Abstract
Protein–ligand interactions, including protein–protein interactions, are ubiquitously essential in biological processes and also have important applications in biotechnology. A wide range of methodologies have been developed for quantitative analysis of protein–ligand interactions. However, most of them do not report direct functional/structural consequence of ligand binding. Instead they only detect the change of physical properties, such as fluorescence and refractive index, because of the colocalization of protein and ligand, and are susceptible to false positives. Thus, important information about the functional state of protein–ligand complexes cannot be obtained directly. Here we report a functional single-molecule binding assay that uses force spectroscopy to directly probe the functional consequence of ligand binding and report the functional state of protein–ligand complexes. As a proof of principle, we used protein G and the Fc fragment of IgG as a model system in this study. Binding of Fc to protein G does not induce major structural changes in protein G but results in significant enhancement of its mechanical stability. Using mechanical stability of protein G as an intrinsic functional reporter, we directly distinguished and quantified Fc-bound and Fc-free forms of protein G on a single-molecule basis and accurately determined their dissociation constant. This single-molecule functional binding assay is label-free, nearly background-free, and can detect functional heterogeneity, if any, among protein–ligand interactions. This methodology opens up avenues for studying protein–ligand interactions in a functional context, and we anticipate that it will find broad application in diverse protein–ligand systems.
Keywords: atomic force microscopy, protein–ligand binding, protein–protein interaction
Protein–ligand interactions, including protein–protein interactions, play crucial roles in almost all biological processes and functions and have important applications in medicine and biotechnology (1). The binding of a ligand to the protein will induce conformational change of the protein, which can be a minute structural perturbation or a large conformation change, and transform the protein into a new functional state that is distinct from the ligand-free form of the protein. This new functional state then can trigger a cascade of biological reactions (2, 3). Many techniques have been developed to characterize protein–ligand interactions and measure their binding affinity in vitro and in vivo (4–6). However, most of the techniques are largely based on colocalization of the proteins and their interacting partners and involve the detection of change of physical properties upon binding of the ligand, such as fluorescence and refractive index, which are not necessarily the structural or functional consequence of ligand binding. However, the functional protein–ligand complexes (ligand-bound functional states) are not merely the colocalization of the two interacting partners. Instead, it is the structural difference, being minute or large, and its functional consequence that distinguish the functional ligand-bound form from the nonfunctional ligand-free form. Hence, it is of critical importance to probe the structural and/or functional consequence of the protein upon binding of ligands and develop functional binding assay to directly report the functional state of the protein–ligand complex.
Mechanical stability is an intrinsic property of a given protein and is governed by specific noncovalent interactions in the key region of the protein (7–9). As such, mechanical stability is susceptible to conformational changes of the proteins caused by external factors, such as ligand binding (10) and point mutation (11). Mechanical stability of proteins can be directly measured using single-molecule atomic force microscopy (AFM) one molecule at a time (7, 12). Therefore, if ligand binding can induce conformational changes in the protein to alter its mechanical stability, mechanical stability of the protein then can serve as an intrinsic reporter to directly report the structural consequence of ligand binding to the protein, thus entailing a functional means to directly identify the functional state of the protein at the single-molecule level without any ambiguity. As a proof of principle, here we use the binding of Fc fragment of human IgG (hFc) to protein G as a model system (13) to report a force-spectroscopy-based, functional single-molecule binding assay that is capable of directly reporting the functional state of protein G upon binding of hFc. In this assay, the mechanical stability of protein G is used as a functional reporter to directly report the functional/structural consequence of the binding of hFc to protein G.
Protein G from streptococci is well known for its ability to bind IgG antibody and has been used as affinity purification matrix for purifying IgG antibody (13, 14). The binding of hFc to protein G domains has been widely studied and used as a model system for a wide range of binding assays (15–19). Protein G contains three IgG binding domains (B1, C2, and B2 domains) arranged in tandem whose sequences only differ from each other by a few amino acid residues [supporting information (SI) Fig. 4]. All three IgG binding domains have similar structures, which are characterized by a four-strand β-sheet packed against an α-helix (SI Fig. 4), and are predicted to bind Fc in an almost identical fashion as C2 domain binds to Fc (20). The three-dimensional structure of Fc/C2 complex shows that Fc binds to C2 domain in the region of the C-terminal part of the α-helix, the N-terminal part of the third β-strand, and the loop between the two structural elements (20) and the binding does not introduce major structural change to protein G (20). The mechanical stability of B1 IgG binding domain (GB1) has been well characterized by using single-molecule AFM techniques (21, 22), and it was shown that its mechanical stability depends on the backbone hydrogen bonds in the β-sheet as well as hydrophobic interactions (23). Thus, we use GB1 and its mutant NuG2 (24) as models to demonstrate the feasibility of the force-spectroscopy-based single-molecule functional binding assay.
Results
Mechanical Stability of NuG2 Is Enhanced by the Binding of hFc.
NuG2 is a GB1 mutant computationally designed by David Baker's group (University of Washington, Seattle, WA), and its three-dimensional structure is very similar to that of wild-type (wt)-GB1 (24). Although the three-dimensional structure of NuG2/hFc complex is not known, it is anticipated that the structure of NuG2/hFc will be very similar to that of wt-GB1/Fc. To characterize the mechanical stability of NuG2, we constructed a polyprotein (NuG2)8, which is composed of eight identical tandem repeats of NuG2 domains. Stretching the polyprotein (NuG2)8 results in force–extension relationships of characteristic sawtooth pattern appearance, where the individual sawtooth peak corresponds to the sequential mechanical unfolding event of individual NuG2 domains in the polyprotein chain (Fig. 1A). The unfolding force peaks are equally spaced. Fits of the worm-like chain (WLC) model of polymer elasticity (25) to the consecutive unfolding force peaks (red lines) measure a contour length increment (ΔLc) of ≈18.0 nm for the mechanical unfolding of NuG2, in good agreement with the expected value for mechanical unfolding of NuG2. ΔLc is an intrinsic structural property of a given protein (7) and serves as a fingerprint for us to identify the mechanical unfolding of NuG2. Because the NuG2 domains in the polyprotein chain are identical to each other, the mechanical unfolding of NuG2 domains occur at similar forces. The average unfolding force of NuG2 domains is 105 ± 20 pN (average ± SD, n = 1,773) at a pulling speed of 400 nm/s (Fig. 2A).
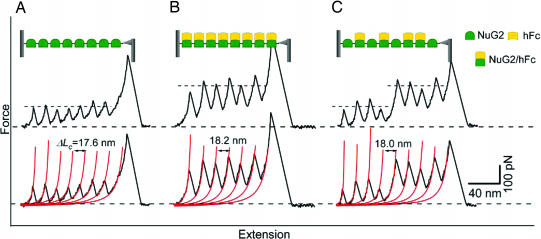
Fig. 1.
Mechanical stability of NuG2 is a functional reporter for the binding of hFc. (A) Stretching polyprotein (NuG2)8 results in typical sawtooth-like force–extension curves that are characterized by unfolding forces of ≈105 pN and contour length increments ΔLc of ≈18 nm. Each individual force peak corresponds to the mechanical unfolding of individual NuG2 domains in the polyprotein. All of the NuG2 domains unfold at a similar force of ≈105 pN, as indicated by the dashed line. Red lines correspond to the WLC fits to the force–extension curve with ΔLc of 17.6 nm. (B) The mechanical stability of NuG2 is enhanced by the binding of hFc. When preequilibrated with 33.3 μM hFc, the majority of NuG2 domains unfold at much higher forces of ≈210 pN, indicating that the unfolding force of NuG2 can be used as an indicator to report effective hFc binding to NuG2. Red lines correspond to the WLC fits to the force–extension curve with ΔLc of 18.2 nm. (C) Force–extension curves of NuG2 at an intermediate concentration of hFc directly identify the hFc-bound and hFc-free forms of NuG2 at the single-molecule level. When preequilibrated with 17.8 μM hFc, the unfolding forces of NuG2 occur at two distinct levels (as indicated by the dashed lines): the first four unfolding events occurred at ≈105 pN and can be ascribed to the unfolding of hFc-free NuG2; the last four unfolding events occurred at ≈210 pN, which corresponds to the unfolding of hFc-bound NuG2. Red lines correspond to the WLC fits to the experimental data. (Insets) Schematic illustration of the stretching of (NuG2)8 polyprotein between an AFM tip and glass substrate in the absence or presence of hFc. The functional states of NuG2 domains in the polyprotein also are indicated.
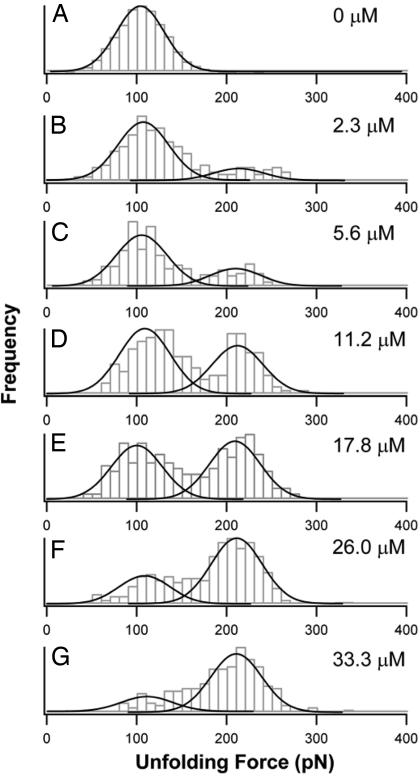
Fig. 2.
Unfolding force histograms of NuG2 in the presence of hFc reveal two distinct populations of NuG2 (hFc-free and hFc-bound forms). (A) Unfolding force histogram of NuG2 in the absence of hFc. The solid line is a Gaussian fit to the experimental data. (B–G) Unfolding force histograms of NuG2 preequilibrated with different concentrations of hFc. The unfolding force histograms of NuG2 show two clear separate peaks in the presence of hFc: one is at 105 pN, which corresponds the unfolding of hFc-free NuG2, and the other is at 210 pN, which corresponds to the unfolding of NuG2 in the complex with hFc. The initial concentration of hFc for each histogram is shown on the right. Each unfolding force histogram was fitted with two Gaussian functions (solid lines), and the relative areas underneath the Gaussian fits directly measure the fraction of hFc-free and hFc-bound of NuG2.
Although the binding of hFc to NuG2 does not introduce major structural changes to NuG2, it enhances the mechanical stability of NuG2 significantly. Stretching (NuG2)8 that is preequilibrated with 33 μM hFc results in sawtooth-like force–extension curves. WLC fits to the consecutive force peaks measure ΔLc of ≈18 nm, indicating that the unfolding force peaks result from the mechanical unfolding of NuG2 domains. However, the majority of NuG2 domains unfold at a much higher average force of 210 ± 20 pN (n = 223) than that of NuG2 in the absence of hFc (Fig. 1B). Because the vast majority of the NuG2 domains are bound to Fc at this concentration of hFc, we attribute the higher unfolding force of 210 pN to the mechanical unfolding of the hFc-bound form of NuG2. Control experiments ruled out the possibility that the higher force peaks were caused by either the mechanical unfolding of Ig domains of hFc (SI Fig. 5) or the Tris/azide buffer used for hFc (SI Fig. 6). These results strongly indicate that the binding of hFc to NuG2 significantly reinforced the mechanical resistance of NuG2 to the force-induced unfolding, despite the small apparent structural changes caused by the binding of hFc.
Mechanical Stability of NuG2 Serves as a Functional Reporter for the Binding of hFc to NuG2.
Because the hFc-bound form of NuG2 is mechanically distinct from that of hFc-free form of NuG2, it becomes possible to use the mechanical stability of NuG2 as a functional reporter to directly report the binding state of NuG2 domains one molecule at a time and determine the relative population of the two forms of NuG2 in the presence of hFc. Indeed, when carrying out single-molecule AFM experiments of NuG2 at various concentrations of hFc, we observed two distinct populations of NuG2. For example, stretching (NuG2)8 preequilibrated with 17.8 μM hFc resulted in force–extension curves with equally spaced unfolding force peaks but with two distinct levels of unfolding forces, one located at ≈105 pN and a second level at ≈210 pN. A typical force–extension curve is shown in Fig. 1C, where four unfolding events occurred at ≈105 pN and the other four occurred at ≈210 pN. The unfolding force histogram (Fig. 2E) shows two clearly separate unfolding force peaks centered at 105 pN and 210 pN, respectively. We can readily identify the NuG2 domains that unfold at ≈105 pN as the hFc-free form of NuG2, whereas the NuG2 domains unfold at ≈210 pN as the hFc-bound forms of NuG2. By counting the number of unfolding events of NuG2 occurring at low and high forces, we can readily determine the distribution of NuG2 among the two distinct populations: hFc-bound and hFc-free forms of NuG2.
By varying the concentration of hFc, we investigated the change of the distribution of the two forms of NuG2. The unfolding force histograms of NuG2 under different concentrations of hFc are shown in Fig. 2. It is evident that, in the presence of hFc, the unfolding force histograms of NuG2 show bimodal distribution, with one peak at ≈105 pN and a second one at ≈210 pN. As expected, upon increasing the concentration of hFc, more unfolding events occur at ≈210 pN and fewer unfolding events occur at ≈105 pN. And eventually the unfolding events at ≈210 pN become dominant. This result clearly demonstrates that the hFc-free NuG2 are converted to hFc-bound form of NuG2 upon increasing the concentration of hFc. The positions of the two unfolding force peaks in the histograms remain unchanged at different hFc concentrations, indicating that the mechanical stability of hFc-bound and hFc-free forms of NuG2 does not depend on hFc concentration, and hence the observed two populations of NuG2 reflect the two intrinsic functional states of NuG2 caused by the specific binding of hFc to NuG2.
Measuring the Dissociation Constant of hFc to NuG2 at the Single-Molecule Level.
The fractions of hFc-bound and hFc-free NuG2 were determined directly from the relative areas under the peaks at 105 pN and 210 pN, respectively (Fig. 2). The fractions of hFc-bound NuG2 are plotted against hFc concentration (Fig. 3, squares). Because NuG2 domains in the polyprotein bind hFc in an independent fashion, as determined by surface plasmon resonance technique (data not shown), we fitted the binding isotherm to a single-site binding model, which takes into account all of the species present, and measured a dissociation constant Kd of 12.6 ± 0.9 μM for the binding of hFc to NuG2.
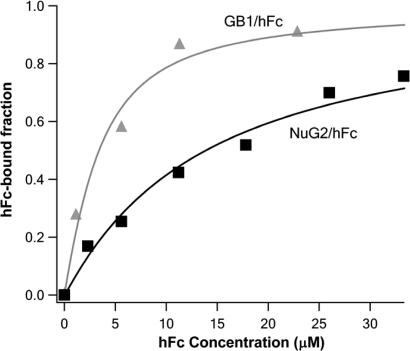
Fig. 3.
Accurate determination of dissociation constant Kd using force-spectroscopy-based single-molecule binding assay. The fractions of hFc-bound NuG2 and wt-GB1 are plotted against the initial concentration of ligand hFc in the binding isotherm (squares, NuG2/hFc; triangles, wt-GB1/hFc). Solid lines are fits to the binding isotherms using a single binding site model that takes into account all of the species present in solution (black line, NuG2/hFc; gray line, wt-GB1/hFc). The measured Kd is 12.6 ± 0.9 μM for the binding of hFc to NuG2 and 2.2 ± 0.4 μM for the binding of hFc to wt-GB1.
To test the sensitivity of this method, we also studied the binding of hFc to wt-GB1. Similar to NuG2, the mechanical stability of GB1 increased significantly upon binding of hFc. The unfolding force of GB1 increased from 180 pN to 265 pN (data not shown). Following similar procedures, we measured the hFc-bound fraction of GB1 as a function of hFc concentration (Fig. 3, triangles) and determined Kd of 2.2 ± 0.4 μM for the binding of GB1 to hFc. Although Kd for NuG2 and GB1 only differ by six times, the force-spectroscopy-based single-molecule binding assay readily detects this difference, demonstrating the high sensitivity of the single-molecule force spectroscopy-based binding assay. It is of note that, although Kd of NuG2 to hFc is approximately six times higher than that of GB1 to hFc, the stabilization effect on the mechanical stability upon binding of a ligand is similar in both cases. Therefore, the sensitivity of force spectroscopy does not depend on the binding strength between protein and its ligand.
However, it is worth noting that the sensitivity and accuracy of this methodology depends on the difference of mechanical stability between ligand-bound and ligand-free forms of proteins. If ligand binding does not result in a measurable difference in mechanical stability (26, 27), such as the binding of ligand E9 to protein Im9 (26), the application of the current methodology may become limited.
Discussion
Developing a functional single-molecule binding assay that can directly probe the structural and functional consequence of ligand binding is the key to eliminating false response in traditional colocalization-based binding assays and revealing possible heterogeneity in protein–ligand interactions. Single-molecule fluorescence resonance energy transfer (FRET) has been used to directly probe the conformational changes of the protein or RNA induced by the binding of a ligand (28, 29) and is one of the few techniques that can directly probe the structural consequence of ligand binding. However, single-molecule FRET requires dual-labeling of the protein with fluorescent reporters and only can be applied to protein–ligand systems involving relatively large conformational changes. Hence, its application in systems involving small conformational changes, such as the GB1/hFc binding, is limited. The force-spectroscopy-based single-molecule binding assay reported here directly probes the functional consequence of ligand binding and does not rely on large conformational changes. Moreover, the force-spectroscopy-based single-molecule assay is label-free and hence effectively eliminates the tedious labeling process and, more importantly, the potential interference of the binding process by fluorescent labels. Hence, this method offers unique advantages for quantitative analysis of protein–ligand interaction and represents an addition to the tool box of powerful single-molecule binding assays.
It is important to note that the force-spectroscopy-based single-molecule binding assay reported here is significantly different from the well established AFM-based protein–ligand unbinding assay (30). The AFM-based protein–ligand unbinding assay measures the force required to unbind the ligand from its protein–ligand complex and is therefore a nonequilibrium method. Equilibrium dissociation constant Kd cannot be determined by AFM-based unbinding assay. In contrast, the force-spectroscopy-based binding assay reported here is an equilibrium binding assay and can directly measure the Kd of the protein–ligand complex.
It is worth noting that the dissociation constant Kd of GB1/hFc we measured here is higher than those reported in the literature, which span a broad range from 0.5 nM to 0.52 μM (18, 31–35) determined by using a wide range of techniques, including isotope labeling, acoustic waveguide, surface plasmon resonance, fluorescence titration, and mass spectrometry. A similar trend is also observed for the Kd for NuG2/hFc. The difference in Kd between our method and traditional methods raises interesting questions. Apart from the intrinsic scatter among different techniques, two possibilities could account for the high Kd measured in the force-spectroscopy-based single-molecule binding assay. The first possibility is that the applied stretching force may change the binding affinity. It was theoretically proposed that the applied force may drive off proteins from DNA, resulting in reduced binding affinity Ka or increased Kd than that measured in the absence of force (36). If this prediction is correct and applies to GB1/hFc, the high Kd we measured here can be readily explained. The measured Kd in our force-spectroscopy-based binding assay would correspond to the Kd in the presence of a stretching force, which will have important implications for a wide range of binding systems that are subject to stretching force under physiological condition, such as ligand binding to muscle protein titin and extracellular matrix proteins: the force-spectroscopy-based binding assay, as the one demonstrated here, will be the only methodology one can use to measure physiologically relevant binding affinities for such binding systems. The second possibility is the heterogeneity in ligand binding. Upon binding of ligands to a protein, it is possible that not all of the ligand–protein complexes are functional. If this is the case for GB1/hFc, our results would indicate that only a small fraction of the GB1/hFc complex are functional, in term of enhancing the mechanical stability, and most of GB1/hFc complexes do not produce functional consequence. This scenario will provide the possibility to decipher the heterogeneity in protein–ligand interactions. However, it is not possible at present to single out the mechanism that accounts for the GB1/hFc system. Future endeavors will be required to investigate into these two interesting possibilities in detail.
In summary, our results demonstrate a label-free, force-spectroscopy-based single-molecule functional binding assay. This is an equilibrium binding assay and can directly determine the equilibrium binding constant based on the ensemble average of single-molecule data. This methodology uses mechanical stability, which changes as a functional consequence of structural changes caused by ligand binding, as a functional reporter to report the binding of ligand and enable the direct identification of the functional binding states of a protein on a single-molecule basis. Compared with traditional nonfunctional binding assays, this method is effectively background-free, providing a potentially much more accurate binding assay to determine binding affinity. Furthermore, it is important to note that the stabilization effect of the ligand on the mechanical stability of the protein depends on the specific interacting partners. Hence, it is feasible to develop this assay into a multiplex detection technique to simultaneously detect multiple protein–ligand systems, as well as use this single-molecule binding assay to investigate any possible functional heterogeneity exhibited in the same protein–ligand system. Although this method is developed based on protein G/IgG binding, we anticipate that this method can be applied to a wide range of protein–ligand interactions, including protein–drug interactions, and hence find broad applications in biotechnology.
Materials and Methods
Protein Engineering.
Plasmids encoding NuG2 and wt-GB1 proteins were generously provided by David Baker. The NuG2 and GB1 monomers, flanked with a 5′ BamHI restriction site and 3′ BglII and KpnI restriction sites, were amplified by PCR and subcloned into the pQE80L expression vector. The (NuG2)8, (GB1)8 polyprotein genes were constructed by iterative cloning monomer into monomer, dimer into dimer, tetramer into tetramer by using a previously described method (7) based on the identity of the sticky ends generated by the BamHI and BglII restriction enzymes (New England Biolabs, Ipswich, MA). The polyproteins were expressed in the DH5α strain and purified by affinity chromatography. The polyprotein was kept at 4°C in PBS buffer with 300 mM NaCl and 150 mM imidazole. The concentrations of stock solutions for (NuG2)8 and (GB1)8 were 210 mg·ml−1 and 740 mg·ml−1, respectively. hFc (catalog no. 16-16-090707-FC) was purchased from Athens Research and Technology (Athens, GA).
Single-Molecule AFM Experiment, and Determination of Dissociation Constant.
Single-molecule AFM experiments were carried out on a custom-built atomic force microscope. All of the force–extension measurements for (NuG2)8 and (GB1)8 were carried out in PBS buffer. For hFc binding studies, we carried out AFM measurements in the presence of different concentrations of hFc. To preequilibrate hFc with polyproteins, we used two methods: premixing and in situ mixing. For premixing, we mixed the hFc with polyprotein solution at least 24 h before the pulling experiments. For the in situ mixing, we first deposited polyprotein onto a glass coverslip and then added hFc solution to mix it with polyprotein. The AFM experiments were carried out after allowing the mixture to equilibrate for ≈30 min. The results for these two mixing methods were identical. The spring constant of each individual cantilever [Si3N4 cantilevers from Veeco Probes (Camarillo, CA), with a typical spring constant of 15 pN·nm−1] was calibrated in PBS buffer by using the equipartition theorem before each experiment. The pulling speed used for all of the pulling experiments was 400 nm·s−1. The spacing between consecutive unfolding events was determined in an unbiased and hands-off fashion by using an algorithm custom-written in Igor Pro 5.0 (WaveMetrics, Lake Oswego, OR).
To estimate the fraction of hFc-bound NuG2 or hFc-bound GB1, each unfolding force histogram was fitted with two Gaussian functions. The binding curves were fitted by using a single binding site model that takes into account all of the species present in solutions:
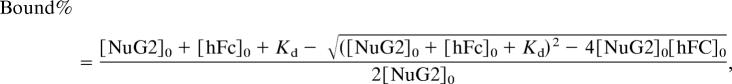 |
where [NuG2]0 is the initial concentration of NuG2 domains in the solution, [hFc]0 is the initial concentration of hFc in the solution, and Kd is the dissociation constant (37).
Supplementary Material
Acknowledgments
We thank David Baker for providing constructs containing proteins NuG2 and GB1. This work was supported by the Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs Program, and the Canada Foundation for Innovation.
Abbreviations
- AFM
atomic force microscopy
- hFc
Fc fragment of human IgG
- wt
wild type
- WLC
worm-like chain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705367104/DC1.
References
- 1.Arkin MR, Wells JA. Nat Rev Drug Discovery. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Swain JF, Gierasch LM. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Changeux JP, Edelstein SJ. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 4.Lakey JH, Raggett EM. Curr Opin Struct Biol. 1998;8:119–123. doi: 10.1016/s0959-440x(98)80019-5. [DOI] [PubMed] [Google Scholar]
- 5.Cooper MA. Anal Bioanal Chem. 2003;377:834–842. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- 6.Piehler J. Curr Opin Struct Biol. 2005;15:4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Carrion-Vazquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernandez JM. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, Schulten K. Biophys J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paci E, Karplus M. Proc Natl Acad Sci USA. 2000;97:6521–6526. doi: 10.1073/pnas.100124597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ainavarapu SR, Li L, Badilla CL, Fernandez JM. Biophys J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 12.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 13.Akerstrom B, Brodin T, Reis K, Bjorck L. J Immunol. 1985;135:2589–2592. [PubMed] [Google Scholar]
- 14.Nilson B, Bjorck L, Akerstrom B. J Immunol Methods. 1986;91:275–281. doi: 10.1016/0022-1759(86)90490-4. [DOI] [PubMed] [Google Scholar]
- 15.Sloan DJ, Hellinga HW. Protein Sci. 1999;8:1643–1648. doi: 10.1110/ps.8.8.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sagawa T, Oda M, Morii H, Takizawa H, Kozono H, Azuma T. Mol Immunol. 2005;42:9–18. doi: 10.1016/j.molimm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Du HN, Hu HY. Biopolymers. 2003;72:116–122. doi: 10.1002/bip.10300. [DOI] [PubMed] [Google Scholar]
- 18.Powell KD, Ghaemmaghami S, Wang MZ, Ma L, Oas TG, Fitzgerald MC. J Am Chem Soc. 2002;124:10256–10257. doi: 10.1021/ja026574g. [DOI] [PubMed] [Google Scholar]
- 19.Sjobring U, Bjorck L, Kastern W. J Biol Chem. 1991;266:399–405. [PubMed] [Google Scholar]
- 20.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Structure (London) 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y, Lam C, Wang M, Li H. Angew Chem Int Ed Engl. 2006;45:642–645. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Li H. Nat Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 23.Li PC, Makarov DE. J Phys Chem B. 2004;108:745–749. [Google Scholar]
- 24.Nauli S, Kuhlman B, Baker D. Nat Struct Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 25.Marko JF, Siggia ED. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 26.Hann E, Kirkpatrick N, Kleanthous C, Smith DA, Radford SE, Brockwell DJ. Biophys J. 2007;92:L79–L81. doi: 10.1529/biophysj.106.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junker JP, Hell K, Schlierf M, Neupert W, Rief M. Biophys J. 2005;89:L46–L48. doi: 10.1529/biophysj.105.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ha T, Zhuang X, Kim HD, Orr JW, Williamson JR, Chu S. Proc Natl Acad Sci USA. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majumdar DS, Smirnova I, Kasho V, Nir E, Kong X, Weiss S, Kaback HR. Proc Natl Acad Sci USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florin EL, Moy VT, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 31.Akerstrom B, Bjorck L. J Biol Chem. 1986;261:10240–10247. [PubMed] [Google Scholar]
- 32.Gulich S, Linhult M, Stahl S, Hober S. Protein Eng. 2002;15:835–842. doi: 10.1093/protein/15.10.835. [DOI] [PubMed] [Google Scholar]
- 33.Malakauskas SM, Mayo SL. Nat Struct Biol. 1998;5:470–475. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 34.Saha K, Bender F, Gizeli E. Anal Chem. 2003;75:835–842. doi: 10.1021/ac0204911. [DOI] [PubMed] [Google Scholar]
- 35.Walker KN, Bottomley SP, Popplewell AG, Sutton BJ, Gore MG. Biochem J. 1995;310:177–184. doi: 10.1042/bj3100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marko JF, Siggia ED. Biophys J. 1997;73:2173–2178. doi: 10.1016/S0006-3495(97)78248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segel IH. Enzyme Kinetics. New York: Wiley; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.