Abstract
Background and aims
There is a need for genetic biomarkers to guide prognosis and management of gastric mucosa‐associated lymphoid tissue (MALT) lymphomas. We assessed the incidence and clinical significance of the MALT lymphoma‐associated genetic abnormalities t(11;18)/API2‐MALT1, t(1;14)/BCL10‐IGH, t(14;18)/IGH‐MALT1, t(3;14)/FOXP1‐IGH, and extra copies of MALT1 and FOXP1 in gastric MALT lymphomas from Japan.
Methods
The presence of translocations and copy number changes involving MALT1, IGH and FOXP1 were assessed in 90 cases of gastric MALT lymphoma using interphase fluorescence in situ hybridisation (FISH). In cases carrying a MALT1 translocation, FISH for API2‐MALT1 was performed, whereas in those carrying an IGH translocation, FISH was performed for BCL10, BCL6, BCL2, c‐MYC and/or CCND1.
Results
t(11;18)/API2‐MALT1 was detected in 18 of 87 (21%) cases and was significantly associated with Helicobacter pylori‐negativity, resistance to H pylori eradication and Bcl10 nuclear expression. Four of 68 (6%) cases carried a translocation involving IGH and FOXP1 (n = 1), BCL2 (n = 1) or an unknown partner (n = 2). Neither t(1;14)/BCL10‐IGH nor t(14;18)/IGH‐MALT1 was detected. Extra copies of MALT1 and FOXP1 were detected in 18 of 71 (25%) cases and 10 of 59 (17%) cases, respectively. The presence of extra copies of MALT1 was significantly associated with progression or relapse of lymphoma, and was an independent adverse prognostic factor for event‐free survival as determined by multivariate analysis.
Conclusions
t(11;18)/API2‐MALT1 is frequent, whereas IGH‐involved translocations are rare in gastric MALT lymphoma in Japan. The presence of extra copies of MALT1, often suggestive of partial or complete trisomy 18, is a frequent genetic aberration in gastric MALT lymphoma, which appears to predict adverse clinical behaviour.
Extranodal marginal zone B‐cell lymphoma of mucosa‐associated lymphoid tissue (MALT lymphoma) of the stomach is the most common low grade lymphoma.1,2 Although most patients with gastric MALT lymphoma experience an indolent clinical course, there is considerable individual variability in the extent of disease, response to treatment, relapse and event‐free survival such that, for many, the optimal management is unclear. There is therefore a need for improved understanding of the genetic abnormalities underlying gastric MALT lymphoma and for new genetic biomarkers able to guide prognosis and management.
MALT lymphoma is genetically characterised by four chromosome translocations.1,2 The t(11;18)(q21;q21) generates a functional API2‐MALT1 fusion transcript, whereas the other three translocations involve translocation to immunoglobulin gene loci and consequently increased expression of BCL10 (1p22), MALT1 (18q21) and forkhead box protein P1 gene (FOXP1) (3p14). Bcl10 and MALT1 proteins are involved in surface immune receptor‐mediated activation of the nuclear factor kappa B (NF‐κB) transcription factor; the chromosomal translocations involving these genes are believed to exert their oncogenic activities through constitutive activation of the NF‐κB pathway, leading to expression of a number of genes important for cell survival and proliferation. FOXP1 is an essential regulator of early B cell development but the molecular mechanism underlying its oncogenic activities remains unknown.3 The above translocations occur at variable frequencies in MALT lymphoma of different sites.4,5,6,7,8 Nonetheless, the majority of MALT lymphomas are negative for these translocations. Instead, these cases often harbour partial or complete trisomies of chromosome 3, 12 and 18.4,9,10
The incidence of chromosome translocations in gastric MALT lymphoma has been described mainly from Western countries.4,5,7,8 In gastric MALT lymphoma, t(11;18)/API2‐MALT1 has been detected frequently in Europe (15–24%),4,5,11 but is relatively rare in the USA (5%).8 However, the incidence of other MALT lymphoma‐associated translocations and numerical abnormalities in gastric MALT lymphoma is less well established, particularly in Asian populations. The clinical impact of t(11;18)/API2‐MALT1 is well characterised: gastric MALT lymphomas with this translocation are usually unresponsive to H pylori eradication, often present with advanced disease and rarely undergo high‐grade transformation.12,13 In contrast, there is little information on the clinical features of gastric MALT lymphomas bearing other translocations or numerical changes affecting chromosomes 3 and 18.
In order to address these issues, we have investigated a large, homogenous, well‐characterised cohort of 90 Japanese cases of gastric MALT lymphoma for all reported MALT lymphoma‐associated translocations and numerical changes at MALT1 and FOXP1 loci, using interphase fluorescence in situ hybridisation (FISH), and examined the clinical impact of such genetic aberrations in detail.
Methods
Subjects
Between 1966 and 2005, 380 consecutive Japanese patients with primary gastric lymphoma were diagnosed at Kyushu University. Among them, 206 cases of MALT lymphoma and/or diffuse large B‐cell lymphoma (DLBCL) had formalin‐fixed paraffin‐embedded tissues and their histological diagnosis was reviewed according to the diagnostic criteria outlined in the World Health Organization classification.14 As a result, 90 cases of MALT lymphoma without any evidence of a DLBCL component were identified and enrolled in the present study. Some of these patients were included in previous studies.15,16,17,18,19
Patients consisted of 47 men and 43 women, ranging in age from 16 to 80 years (mean 58 years). Depth of tumour invasion was determined by histological examination of the resected specimens or by endoscopic ultrasonography;16 37 lymphomas were considered to be restricted to the mucosa and the superficial portion of the submucosa,15,16 27 invaded the deep layer of the submucosa, 14 involved the muscularis propria and 12 reached the subserosa or serosa. A physical examination to determine the staging included an inspection of Waldeyer's ring, chest radiographs, abdominal ultrasound, computed tomography of the chest and abdomen, colonoscopy, small bowel barium radiography, bone marrow aspiration or biopsy, and gallium scintigraphy or fluorine‐18 fluorodeoxyglucose positron emission tomography. According to the Lugano International Conference classification for clinical staging of gastrointestinal lymphoma,20 61 cases were classified as stage I, 19 as stage II1, 6 as stage II2, none as stage IIE and 4 as stage IV (involving bone marrow in 2, cervical lymph nodes in 1 and lung in 1). H pylori infection was judged as positive if one of any pretreatment tests for H pylori, including both serology and histology, showed a positive result, and as negative only when all tests were negative. A total of 69 of 78 patients (88%) were positive and 9 patients (12%) were negative for H pylori. The remaining 12 patients were not evaluated for H pylori.
Thirty‐five (39%) patients underwent antibiotic treatment for eradication of H pylori with a proton pump inhibitor (omeprazole, lansoprazole or rabeprazole) plus a combination of antibiotics (clarithromycin and amoxicillin with or without metronidazole). Follow up endoscopic examinations with biopsies after eradication were performed every 4 to 8 weeks until confirmation of complete remission (CR), and they were repeated every 3 to 6 months after CR. CR was defined as a complete disappearance of clinical evidence of lymphoma and an absence of histological evidence of lymphoma on biopsy specimens. Partial remission was defined as a tumour reduction of at least 50%. Twenty‐one of the 35 patients underwent eradication therapy alone, whereas 14 non‐responsive patients then received radiotherapy (n = 6), oral monochemotherapy with cyclophosphamide (n = 6),17 multi‐agent chemotherapy with cyclophosphamide, doxorubicin, vincristine, prednisolone plus rituximab (n = 1) or gastrectomy (n = 1). The remaining 55 patients (61%) were initially treated by gastrectomy with (n = 15) or without (n = 40) chemotherapy.
This study was carried out in accordance with the Helsinki Declaration as revised in 1989 and with the ethical guidelines of the participating centres and countries. Informed consent was obtained from each patient on the aims and the protocol of the study.
Interphase FISH
MALT lymphoma‐associated chromosome translocations were investigated by interphase FISH on formalin‐fixed paraffin‐embedded tissues as described previously.10,21 In each case, the hybridisation signals for each probe were evaluated in at least 100 nuclei. Initially, translocations were screened by FISH with dual‐colour break‐apart probes for MALT1, IGH (Vysis/Abbott Laboratories Ltd, UK) and FOXP1. The genomic bacterial artificial chromosome clones used as the FOXP1 probe were described previously.22 In cases positive for a MALT1 gene break, additional FISH with dual‐colour, dual‐fusion translocation probe for API2/MALT1 (Vysis/Abbott) was performed, whereas in those positive for an IGH gene break, further FISH was carried out with dual‐colour break‐apart probes for BCL6, BCL2, cyclin (CCN) D1, c‐MYC, (Vysis/Abbott) and BCL10 (provided by Reiner Siebert, Kiel, Germany). A FOXP1/IGH dual‐colour, dual‐fusion translocation probe was developed in‐house. For FISH with MALT1 and FOXP1 break‐apart probes, numerical gains at chromosomes 18q21 and 3p14 were also evaluated. The threshold for positivity using each probe was the mean percentage of cells with false positive signals plus 3 standard deviations, as assessed on sections from 8 to 10 reactive tonsils.10
Reverse‐transcription polymerase chain reaction
Reverse‐transcription polymerase chain reaction (RT‐PCR) for t(11;18)(q21;q21) was carried out to assess samples showing no signals by FISH with MALT1 probe and to confirm FISH detected API2‐MALT1 translocations. Total RNA extraction from formalin‐fixed paraffin‐embedded tissues, complementary DNA synthesis and PCR for the API2‐MALT1 fusion transcript were performed as described previously.12 RNA quality was assessed by RT‐PCR of the control gene glucose‐6‐phosphate dehydrogenase. PCR products were analysed by electrophoresis on 10% polyacrylamide gels. We have previously shown that RT‐PCR and FISH have similar sensitivity and specificity for the detection of t(11;18)/API2‐MALT1.5
Immunohistochemistry
Immunohistochemical staining for Bcl10 was performed by the streptavidin–biotin complex method using a mouse monoclonal antibody (clone 151) on formalin‐fixed paraffin‐embedded tissues in all 90 cases as described previously.23
Prognosis and statistical analysis
Overall survival (OS) was measured from the date of diagnosis to death from any cause. Event‐free survival (EFS) was measured from the date of diagnosis to disease progression, relapse or death from any cause. Probabilities of OS and EFS for patients were calculated by the Kaplan‐Meier method and the value was compared using the log‐rank test. All variables with p<0.1 were put into a multivariate analysis using the Cox proportional hazards regression model. Model selection for identifying the subset of significant variables was based on a forward stepwise procedure using the maximum likelihood ratio test. Other statistical differences were evaluated by Fisher's exact probability test, the χ2 test or the Mann‐Whitney U test. Probabilities of less than 0.05 were statistically significant.
Results
Incidence of MALT lymphoma‐associated genetic aberrations
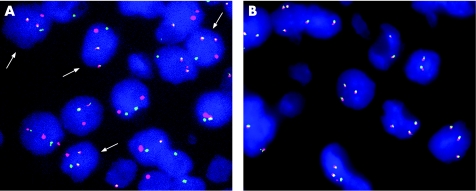
Using a combination of FISH and RT‐PCR, 87 cases were of sufficient quality for assessment of the presence of t(11;18)/API2‐MALT1. Nine t(11;18)/API2‐MALT1‐positive cases were identified by FISH using a MALT1 break‐apart probe and an API2‐MALT1 dual‐colour dual‐fusion translocation probe (fig 1A); in all nine cases the presence of API2‐MALT1 fusion transcripts was also confirmed by RT‐PCR. In addition, API2‐MALT1 fusion transcripts were also identified by RT‐PCR in nine cases in which FISH signals were poor. Thus overall, 18 of 87 (21%) cases were positive for t(11;18)/API2‐MALT1.
Figure 1 Interphase fluorescence in situ hybridisation (FISH) patterns in representative cases. (A) t(11;18)/API2‐MALT1. FISH with an API2/MALT1 dual‐colour, dual‐fusion translocation probe shows co‐localisation of the red and green signals (arrows). (B) Extra copies of MALT1. FISH with a MALT1 dual‐colour break‐apart probe demonstrates three or four copies of MALT1 gene in 40% of tumour cells.
Evidence of IGH breakage was found in 4 (6%) of 68 cases in which adequate signals were obtained. The remaining 22 cases showed no signals for FISH with IGH probe. Among the four cases with IGH breakage, one showed FOXP1 breakage; the presence of t(3;14)/FOXP1‐IGH was confirmed by FISH with a FOXP1/IGH dual‐colour, dual‐fusion translocation probe. In another case with IGH breakage, t(14;18)(q32;q21) involving IGH and BCL2 was detected by FISH with BCL2 dual‐colour break‐apart probe and IGH/BCL2 dual‐colour, dual‐fusion translocation probe; the possibility of follicular lymphoma was excluded by extensive histological and immunohistochemical investigation.24 In the remaining two cases with potential IGH‐involved translocations, the partner genes were unknown; neither case showed evidence of a split signal with MALT1, FOXP1, BCL10, BCL6, BCL2, CCND1 or c‐MYC dual‐colour break‐apart probes. None of the cases showed t(1;14)/BCL10‐IGH or t(14;18)/IGH‐MALT1 in the present study. In addition, no cases showed strong Bcl10 nuclear staining as seen in cases with t(1;14)/BCL10‐IGH.
Extra copies of MALT1 (three and occasionally four copies), suggestive of partial or complete trisomy (or occasionally tetrasomy) 18, were detected in 18 (25%) of 71 cases in which adequate signals were obtained (fig 1B). Extra copies of MALT1 were not detected in any of the translocation‐positive cases, except for one case with a translocation involving IGH and an unknown partner gene. Adequate signals were obtained with the FOXP1 dual‐colour break‐apart probe in 59 of 90 cases, and an extra copy of FOXP1 suggestive of partial or complete trisomy 3 was detected in 10 of 59 (17%) cases. None of the 10 cases with an extra copy of FOXP1 had a translocation involving MALT1 or IGH. Extra copies of both MALT1 and FOXP1 were observed in 4 of 56 (7%) cases in which adequate signals were observed using both FISH probes. Overall, 24 of 74 (32%) cases contained extra copies of MALT1, FOXP1 or both. In total, at least 43 of 87 (49%) cases had structural or numerical genetic aberrations.
Relation between genetic aberrations and clinicopathological features
Clinicopathological findings in the t(11;18)/API2‐MALT1 positive and negative groups are summarised in Supplemental table A. Cases negative for H pylori (38% vs 3%) and those not responding to H pylori eradication treatment (83% vs 32%) were more frequent in the t(11;18)/API2‐MALT1‐positive group than in the negative group (p<0.001 and p<0.05, respectively). Bcl10 nuclear expression as shown by immunohistochemistry was observed more frequently in the t(11;18)/API2‐MALT1‐positive group than in the negative group (73% vs 39%, p<0.05). Other factors, including depth of tumour invasion, clinical stage, overall CR induction, and disease progression or relapse, did not differ between the two groups.
Clinicopathological findings in the IGH‐involved translocation positive and negative groups are also shown in Supplemental table A. No differences were observed between the two groups in any of the factors analysed.
Supplemental table A (available at http://gut.bmj.com/supplemental) compares the clinicopathological findings between cases with and without extra copies of MALT1 or FOXP1. Cases with extra copies of MALT1 were older than those without extra copies of MALT1 (p<0.05). Disease progression or relapse of lymphoma was more frequently observed in cases with extra copies of MALT1 than in those without (22% vs 0%, p<0.005). Cases with an extra copy of FOXP1 had tumours invading the submucosa or beyond more frequently than cases with only two copies (90% vs 55%, p<0.05). All other factors, including clinical stage or overall CR induction were not different.
Prognostic impact of genetic aberrations
The follow‐up after the diagnosis in 90 patients ranged from 2 to 240 months (mean 65 months). The OS and EFS after 5 years were 88% and 85%, respectively.
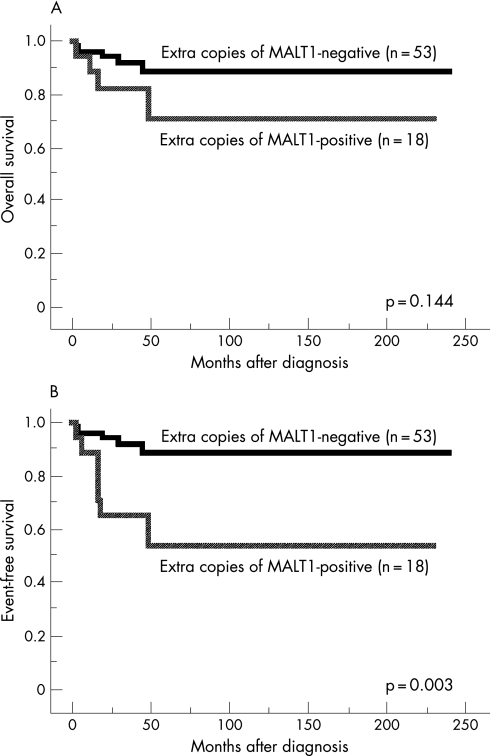
Table 1 summarises the results of a univariate analysis for prognosis of patients as assessed by the Kaplan‐Meier method with the log‐rank test. Cases with macroscopically superficial type tumours showed significantly better OS and EFS (both p<0.05). Cases with deeper invasion of the gastric wall showed a significantly worse OS (p<0.05), but the difference in EFS was insignificant (p<0.1). The presence of extra copies of MALT1 was significantly associated with worse EFS (p<0.005), although it did not correlate with OS (fig 2A and B). Older patients tended to show worse OS and EFS, and male patients also tended to show worse OS, but these differences were not statistically significant (all p<0.1). Other factors, including t(11;18)/API2‐MALT1, IGH‐involved translocations and extra copies of FOXP1, did not affect OS or EFS.
Table 1 Results of univariate analysis for prognosis evaluated by Kaplan‐Meier method.
| Factors | Overall survival | Event‐free survival | ||
|---|---|---|---|---|
| 5 years | p Value* | 5 years | p Value* | |
| Age (y) | ||||
| ⩽58 or younger (n = 39) | 0.94 | 0.072 | 0.91 | 0.069 |
| ⩾59 or older (n = 51) | 0.84 | 0.79 | ||
| Gender | ||||
| Male (n = 47) | 0.82 | 0.071 | 0.80 | 0.197 |
| Female (n = 43) | 0.95 | 0.90 | ||
| Macroscopic type | ||||
| Superficial (n = 71) | 0.93 | 0.019 | 0.90 | 0.017 |
| Others (n = 19) | 0.71 | 0.65 | ||
| Depth of tumour invasion | ||||
| Mucosa (n = 37) | 1.00 | 0.013 | 0.94 | 0.083 |
| SM or beyond (n = 53) | 0.81 | 0.79 | ||
| Clinical stage | ||||
| I (n = 61) | 0.87 | 0.369 | 0.83 | 0.423 |
| II1 or more (n = 29) | 0.92 | 0.88 | ||
| t(11;18)/API2‐MALT1 | ||||
| Positive (n = 18) | 1.00 | 0.370 | 1.00 | 0.229 |
| Negative (n = 69) | 0.85 | 0.80 | ||
| IGH‐involved translocation | ||||
| Positive (n = 4) | 1.00 | 0.507 | 0.75 | 0.583 |
| Negative (n = 64) | 0.87 | 0.84 | ||
| Extra copies of MALT1 | ||||
| Positive (n = 18) | 0.71 | 0.144 | 0.54 | 0.003 |
| Negative (n = 53) | 0.89 | 0.89 | ||
| Extra copies of FOXP1 | ||||
| Positive (n = 10) | 0.90 | 0.949 | 0.79 | 0.647 |
| Negative (n = 49) | 0.87 | 0.83 | ||
FOXP1, forkhead box protein P1; MALT, mucosa‐associated lymphoid tissue. *Log‐lank test.
Figure 2 Survival curves of patients of gastric mucosa‐associated lymphoid tissue (MALT) lymphoma with (solid line, n = 18) and without (dotted line, n = 53) extra copies of MALT1. (A) Overall survival (p = 0.144). (B) Event‐free survival (p = 0.003).
The results of a multivariate analysis are shown in table 2. Significant independent prognostic factors for worse OS were deeper invasion of the gastric wall and older age, whereas those for worse EFS were extra copies of MALT1 and macroscopic appearance other than superficial type.
Table 2 Independent prognostic factors determined by multivariate analysis*.
| Variables | Coefficient | Standard error | Relative risk | p Value |
|---|---|---|---|---|
| Overall survival | ||||
| Depth of invasion | 0.639 | 0.278 | 2.294 | 0.0218 |
| Age | 0.073 | 0.033 | 2.245 | 0.0248 |
| Gender | 0.956 | 0.803 | 1.190 | 0.2340 |
| Event‐free survival | ||||
| Extra copies of MALT1 | 1.448 | 0.590 | 2.454 | 0.0141 |
| Macroscopic type | 1.351 | 0.582 | 2.321 | 0.0203 |
| Age | 0.046 | 0.031 | 1.487 | 0.1371 |
*Cox proportional hazards regression model.
Discussion
Although there are a number of publications about the incidence and clinical impact of t(11;18)/API2‐MALT1 in gastric MALT lymphoma, comprehensive investigation of all MALT lymphoma‐associated translocations, including t(11;18)/API2‐MALT1, t(1;14)/BCL10‐IGH, t(14;18)/IGH‐MALT1 and t(3;14)/FOXP1‐IGH, has been carried out in only a few studies.4,5,7,8 So far, such a comprehensive study has not been done in Asian cases. In the present study, we investigated the incidence of all the four reported chromosome translocations and of extra copies of MALT1 and FOXP1 in Japanese cases of gastric MALT lymphoma.
The reported frequency of t(11;18)/API2‐MALT1 in gastric MALT lymphoma has varied from 5% to 40%.4,5,8,11,12,13,25 Such a variation in frequency among these studies is probably due to bias in case selection12,25 or small sample sizes.8 In four large studies comprising more than 60 gastric cases, the frequency of t(11;18)/API2‐MALT1 ranged from 15 to 24%,4,5,11,13 which was compatible with our present data (21%). All of the four studies and our present one examined a large cohort of unselected cases, and thus the frequency of 20% likely represents its true incidence in gastric MALT lymphoma.
Our present study confirmed that t(11;18)/API2‐MALT1 was significantly associated with H pylori‐negative cases, resistance to H pylori eradication and Bcl10 nuclear expression (Supplemental table A; available at http://gut.bmj.com/supplemental), as reported previously.12,13,19,25,26 In our study, however, the translocation did not correlate to the clinical stage; this was discordant from several previous studies in which t(11;18)/API2‐MALT1 was associated with advanced stage MALT lymphoma.13,25 The reason for this discrepancy is unclear.
A translocation involving IGH was detected in four cases (6%) in our study. Similarly, Remstein et al8 reported that IGH‐involved translocations were present in 8% of MALT lymphomas from various organs of patients in North America. In our study, however, none of the cases showed t(1;14)/BCL10‐IGH or t(14;18)/IGH‐MALT1. In a large study by Ye et al,5 t(1;14)/BCL10‐IGH was detected in 8 of 185 (4%) gastric MALT lymphomas, while three other studies did not detect this translocation in any of the gastric cases examined (n = 71, 24 and 22, respectively).4,7,8 t(14;18)/IGH‐MALT1 has been found predominantly in extra‐gastric MALT lymphoma, whereas gastric cases with this translocation are very rare.4,5,8
In our study, t(3;14)/FOXP1‐IGH was detected in 1 of 59 (1.7%) cases. This translocation has been occasionally seen in extra‐gastric MALT lymphoma and extra‐nodal DLBCL.6,22,27 Recent data have shown that overexpression of FOXP1 protein is associated with inferior survival in DLBCL28 and MALT lymphoma.7 In gastric MALT lymphoma without DLBCL component, t(3;14)/FOXP1‐IGH has been previously found in only one case in a study from Belgium.27 These observations suggest that t(3;14)/FOXP1‐IGH rarely occurs in gastric MALT lymphoma and such lymphomas may be at risk of poor clinical outcome and/or transformation to DLBCL.7
In another case with an IGH‐involved translocation, we found t(14;18)/IGH‐BCL2.24 Although t(14;18)/IGH‐BCL2 is the genetic hallmark of follicular lymphoma, this translocation has also been described in a few cases of MALT lymphoma.29 In the remaining two cases (3%), we could not identify the partner gene for IGH. Both the cases were negative for FISH with almost all of the known partner genes. Although t(6;14)/CCND3‐IGH has been detected in several cases of DLBCL transformed from MALT lymphoma,30 probes for CCND3 were not available in our study. Remstein et al8 also described that 4 of 131 (3%) MALT lymphomas showed translocations involving IGH and unknown partner gene. Thus, other as yet undiscovered partner genes for IGH‐involved translocations are considered to exist among gastric MALT lymphomas. When assessed together in our cohort, IGH‐involved translocations did not affect any of the clinicopathological features studied, including prognosis (Supplemental table A (available at http://gut.bmj.com/supplemental) and table 2); however, cases with this translocation were few and further analyses of a larger number of such translocation‐positive cases are warranted.
Aneuploidy, most commonly trisomy 18 and trisomy 3 or both, are frequently observed in t(11;18)/API2‐MALT1‐negative MALT lymphomas.4,9,10,31 We identified extra copies of MALT1 and FOXP1 genes, most likely partial or complete trisomies 18 and 3, in 25% and 17% of cases, respectively. The most important finding in the present study was that the presence of extra copies of MALT1 was significantly associated with progression or relapse of lymphoma (table 1), and it was an independent prognostic factor for worse event‐free survival as determined by Cox multivariate analysis (table 2 and fig 2). Previous studies suggested such numerical gains as trisomies 3 or 18 to be associated with high‐grade transformation of MALT lymphoma.9,32 Krugmann et al33 reported that trisomy 18q21 (extra copy of MALT1) was significantly associated with worse disease‐specific survival in 19 cases of gastrointestinal DLBCL with or without MALT lymphoma, although such a difference was not observed among 11 cases of MALT lymphoma studied. In a recent study by Tanimoto et al,34 trisomy 18 determined by FISH with a centromere‐specific probe was significantly associated with lymphoma recurrence in 34 cases of ocular adnexal MALT lymphoma. Together, these results suggest that partial or complete trisomy 18 may be associated with an adverse clinical course of gastric MALT lymphoma. We thus believe that assessment of copy number of MALT1, in addition to other known clinical parameters, is valuable for predicting the clinical course of patients with gastric MALT lymphoma and may help to identify patients for whom closer clinical follow‐up is appropriate. However, these findings were based on retrospective studies of cases accrued over relatively long periods and receiving heterogeneous treatment. Therefore, further prospective studies are clearly indicated.
In conclusion, our comprehensive study revealed that t(11;18)/API2‐MALT1 is frequent, whereas IGH‐involved translocations are rare in gastric MALT lymphoma in Japan. The presence of extra copies of MALT1 is a frequent genetic aberration in gastric MALT lymphoma, which appears to predict an adverse clinical course. Further clinical studies of a large number of cases with longer follow up are warranted to clarify whether this numerical change has a major influence on the prognosis of gastric MALT lymphoma.
Table A can be viewed on the Gut website at http://www.gutjnl.com/supplemental
Copyright © 2007 BMJ Publishing Group & British Society of Gastroenterology
Supplementary Material
Acknowledgements
We thank Professor Reiner Siebert, Institute of Human Genetics, University Hospital Schleswig‐Holstein, Kiel, Germany, for providing the BCL10 FISH probe. This work was supported by research grants from Leukaemia Research Fund, UK. Chris Bacon was supported by a Senior Clinician Scientist Fellowship from The Health Foundation, The Royal College of Pathologists and The Pathological Society of Great Britain and Ireland. Alison Goatly was supported by the Wellcome Trust, UK.
Abbreviations
CR - complete remission
DLBCL - diffuse large B‐cell lymphoma
EFS - event‐free survival
FISH - fluorescence in situ hybridisation
FOXP1 - forkhead box protein P1
MALT - mucosa‐associated lymphoid tissue
NF‐κB - nuclear factor kappa B
OS - overall survival
RT‐PCR - reverse‐transcription polymerase chain reaction
Footnotes
Competing interests: None.
Table A can be viewed on the Gut website at http://www.gutjnl.com/supplemental
References
- 1.Isaacson P G, Du M Q. MALT lymphoma: from morphology to molecules. Nat Rev Cancer 20044644–653. [DOI] [PubMed] [Google Scholar]
- 2.Du M Q, Atherton J C. Molecular subtyping of gastric MALT lymphomas: implications for prognosis and management. Gut 200655886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu H, Wang B, Borde M.et al Foxp1 is an essential transcriptional regulator of B cell development. Nature Immunol 2006719–26. [DOI] [PubMed] [Google Scholar]
- 4.Streubel B, Simonitsch‐Klupp I, Müllauer L.et al Variable frequencies of MALT lymphoma‐associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004181722–1726. [DOI] [PubMed] [Google Scholar]
- 5.Ye H, Gong L, Liu H.et al MALT lymphoma with t(14;18)(q32;q21)/IGH‐MALT1 is characterized by strong cytoplasmic MALT1 and BCL10 expression. J Pathol 2005205293–301. [DOI] [PubMed] [Google Scholar]
- 6.Streubel B, Vinatzer U, Lamprecht A.et al T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 200519652–658. [DOI] [PubMed] [Google Scholar]
- 7.Sagaert X, de Paepe P, Libbrecht L.et al Forkhead box protein P1 expression in mucosa‐associated lymphoid tissue lymphoma predicts poor prognosis and transformation to diffuse large B‐cell lymphoma. J Clin Oncol 2006242490–2497. [DOI] [PubMed] [Google Scholar]
- 8.Remstein E D, Dogan A, Einerson R R.et al The incidence and anatomic site specificity of chromosomal translocations in primary extranodal marginal zone B‐cell lymphoma of mucosa‐associated lymphoid tissue (MALT lymphoma) in North America. Am J Surg Pathol 2006301546–1553. [DOI] [PubMed] [Google Scholar]
- 9.Remstein E D, Kurtin P J, James C D.et al Mucosa‐associated lymphoid tissue lymphomas with t(11;18)(q21;q21) and mucosa‐associated lymphoid tissue lymphomas with aneuploidy develop along different pathogenetic pathways. Am J Pathol 200216163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Ye H, Martin‐Subero J I.et al Distinct comparative genomic hybridisation profiles in gastric mucosa‐associated lymphoid tissue lymphomas with and without t(11;18)(q21;q21). Br J Haematol 200613335–42. [DOI] [PubMed] [Google Scholar]
- 11.Wündisch T, Thiede C, Morgner A.et al Long‐term follow‐up of gastric MALT lymphoma after Helicobacter pylori eradication. J Clin Oncol 2005238018–8024. [DOI] [PubMed] [Google Scholar]
- 12. Liu H , Ye H , Ruskoné‐Fourmestraux A , et al T(11; 18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 20021221286–1294. [DOI] [PubMed] [Google Scholar]
- 13.Inagaki H, Nakamura T, Li C.et al Gastric MALT lymphomas are divided into three groups based on responsiveness to Helicobacter pylori eradication and detection of API2‐MALT1 fusion. Am J Surg Pathol 2004281560–1567. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson P G, Müller‐Hermelink H K, Piris M A.et al Extranodal marginal zone B‐cell lymphoma of mucosa‐associated lymphoid tissue (MALT lymphoma). In: Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. World Health Organization classification of tumours. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press, 2001157–160.
- 15.Nakamura S, Yao T, Aoyagi K.et alHelicobacter pylori and primary gastric lymphoma: a histopathologic and immunohistochemical analysis of 237 patients. Cancer 1997793–11. [PubMed] [Google Scholar]
- 16.Nakamura S, Matsumoto T, Suekane H.et al Predictive value of endoscopic ultrasonography for regression of gastric low grade and high grade MALT lymphomas after eradication of Helicobacter pylori. Gut 200148454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura S, Matsumoto T, Suekane H.et al Long‐term clinical outcome of Helicobacter pylori eradication for gastric mucosa‐associated lymphoid tissue lymphoma with a reference to second‐line treatment. Cancer 2005104532–540. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura S, Nakamura S, Matsumoto T.et al Overexpression of caspase recruitment domain (CARD) membrane‐associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric lymphoma. Cancer 20051041885–1893. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura S, Matsumoto T, Ye H.et alHelicobacter pylori‐negative gastric mucosa‐associated lymphoid tissue lymphoma: a clinicopathologic and molecular study with reference to antibiotic treatment. Cancer 20061072770–2778. [DOI] [PubMed] [Google Scholar]
- 20.Rohatiner A, d'Amore F, Coiffier B.et al Report on a workshop convened to discuss the pathological and staging classifications of gastrointestinal tract lymphoma. Ann Oncol 19945397–400. [DOI] [PubMed] [Google Scholar]
- 21.Ye H, Liu H, Attygalle A.et al Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood 20031021012–1018. [DOI] [PubMed] [Google Scholar]
- 22.Haralambieva E, Adam P, Ventura R.et al Genetic rearrangement of FOXP1 is predominantly detected in a subset of diffuse large B‐cell lymphomas with extranodal presentation. Leukemia 2006201300–1303. [DOI] [PubMed] [Google Scholar]
- 23.Ye H, Dogan A, Karran L.et al BCL10 expression in normal and neoplastic lymphoid tissue: nuclear localization in MALT lymphoma. Am J Pathol 20001571147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura S, Ye H, Bacon C M.et al Gastric MALT lymphoma with t(14;18)(q32;q21) involving IGH and BCL2 genes that responded to Helicobacter pylori eradication. J Clin Pathol. in press [DOI] [PMC free article] [PubMed]
- 25.Liu H, Ye H, Dogan A.et al T(11;18)(q21;q21) is associated with advanced mucosa‐associated lymphoid tissue lymphoma that express nuclear BCL10. Blood 2001981182–1187. [DOI] [PubMed] [Google Scholar]
- 26.Ye H, Liu H, Raderer M.et al High incidence of t(11;18)(q21;q21) in Helicobacter pylori‐negative gastric MALT lymphoma. Blood 20031012547–2550. [DOI] [PubMed] [Google Scholar]
- 27.Wlodarska I, Veyt E, De Paepe P.et al FOXP1, a gene highly expressed in a subset of diffuse large B‐cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia 2005191299–1305. [DOI] [PubMed] [Google Scholar]
- 28.Banham A H, Connors J M, Brown P J.et al Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B‐cell lymphoma. Clin Cancer Res 2005111065–1072. [PubMed] [Google Scholar]
- 29.Libra M, De Re V, Gloghini A.et al Detection of bcl‐2 rearrangement in mucosa‐associated lymphoid tissue lymphomas from patients with hepatitis C virus infection. Haematologica 200489873–874. [PubMed] [Google Scholar]
- 30.Sonoki T, Harder L, Horsman E D.et al Cyclin D3 is a target gene of t(6;14)(p21.1;q32.3) of mature B‐cell malignancies. Blood 2001982837–2844. [DOI] [PubMed] [Google Scholar]
- 31.Wotherspoon A C, Finn T M, Isaacson P G. Trisomy 3 in low‐grade B‐cell lymphomas of mucosa‐associated lymphoid tissue. Blood 1995852000–2004. [PubMed] [Google Scholar]
- 32.Hoeve M A, Gisbertz I A M, Schouten H C.et al Gastric low‐grade MALT lymphoma, high‐grade MALT lymphoma and diffuse large B cell lymphoma show different frequencies of trisomy. Leukemia 199913799–807. [DOI] [PubMed] [Google Scholar]
- 33.Krugmann J, Tzankov A, Dirnhofer S.et al Unfavourable prognosis of patients with trisomy 18q21 detected by fluorescence in situ hybridisation in t(11;18) negative, surgically resected, gastrointestinal B cell lymphomas. J Clin Pathol 200457360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanimoto K, Sekiguchi N, Yokota Y.et al Fluorescence in situ hybridization (FISH) analysis of primary ocular adnexal MALT lymphoma. BMC Cancer 20066249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.