for H7 were H7S: 5′-ACGTTCAAACTAAATACGGAGCTGGT-3′ (nucleotides 5–30) and H7AS: 5′-TTTGAGTGGTTTGCCAATTTCTTTT-3′ (nucleotides 288–312).
Immunoprecipitation
Total cellular membranes and cytosol were prepared as described previously (Senda et al., 2001). Total membranes were diluted to the volume of the cytosolic fraction with immunoprecipitation lysis buffer (ILB; 25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 10 mM Mg-ATP) and 1% Triton X-100 (Anatrace) and protease inhibitors (PIs; 1 mM Pefablock, 100 μM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone), 100 μM TPCK (N-tosyl-l- phenylalanine chloromethyl ketone), 333 μM E64, and 0.4 μM ALLN) were added to both fractions. To remove proteins that nonspecifically bind the beads, both fractions were incubated sequentially with three separate solutions of protein A Sepharose Fast Flow beads (Amersham Biosciences, Piscataway, NJ). For each incubation, fractions were added to 100 μl of protein A beads that had been washed twice with ILB (25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 10 mM Mg-ATP). The sample was gently mixed at 4°C for 1 h, and then the beads were pelleted by centrifugation at low speed for 3 min. The resulting supernatant was collected for the next incubation with beads. A rabbit polyclonal anti-GFP antibody (Molecular Probes) was added to the precleared supernatant, at a final concentration of 12 μg/ml, and the sample was incubated with gentle mixing at 4°C for 1–12 h. The antibody-supernatant solution was then added to 100 μl of protein A beads that had been washed with ILB, and the slurry was incubated with gentle mixing at 4°C for 6 h. The beads were then collected by gentle centrifugation, and the supernatant was saved for analysis. The bead complex was then washed several times with ILB containing PIs, and the final bead pellet was carefully drained of all excess liquid. A total of 30 μl of ULSB was added to the beads, and the samples were heated at 100°C for 3 min and then applied to a 6% SDS-PAGE gel. The gels were then either transferred to PVDF membrane (Millipore) for immunoblotting or stained with silver.
Phagocytosis and Adhesion Assays
A modified small-scale version of the standard phagocytosis assay (Vogel et al., 1980; Tuxworth et al., 2001) using 1-μm fluorescent latex beads (Polysciences, Warrington, PA) was performed. In brief, a total of 1 × 106 cells in a 900 μl volume were shaken in phosphate buffer (16.6 mM phosphate, pH 6.1) at 150 rpm in each well of a 24-well plate at room temperature for 1 h. A 200-fold excess of 1.0-μm fluorescent latex beads in 100 μl was then added; this point defined as time = 0, and 75 μl of cell suspension was removed at various times and added immediately to 5 ml of ice-cold stop solution (0.04% sodium azide in phosphate buffer). After sample collection, cell suspensions were spun at 1140 × g for 10 min, and all but 200 μl of supernatant was removed. Cells were resuspended in the remaining liquid and quantified using a FACSCalibar flow cytometer (Benton-Dickson, Franklin Lakes, NJ).
Bead binding was measured using a slightly modified bead adhesion assay (Tuxworth et al., 2001). Equal numbers of axenically growing wild-type, M7 null, or cP-expressing cells were seeded at subconfluent density for 30 min onto glass coverslips at 20°C. The coverslips were then transferred to 4°C for 30 min. Latex beads of 4.0-μm diameter were washed once and diluted to a density of 6.0 × 106 particles/ml in ice-cold HL5. After the cells were chilled, the media covering them was replaced with 300 μl bead suspension and incubated at 4°C for 15 min. Cells were then fixed for 10 min by replacement of the bead suspension with 300 μl picric acid fixative (Humbel and Biegelmann, 1992). No wash was necessary to remove nonadherent particles. The cells with adhered beads were visualized using a 63× DIC (differential interference contrast) objective mounted on a Zeiss Axiovert microscope (Carl Zeiss MicroImaging, Thornwood, NY). At least nine areas were selected at random in each experiment, and every cell in the field of view was analyzed. A modified plate adhesion assay was performed as described (Fey et al., 2002; Bukharova et al., 2005). In brief, a total of 2.8 × 106 cells total were seeded into each well of a six-well plate and allowed to attach for 15 min at room temperature. The growth medium was replaced with 2 ml of starvation buffer, and the samples were incubated for another 1 h before placing on a shaker at 150 rpm and counting the number of detached cells over time using a Coulter counter (Beckman Coulter, Fullerton, CA).
Photobleaching
Growth-phase cells were seeded into a custom stainless steel chamber (Tuxworth et al., 2001) at a density of 0.5–2 × 106 cell/ml in starvation buffer (16.6 mM phosphate, pH 6.1) for 1–2 h before each experiment. The chamber was inverted, and cells were visualized on an inverted Nikon TE-200 microscope equipped with a 63× 1.4 NA TIRF (total internal reflection fluorescence) objective and a 20 mW argon laser (Melles Griot, Carlsbad, CA) with 488-nm and 514-nm lines. A 488-nm filter was used to remove the 514-nm line, and all experiments were performed at full power, with measured laser intensity at the focal plane of 1.2–1.5 mW. Single pulses of 10 ms were applied to cells and changes in fluorescence intensity in the bleached area (an ∼0.5-μm spot) monitored with respect to time after background subtraction. Images were captured every 100 ms using a Cascade 2 Digital camera (Photometrics, Tucson, AZ), and all data were collected at 14 bits gray depth. The data were fit with curves using Origin graphical analysis software (Originlab, Northhampton, MA). The time course of recovery was fitted to the following equation for a single exponential rise to maximum:
where A is the amplitude of the exponential, k is the rate constant, t is time (in milliseconds), and t0 is the time of the bleach. For double exponential rises to maximum, the following equation was used for fitting:
where A and B are respective amplitudes and k1 and k2 are rate constants. The half-time for recovery (t1/2) for a given rate constant k was calculated using the following equation:
 |
The fractional recovery was determined using the following:
RESULTS
TalinA Protein Levels Are Directly Correlated to M7 Tail Expression
TalinA and M7 form a complex in the Dictyostelium cytosol (Tuxworth et al., 2005) and, consistent with an intimate relationship between these two proteins, there is an observed decrease in the overall levels of talinA in the absence of M7 (Gebbie et al., 2004; Tuxworth et al., 2005). The extent of talinA decrease in the M7 null mutant was investigated as a first step toward understanding the dependence of talinA levels on M7. Quantitative immunoblotting of talinA and M7 using whole cell lysates from wild-type and M7 null mutants was carried out and the levels of each protein relative to myoB, a class I myosin that served as a loading control and then compared (Figure 1A). Cells lacking M7 have a significantly reduced amount of talinA, down to 17% of wild-type levels (Figure 1A, Table 1).
Figure 1.
The level of M7 expression affect TalinA levels. The levels of M7 and talinA, relative to the loading control myoB, were determined by quantitative Western blotting of total cell lysates. The antibody used to probe each set of blots in indicated on the left. (A) Comparison of talinA levels in wild-type Ax2 and M7 null cells. Numbers above each lane indicate sample volumes loaded (in μl). (B) TalinA levels in cell lines expressing different levels of either GFP-tagged full-length M7 (GFP-M7), or tail in wild-type or M7 null cells or in talinA null cells. Note that the top panel shows immunoblotting with the M7 antibody.
Table 1.
TalinA levels are linked to M7 levels
| M7 levels (%) | TalinA levels (%) | |
|---|---|---|
| Wild type | 100 | 100 |
| M7 null | 0 | 17 |
| M7 tail in M7 null | 185 | 108 |
| GFP-M7 in M7 null | 315 | 185 |
| GFP-M7 in wild type | 679 | 571 |
| M7 tail in wild type | 315 | 302 |
| TalinA null | 108 | 0 |
Average levels of M7 and talinA protein in different cell lines. In all cases, wild-type levels are defined as 100%, and all protein measurements were standardized to levels of myoB as a loading control. Percentages are the average of three independent experiments.
The reduction in talinA levels could occur as a result of feedback mechanism that controls talinA gene expression or causes increased susceptibility to degradation. TalinA gene expression was first examined using real-time qPCR to measure steady-state talA mRNA levels. The talA mRNA levels in wild type and in the M7 null mutant are identical (99 vs. 100%, n = 3), demonstrating that the observed decrease in cellular talinA levels is not simply due to decreased gene expression. If M7 were required to directly stabilize talinA, then the turnover rate of talinA should be increased in the M7 null cells. It was not possible to assess this directly using a standard pulse-chase analysis because of the lack of an antibody suitable for talinA immunoprecipitation. An alternate approach of examining talinA degradation kinetics over the course of one hour in whole cell detergent lysates from wild-type and M7 null cells was instead used (Figure 2). Nearly all of the talinA was lost by 60 min in wild-type lysates, with a 50% decrease observed at 18 min. In contrast, talinA levels in the M7 null cell lysates were reduced significantly faster, with a nearly complete loss by 10 min. The decay of talinA levels was fit to a single exponential decay curve (Figure 2), and the calculated rate of decay for talinA in wild-type cells was 0.04% min−1 ± 0.01% and 0.30% min−1 ± 0.04% for M7 null cells. Incubation of M7 null cells with either inhibitors of the proteasome or calpain for several hours did not result in an increase of talinA levels (Galdeen and Titus, unpublished observations), suggesting that talin is degraded by a general proteolytic mechanism, possibly due to protein instability. Together, these data indicate that the association of M7 with talinA is essential for maintaining wild-type levels of this protein by protecting it from degradation.
Figure 2.
TalinA degradation occurs faster in M7 null cell lysates than in wild-type lysates. (A) Representative Western blots showing changes in talinA levels in wild-type Ax2 and M7 null detergent lysates incubated at 4°C for up to 1 h. (B) Time course of talinA degradation in total cell lysates. The levels in each sample were standardized to time = 0 (100%). Data points from seven separate experiments were plotted together and exponential decay regression analysis was performed for wild-type Ax2 (dashed line) and M7 null (solid line) samples. □, levels of talinA in wild-type Ax2 cells (WT); ○, levels in M7 null cells.
TalinA Levels Are Closely Linked to M7 Levels
The finding that cells lacking M7 have significantly reduced levels of talinA suggested that the overall levels of talinA are limited by the amount of available M7. A series of cell lines expressing increased amounts of either full-length M7 or the tail region alone were generated, and talinA levels in each were quantified, as above. Increases in M7 levels were accompanied by increases of talinA levels (Figure 1B, Table 1). Wild-type cells expressing about a sevenfold excess of M7 were found to have an approximately sixfold increase in talinA. Expression of the tail domain of M7 alone in wild-type cells also resulted in an increase in total levels of talinA (Figure 1B, Table 1), indicating that this region was sufficient to stabilize talinA. Expression of either the full-length M7 or the tail alone in M7 null cells resulted in a restoration of near wild-type levels of talinA (Table 1). The dependence of talinA levels on M7 contrasted with the observation that the talinA null cells have wild-type levels of M7 (Table 1), clearly showing that costabilization was not occurring. In fact, a slight excess of M7 was observed in cells expressing GFP-M7. These results revealed that M7 levels determined the total cellular concentration of talinA and when combined with previous observations that cytosolic M7 and talinA are fully complexed with each other (Tuxworth et al., 2005) suggested that the cell regulates the amount of talin necessary to maintain the correct stoichiometric ratio with M7.
A Minimal Portion of the M7 Tail Responsible for talinA Stabilization
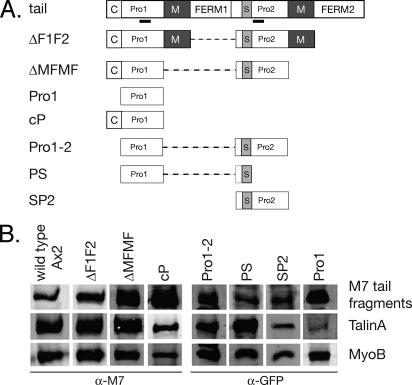
Prevention of talinA turnover by M7 could occur by masking of protease-sensitive sites, either by directly binding to these sites or through steric blocking by adjacent M7 domains. Protection of talinA could alternatively be accomplished by M7 binding, inducing a conformational change in talinA that stabilizes the protein. A series of cell lines were generated to identify the region of the M7 tail that confers proteolytic protection and gain information concerning the prevention of talinA degradation by M7.
The talinA binding site on M7 resides in the N-terminal region of the tail (Tuxworth et al., 2005), a 253-amino acid proline-rich region (Pro1) located between the coil domain and the first MyTH4/FERM repeat (Figure 3A). Quantitative Western blotting of M7 null cells expressing GFP-tagged Pro1 reveals that talinA levels are not restored (Figure 3, Table 2). A series of different GFP-tagged M7 tail fragments were then expressed in the M7 null strain to identify the minimal region necessary for effective talinA stabilization. Expression of a tail lacking both MyTH4/FERM domains was capable of restoring near wild-type levels of talinA (Table 2). The N-terminal region of the tail comprising both the region of predicted coil and Pro1 (cP) or Pro1 fused to the centrally located SH3 and Pro2 domains (Pro1–2) restored almost wild-type levels of talinA (Table 2, Figure 3B). However, the SH3-Pro2 domain alone does not stabilize talinA. These results suggest that steric hindrance is responsible for talinA stabilization and that the precise sequence required is somewhat promiscuous but requires the minimal talinA-binding region, Pro1 (Figure 3, Table 2).
Figure 3.
Identification of the region of the M7 tail that stabilizes TalinA levels. (A) Schematic illustration of full-length M7 and expressed tail fragments indicating major protein domains. Note that all of the fragments were fused to GFP. The region of predicted coiled-coil is labeled “C,” and the MyTH4 domains are indicated by the dark gray box labeled “M.” The two proline-rich regions are labeled “Pro1” and “Pro2,” and the SH3 domain is represented by the light gray box labeled “S.” (B) Immunodetection of M7, talinA, and myoB levels in tail deletion constructs. The antibody used to detect the M7 tail fragments is indicated at the bottom of each set of blots.
Table 2.
Extent of talinA stabilization by M7 tail fragments
| M7 levels (%) | TalinA levels (%) | |
|---|---|---|
| Wild type | 100 | 100 |
| ΔF1F2 | 193 | 87 |
| ΔMFMF | 318 | 90 |
| Pro1 | 160 | 10 |
| cP | 1362 | 81 |
| Pro1–2 | 146 | 148 |
| PS | 59 | 116 |
| SP2 | 160 | 10 |
Average levels of M7 and talinA protein expression in the M7 null strain expressing various M7 tail fragments. In all cases, wild-type levels are defined as 100%, and all protein measurements for the ΔF1F2, ΔMFMF and cP fragments are standardized to myoB. The Pro1–2, PS and SP2 fragments lack the M7 epitope and were detected using an anti-GFP antibody antibody. Relative levels were then determined by correlating the GFP signal to both endogenous M7 and myoB in a wild-type cell line expressing a GFP-M7 tail and comparing that to the GFP-fusions of interest. Percentages are the average of three independent experiments.
M7 Adhesion Defects Are Not Dependent on the Loss of talinA
The finding that cells lacking M7 have significantly lower levels of talinA and the observed differences between the M7 and talinA null phenotypes (e.g., M7 nulls do produce filopodia, whereas talinA nulls do; Tuxworth et al., 2001) raises the question of whether the M7 null mutant adhesion defects can be attributed to the loss of talinA. M7 null cells expressing either the full-length M7 tail or a minimal stabilization domain, cP, were first tested for phagocytic activity because loss of adhesion results in impaired particle uptake (Niewöhner et al., 1997; Tuxworth et al., 2001). M7 nulls expressing the full-length tail have slightly increased phagocytic activity compared with the M7 null mutant but restoration of wild-type levels of talinA (Table 1) does not result in wild-type levels of phagocytosis (Figure 4A). Because previously published data showed that a full-length M7 construct is fully able to rescue the phagocytic defect in M7 null cells (Tuxworth et al., 2001), these data indicate that the M7 motor domain has a critical role in phagocytosis and that M7 phagocytic defects are not solely due to loss of talinA. Similarly, M7 null cells expressing the talinA-stabilizing cP domain exhibit a small recovery of phagocytic activity but are still defective in particle uptake (Figure 4B).
Figure 4.
Restoration of wild-type talinA levels in M7 null cells does not rescue the M7 null mutant phenotype. (A) The uptake of 1-μm fluorescent latex beads was assayed. Cells were incubated with a 200-fold excess of beads, samples were taken at the times indicated, and uptake was measured by flow cytometry. Error bars, SD from three experiments. ■, wild-type Ax2; •, M7 null; ♦, M7 tail in M7 null. (B) The uptake of 1-μm fluorescent latex beads by M7 null cells expressing the cP fragment is shown. Error bars, SD from three experiments. ■, wild-type Ax2; •, M7 null; ♦, cP in M7 null. (C) Binding of 4-μm latex beads at 4°C. The number of beads attached per cell was measured for wild-type (WT; black), M7 null (gray), or cP in M7 null (light gray) cells. The average number of beads/cell was 0.421 (WT; n = 95), 0.09 (M7 null; n = 220), and 0.05 (cP in M7 null; n = 365). (D) A representative assay showing the percentage of cells detached from a six-well plate after shaking at 150 rpm. Shown are the results for wild-type Ax2 (■), M7 null (▩), or cP in M7 null (□) cells.
The reduced phagocytic activity observed in cells that lack full-length M7, yet retain wild-type levels of talinA could be due to reduced adhesion or to another defect, such as dysfunction of phagosome internalization or trafficking. The ability of cP cells to adhere to substrata was measured using two distinct assays: bead binding and plate adhesion. Wild type, M7 null, and cP cells were adhered to a coverslip and incubated with an excess of beads at 4°C, a condition that permits bead binding but not engulfment. The M7 null and cP cells exhibited a significant reduction in bead binding when compared with wild-type cells (Figure 4C). Similarly, M7 null and cP cells attached to tissue culture plastic were more readily released from the plate when subjected to shaking (Figure 4D). These data establish that restoration of wild-type talinA levels in the M7 null cells is not accompanied by a recovery of substrate adhesion and affirm the role of M7 in cell–substrate adhesion.
TalinA Modulates M7 Membrane Dynamics
The similar adhesion defect of the M7 and talinA null mutants suggests that M7 and talinA may be associated with each other at the plasma membrane where their interaction could be required for proper function of an adhesion complex. The existence of a M7-talinA complex in membrane fractions was investigated using an immunoprecipitation (IP) approach. Cytosol and total membrane fractions were isolated from GFP-M7–expressing cells using a sucrose step gradient. The membrane fraction was solubilized with Triton X-100, and the M7 was immunoprecipitated from both fractions using an anti-GFP antibody. Two high-molecular-weight bands are present in both the cytosol and membrane IP pellets, one at the predicted ∼300 kDa size of GFP-M7 and a second band migrating slightly below that, at the ∼260 kDa size of talinA. Immunoblotting with either a M7 or talinA antibody confirms that the higher molecular weight band is M7 and the lower one is talinA (Figure 5). Additional IP experiments using a variety of detergents to solubilize the membrane fraction were performed and the same results obtained: talinA is the only protein that coprecipitates with GFP-M7 (Stephens and Titus, unpublished observations). Taken together with the finding that GFP-M7 is localized only to the plasma membrane of cells (Tuxworth et al., 2001), this result suggests the existence of a plasma membrane–associated M7-talinA complex.
Figure 5.
M7 and talinA coprecipitate from isolated total membranes. Detergent lysates of a total membranes fraction from cells expressing GFP-M7 were incubated with a GFP-specific antibody, and the bound protein precipitated with protein A beads. The immunoprecipitated pellets were analyzed on a 6% SDS-PAGE gel stained with silver or by immunoblotting with specific M7 and talinA antibodies (shown at the bottom of the panel). The positions of known molecular mass standards, in kDa, are indicated to the left of the gel.
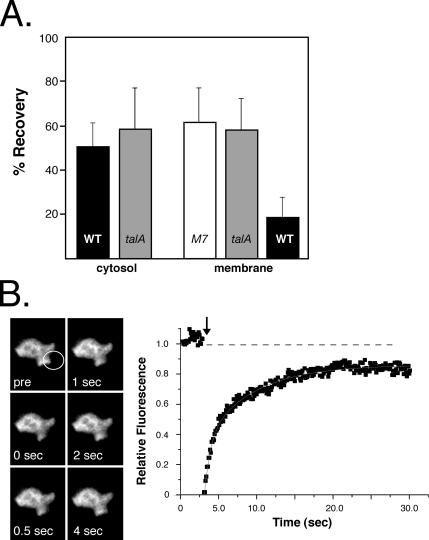
M7 and talinA are both localized to the leading edge of migrating cells, but independently of each other (Niewöhner et al., 1997; Tuxworth et al., 2001, 2005). The finding that talinA and M7 are complexed with each other in membrane fractions suggests the possibility that the association of M7 with talinA at the plasma membrane affects the dynamics of membrane association. Fluorescence recovery after photobleaching (FRAP) is a useful method for assessing protein dynamics both in the cytosol as well as on intracellular membranes. FRAP experiments were undertaken to determine if the intracellular mobility or turnover on the plasma membrane of M7 is influenced by association with talinA. The recovery of cytosolic GFP-M7 was first examined to determine if association with talinA affects the free diffusion of M7. An ∼0.5-μm spot in the cytosol of either wild-type or talinA null cells expressing GFP-M7 was photobleached, and the extent and half-time of fluorescence recovery was determined. The fluorescence level in neighboring, nonbleached cells was also measured to determine if general bleaching occurred during the course of the analysis because of exposure to fluorescence during the observation period. Quantification of cytosolic fluorescence levels in nonbleached cells did not reveal any decrease in total fluorescence levels. Bleach recovery in the cytosol of both strains is ∼49% and ∼59% of prebleach levels, respectively (Figure 6A). The data were fit with a single exponential and the t1/2 of recovery for cytosolic GFP-M7 in wild-type (1.0 ± 0.4 s) and talinA null (0.7 ± 0.4 s) do not significantly differ, indicating that the cytosolic diffusion of M7 is essentially the same regardless of whether it is in a complex with talinA (Table 3).
Figure 6.
Recovery of GFP-M7 fluorescence at the leading edge. The overall recovery levels for each cells line and representative images and curves of a FRAP analysis of GFP-M7 membrane dynamics are presented. (A) Comparison of the percent recovery of fluorescence in either the cytosol or at the membrane in wild-type Ax2 (WT), M7 null (M7), and talinA (talA) null cells. The number of samples ranged from 8 to 15 per cell line, and the SD is shown. (B) FRAP of GFP-M7 expressed in M7 null cells. Shown are selected images demonstrating recovery of fluorescence at the leading edge before (pre) and at select times after the bleach (left). A time course of fluorescence recovery at the leading edge of the same cell is presented. The initial value for the unbleached leading edge is set to 1.0 (dashed line), and the value immediately after bleaching is set to 0. The arrow indicates the time of the bleach. The light gray line is the calculated curve that fits the experimental data.
Table 3.
Recovery rates of GFP-M7 on the plasma membrane are affected by TalinA levels
| Fast t1/2 | Slow t1/2 | |
|---|---|---|
| Wild-type cytosol (n = 8) | 1.0 ± 0.4 | n.a. |
| talinA null cytosol (n = 8) | 0.7 ± 0.4 | n.a. |
| GFP-M7 in M7 null (n = 15) | 5.2 ± 1.6 | 45.2 ± 10.7 |
| GFP-M7 in talinA null (n = 10) | 3.6 ± 1.5a | 26.7 ± 13.1 |
| GFP-M7 in wild type (n = 8) | 2.8 ± 1.7b | n.a. |
Values are average half-times (t1/2, expressed in seconds) of maximal fluorescence recovery in wild-type and M7 null cells. n.a., not applicable.
a p = ≤ 0.018 when compared with GFP-M7 in M7 null.
b p = ≤ 0.001 when compared with GFP-M7 in M7 null.
GFP-M7 is recruited to the leading edge of migrating cells and this localization is maintained in cells moving in a persistent manner (Tuxworth et al., 2001), making it possible to determine the dynamics of M7 membrane association. As described above, an ∼0.5-μm spot that included the leading edge was photobleached, and the extent and rate of fluorescence recovery only at the membrane—measured in GFP-M7 cells expressing different levels of talinA and M7-talinA null cells (M7 alone), M7 nulls (1.8× talinA and M7), and wild-type cells (∼6× talinA and M7; Table 1)—were examined (Figures 6 and 7). The total fluorescence recovery at the membrane in M7 null cells expressing GFP-M7 occurs to a similar overall extent as that of the cytosol (62.3 ± 15.5%; n = 15; Figure 6A). Fitting of the recovery curve revealed that it is biphasic with a fast phase t1/2 of 5.2 ± 1.6 s and a slow phase t1/2 of 45.2 ± 10.7 s (Table 3). The majority of fluorescence recovery occurred uniformly within the bleached region, suggesting that it occurs via recruitment of cytosolic M7 instead of lateral diffusion from the membrane adjacent to the spot. The initial fast rate of recovery accounts for the bulk of recovery and most likely reflects this rebinding of cytosolic M7. There may also be a relatively small amount of recovery occurring by lateral diffusion from the adjacent membrane, and this would be consistent with the second observed, slower t1/2 (Table 3). Interestingly, the same extent of recovery (58.4 ± 13.3%; n = 10) is also observed for GFP-M7 in a talinA null background (Figure 6A), yet these cells exhibit a more rapid biphasic recovery of M7 at the membrane when compared with M7 null cells. A fast phase t1/2 of 3.6 ± 1.5 s, and a slow phase t1/2 of 26.7 ± 13.1 s are observed (Table 3). These results show that the presence of talinA slows the turnover of M7 at the plasma membrane.
Figure 7.
TalinA influences GFP-M7 membrane dynamics. The membrane dynamics of GFP-M7 expressed in wild-type cells and talinA null cells was analyzed by FRAP. (A) An example of the FRAP of GFP-M7 expressed in talinA null cell. (B) Representative FRAP experiment of wild-type cells expressing GFP-M7 showing selected images that illustrate the minimal recovery after bleach (left). Quantitative analysis of the time course of fluorescence recovery at the leading edge of the same cell is presented. The initial value for the unbleached leading edge is set to 1.0, and the value immediately after bleaching is set to 0. The arrow indicates the time of the bleach and the dashed line 1.0. The light gray line is the calculated curve that fits the experimental data.
Interestingly, FRAP of GFP-M7 expressed in wild-type cells that have a sixfold excess of talinA and M7 compared with control cells (Table 1) reveals minimal recovery of the bleached spot. Only an average 19.3 ± 7.8% (n = 8) recovery of fluorescence was observed (Figure 6A), and the bleached area clearly persists over the course of the observation (Figure 7B). The modest recovery observed is rapid, and the resulting curve is fit only by a single exponential with a t1/2 of 2.8 ± 1.7 s (Table 3). The recovery time is similar to that of the fast phase observed in the talinA nulls and could be due to binding of M7 to the membrane in the absence of talinA, consistent with the observation that there is a slight excess of M7 over talinA in this cell line (Table 1). Note that it was not possible to follow the behavior of the bleached spot for extended periods of time because the leading edge containing M7 is often retracted within 20–30 s. Taken together, these data reveal that talinA, a protein that is protected by M7, contributes to increasing the residence time of M7 at the plasma membrane.
DISCUSSION
The interaction of migrating cells with the substrate must be finely tuned to achieve optimal translocation. Talin is a key player in adhesion in multicellular organisms where it has critical roles in linking integrins to the actin cytoskeleton in migrating cells, acting as both a tension sensor and a mediator of “inside-out” signaling (Nayal et al., 2004). Talin is an ancient and ubiquitous protein (Senetar and McCann, 2005) and in simpler systems such as Dictyostelium, it also has a significant role in cell–substrate adhesion (Niewöhner et al., 1997). Here it is shown that talinA influences the membrane dynamics of another conserved adhesion protein, M7, in Dictyostelium. These two proteins are associated with each other both in the cytosol and on the membrane, as revealed by coIP experiments (Tuxworth et al., 2005; Figure 5). FRAP analysis of cells expressing various levels of M7 and talinA reveals that the lifetime of GFP-M7 on the membrane is significantly increased when an excess of M7 and talinA is present (Figures 6A and 7B, Table 3). This decreased recovery rate is unlikely to be due simply to an increase in the levels of GFP-M7 as recovery is actually faster in talinA cells that have endogenous levels of M7 plus ectopically expressed GFP-M7. Thus, decreased exchange correlates with increased amounts of the talinA/M7 complex.
Talin regulates adhesion protein complex dynamics at the plasma membrane. In mammalian cells, talin1 is specifically cleaved by calpain 2 at a site C-terminal to the FERM domain, and mutations in the site result in the increased lifetime of talin in focal adhesion complexes in cultured cells, indicating that this cleavage releases talin1 from focal complexes (Franco et al., 2004). The persistence of talin1 in adhesion complexes is accompanied by reduction in the turnover of adhesion complex components such as zyxin (Huang et al., 2003). The observation that M7 turnover at the plasma membrane is slowed in cells that have increased levels of the talinA is consistent with a conserved role for talins in controlling the dynamics of adhesion complex components in both simple and complex organisms. It will be interesting to determine if the mechanism of controlling talinA dynamics is shared with other systems or if the requirement for calpain cleavage is found only in higher eukaryotes. Although the molecular details of how talin controls the dynamics of cellular adhesion may depend on the system or cell type, it seems likely that the basic mechanism will be conserved.
The role of talin in clustering and activating adhesion receptors in a number of cell types has been well described (Nayal et al., 2004), yet little is known about control of the cytosolic pool of available talin. The work described here reveals that steady-state levels of this critical adhesion protein are tightly regulated by its association with M7, its major binding partner in Dictyostelium. It interesting to note that there is no detectable free talinA in the Dictyostelium cytosol of wild-type cells—all of it appears to be present in a complex with M7 (Tuxworth et al., 2005). In addition to determining the overall levels of talinA, it is possible that binding to M7 in the cytosol may prevent potentially deleterious effects of free talin. For example, uncontrolled binding of the I/LWEQ region to actin may cause general disruption of the cytoskeleton (Weber et al., 2002). The association with M7 in the cytosol could be required to maintain talinA in the “off” state to prevent this possibility. Alternatively, keeping talinA together in a complex with M7 may coordinate the recruitment of these two functionally related linker proteins to the membrane as a means of efficiently initiating adhesion and subsequently recruiting the additional, necessary adhesion complex components.
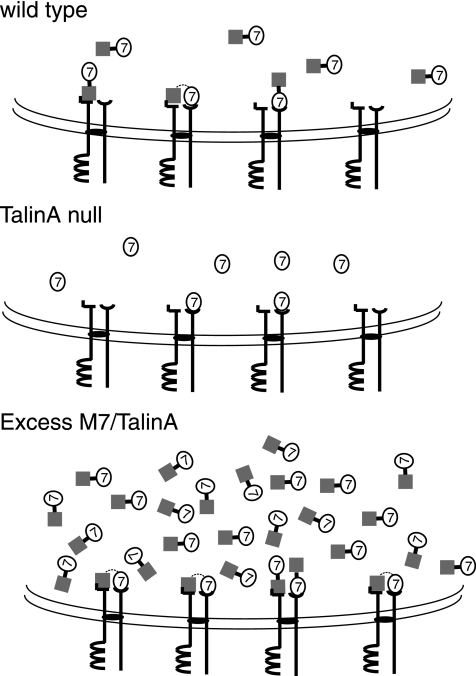
The observed differences in the dynamics of membrane-associated M7 in cells with different levels of M7 and talinA cannot be accounted for by proposing that talinA is simply required to tether M7 to the membrane. In talinA null cells, no gross change in the amount of M7 in total membrane fractions is observed and M7 localizes to the plasma membrane correctly (Tuxworth et al., 2005). A model that might account for both these observations as well as the FRAP results is presented in Figure 8. Membrane-binding sites for talinA and M7 could be physically associated with each other, bringing these into close proximity of each other as part of a large adhesion receptor complex. The existence of such a big complex might explain the failure to coprecipitate any protein other than talinA with M7 from membrane fractions treated with various detergents (Stephens, Galdeen, and Titus, unpublished observations). On binding to its own receptor, M7 would also be quite near to a talinA-binding site that its associated talinA could readily bind to. M7 binding to its receptor would not alter the affinity of M7 for talinA and, similarly, M7 binding to talinA would not change the affinity of M7 for its receptor. When M7 is released from its receptor, it can either bind to the nearby talinA or rebind to its receptor or diffuse away. Association with talinA might even increase the likelihood of M7 rebinding to a neighboring M7 receptor. Such a model could explain the observed changes in recovery rates in each cell line. For example, the presence of a large excess of M7 and talinA could result in occupation of all of the talinA and M7 membrane-binding sites, creating a relative abundance of M7 binding sites—both M7 receptors and talinA bound to neighboring receptors, and this slows the observed exchange of M7. Conversely, lack of talinA would not affect the ability of M7 to bind to the membrane, but its likelihood of its being retained at the membrane through combined interactions with the receptor and talinA would be reduced.
Figure 8.
Speculative model of the dynamics of the M7-talinA association with the membrane. Schematic illustration of M7 (circled 7) and talinA (gray square) interactions with each other and with transmembrane receptors. The double curved lines indicate that plasma membrane. In the case of wild-type cells (top), M7 and talinA bind to each other (indicated by black line linking the two). They can also interact with their respective transmembrane adhesion receptors that are complexed together (joined by black oval). Note that the interaction between either talinA and/or M7 with adhesion receptors may be direct as illustrated or could occur via an adapter protein. Three binding states are illustrated: talinA only binds to its receptor, M7 binds only to its receptor, and M7 and talinA bind to their receptors that are adjacent to each other and at the same time they maintain an affinity for each other (dashed line). In talinA null cells (middle), M7 interacts with its membrane receptor. In cells expressing an excess of both M7 and talinA (bottom), there is an increased probably of M7 binding to both its receptor and talinA on the membrane, because it now has two binding sites at the membrane its residence time is increased.
The adhesion defects of the Dictyostelium M7 and talinA null mutants are quite similar: both exhibit a significant reduction in adhesion to a range of substrata (glass, plastic, and bacteria) as well as in calcium-dependent cell–cell cohesion (Niewöhner et al., 1997; Tuxworth et al., 2001). These common deficits are consistent with they hypothesis that talinA and M7 work in concert to effectively link a range of cell surface receptors to the underlying actin cytoskeleton. Recent work has identified several adhesion receptors that may associate (directly or indirectly) with talinA and M7. Among these is SibA, a member of a family of likely adhesion molecules that have extracellular motifs typical of cell surface receptors in higher eukaryotes (Cornillon et al., 2006). The sibA mutant is defective in substrate adhesion and in the uptake of latex beads, but not bacteria. Interestingly, talinA coprecipitates with GST fusions of the C-terminal cytoplasmic domain of several Sib family incubated with cell lysates. TalinA does not coprecipitate with the SibA cytoplasmic tail when the SibA NPxY motif is mutated, suggesting that the interaction between the two proteins is direct. SadA is an adhesion receptor that has nine-membrane spanning domains and extracellular EGF repeats. The sadA null mutant is defective in adhesion to plates and exhibits a phagocytosis defect consistent with a decrease in particle binding (Fey et al., 2002). It is not yet known if SadA is associated with either talinA or M7, but the similarity between the talinA, M7, and SadA null mutant phenotypes suggests that it is quite possible. Thus, talinA and M7 have the potential to interact with at least two distinct classes of adhesion receptors. It should be noted that this association could be direct, as may be the case for the talinA-SibA interaction or indirect, via a receptor associated adaptor protein. Regardless of the exact mechanism, the timely recruitment of both talinA and M7 to adhesion receptors such as Sib family members or SadA would serve to provide two different types of connections between these receptor and actin. Talin may serve as a critical initial link to actin that also helps to recruit other proteins involved in strengthening the association to the cytoskeleton (Giannone et al., 2003). M7 may enhance or reinforce the initial adhesin by the generation of tension and thus linking M7 membrane dynamics to talinA may provide a mechanism for precisely determining the establishment and/or maintenance of optimal adhesion.
ACKNOWLEDGMENTS
The authors thank Dr. Günther Gerisch (Max-Planck Institut für Biochemie, Munich, Germany) for generously providing both the talinA null mutant strain HG1666 and Dictyostelium talinA hybridoma. The talinA mAb was obtained from the Developmental Studies Hybridoma Bank (DSHB; Iowa City, IA), which is supported by the National Institute of Child Health and Human Development. We thank Karen Jensen of the DSHB for her invaluable help with the talinA antibody. We also thank also Dr. Gaku Ashiba and Dr. E. Michael Ostap (University of Pennsylvania), Dr. John Cooper (Washington University), and Laura Breshears for stimulating discussions; Dr. Michael O'Connor for the use of a LI-COR fluorescent imaging workstation; and Casey Dorr for help with early real-time qPCR experiments. Finally, we thank to Seth Robia (Loyola University) and Ji Li, Mohac Tecman, Dave Kast, and other members of the Thomas lab for their invaluable assistance in initiating the FRAP experiments. This work was supported by a grant from the National Institutes of Health and a shared equipment grant from the Minnesota Medical Foundation for purchase of the CCD camera used in the FRAP experiments.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-07-0586) on August 1, 2007.
REFERENCES
- Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. Talin is essential for integrin function in Drosophila. Dev. Cell. 2002;3:569–579. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Bukharova T., Weijer G., Bosgraaf L., Dormann D., van Haastert P. J., Weijer C. J. Paxillin is required for cell-substrate adhesion, cell sorting and slug migration during Dictyostelium development. J. Cell Sci. 2005;118:4295–4310. doi: 10.1242/jcs.02557. [DOI] [PubMed] [Google Scholar]
- Cornillon S., Gebbie L., Benghezal M., Nair P., Keller S., Wehrle-Haller B., Charette S. J., Brückert F., Letourneur F., Cosson P. An adhesion molecule in free-living Dictyostelium amoebae with integrin beta features. EMBO Rep. 2006;7:617–621. doi: 10.1038/sj.embor.7400701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amraoui A., Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J. Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- Fey P., Stephens S., Titus M. A., Chisholm R. L. SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- Gebbie L., et al. Phg2, a kinase involved in adhesion and focal site modeling in Dictyostelium. Mol. Biol. Cell. 2004;15:3915–3925. doi: 10.1091/mbc.E03-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Jiang G., Sutton D. H., Critchley D. R., Sheetz M. P. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol. 2003;163:409–419. doi: 10.1083/jcb.200302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann W. H., Guttenberg Z., Kaufmann S., Hess D., Ezzell R. M., Isenberg G. Examining F-actin interaction with intact talin and talin head and tail fragment using static and dynamic light scattering. Eur. J. Biochem. 1997;250:447–450. doi: 10.1111/j.1432-1033.1997.0447a.x. [DOI] [PubMed] [Google Scholar]
- Huang X., Czerwinski E., Mellgren R. L. Purification and properties of the Dictyostelium calpain-like protein, Cpl. Biochem. 2003;42:1789–1795. doi: 10.1021/bi026461+. [DOI] [PubMed] [Google Scholar]
- Humbel B. M., Biegelmann E. A preparation protocol for postembedding immunoelecron microscopy of Dictyostelium discoideum cells with monoclonal antibodies. Scanning Microsc. 1992;6:817–825. [Google Scholar]
- Kreitmeier M., Gerisch G., Heizer C., Müller-Taubenberger A. A talin homologue of Dictyostelium rapidly assembles at the leading edge of cells in response to chemoattractant. J. Cell Biol. 1995;129:179–188. doi: 10.1083/jcb.129.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S., Polyakov M., Egelhoff T. T. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Marchisio P. C., Bergui L., Corbascio G. C., Cremona O., D'Urso N., Schena M., Tesio L., Caligaris-Cappio F. Vinculin, talin, and integrins are localized at specific adhesion sites of malignant B lymphocytes. Blood. 1988;72:830–833. [PubMed] [Google Scholar]
- Monks C. R., Freiberg B. A., Kupfer H., Sciaky N., Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Nayal A., Webb D. J., Horwitz A. F. Talin: an emerging focal point of adhesion dynamics. Curr. Opin. Cell Biol. 2004;16:94–98. doi: 10.1016/j.ceb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Niewöhner J., Weber I., Maniak M., Müller-Taubenberger A., Gerisch G. Talin-null cells of Dictyostelium are strongly defective in adhesion to particle and substrate surfaces and slightly impaired in cytokinesis. J. Cell Biol. 1997;138:349–361. doi: 10.1083/jcb.138.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak K. D., Peterson M. D., Reedy M. C., Titus M. A. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol. 1995;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuckolls G. H., Romer L. H., Burridge K. Microinjection of antibodies against talin inhibits the spreading and migration of fibroblasts. J. Cell Sci. 1992;102:753–762. doi: 10.1242/jcs.102.4.753. [DOI] [PubMed] [Google Scholar]
- Priddle H., Hemmings L., Monkley S., Woods A., Patel B., Sutton D., Dunn G. A., Zicha D., Critchley D. R. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol. 1998;142:1121–1133. doi: 10.1083/jcb.142.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Senda S., Lee S. F., Côté G. P., Titus M. A. Recruitment of a specific amoeboid myosin I isoform to the plasma membrane in chemotactic Dictyostelium cells. J. Biol. Chem. 2001;276:2898–28904. doi: 10.1074/jbc.M008059200. [DOI] [PubMed] [Google Scholar]
- Senetar M. A., McCann R. O. Gene duplication and functional divergence during evolution of the cytoskeletal linker protein talin. Gene. 2005;362:141–152. doi: 10.1016/j.gene.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Simonson W. T., Franco S. J., Huttenlocher A. Talin1 regulates TCR-mediated LFA-1 function. J. Immunol. 2006;177:7707–7714. doi: 10.4049/jimmunol.177.11.7707. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Manning S. S., Feng Y. Effect of protein synthesis inhibition on gene expression during early development of Dictyostelium discoideum. Mol. Cell Biol. 1988;8:10–16. doi: 10.1128/mcb.8.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Titus M. A. A class VII unconventional myosin is required for phagocytosis. Curr. Biol. 1999;9:1297–1303. doi: 10.1016/s0960-9822(00)80051-2. [DOI] [PubMed] [Google Scholar]
- Tsujioka M., Yoshida K., Inouye K. Talin B is required for force transmission in morphogenesis of Dictyostelium. EMBO J. 2004;23:2216–2225. doi: 10.1038/sj.emboj.7600238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth R. I., Stephens S., Ryan Z. C., Titus M. A. Identification of a myosin VII/talin complex. J. Biol. Chem. 2005;280:26557–26564. doi: 10.1074/jbc.M503699200. [DOI] [PubMed] [Google Scholar]
- Tuxworth R. I., Weber I., Wessels D., Addicks G. C., Soll D. R., Gerisch G., Titus M. A. A role for myosin VII in dynamic cell adhesion. Curr. Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- Vogel G., Thilo L., Schwarz H., Steinhart R. Mechanism of phagocytosis in Dictyostelium discoideum: phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J. Cell Biol. 1980;86:456–465. doi: 10.1083/jcb.86.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I., Niewöhner J., Du A., Rohrig U., Gerisch G. A talin fragment as an actin trap visualizing actin flow in chemotaxis, endocytosis, and cytokinesis. Cell Motil. Cytoskelet. 2002;53:136–149. doi: 10.1002/cm.10065. [DOI] [PubMed] [Google Scholar]
- Zhang J., Robson R. M., Schmidt J. M., Stromer M. H. Talin can crosslink actin filaments into both networks and bundles. Biochem. Biophys. Res. Commun. 1996;218:530–537. doi: 10.1006/bbrc.1996.0095. [DOI] [PubMed] [Google Scholar]