Abstract
Human colonic epithelial cell renewal, proliferation, and differentiation are stringently controlled by numerous regulatory pathways. To identify genetic programs of human colonic epithelial cell differentiation in vivo as well as candidate marker genes that define colonic epithelial stem/progenitor cells and the stem cell niche, we applied gene expression analysis of normal human colon tops and basal crypts by using expression microarrays with 30,000 genes. Nine hundred and sixty-nine cDNA clones were found to be differentially expressed between human colon crypts and tops. Pathway analysis revealed the differential expression of genes involved in cell cycle maintenance and apoptosis, as well as genes in bone morphogenetic protein (BMP), Notch, Wnt, EPH, and MYC signaling pathways. BMP antagonists gremlin 1, gremlin 2, and chordin-like 1 were found to be expressed by colon crypts. In situ hybridization and RT-PCR confirmed that these BMP antagonists are expressed by intestinal cryptal myofibroblasts and smooth muscle cells at the colon crypt. In vitro analysis demonstrated that gremlin 1 partially inhibits Caco-2 cell differentiation upon confluence and activates Wnt signaling in normal rat intestinal epithelial cells. Collectively, the expression data set provides a comprehensive picture of human colonic epithelial cell differentiation. Our study also suggests that BMP antagonists are candidate signaling components that make up the intestinal epithelial stem cell niche.
Keywords: gremlin, expression profiling, microarray, crypt maturation program, myofibroblast
The human adult colonic epithelium undergoes perpetual regeneration fueled by intestinal epithelial stem and progenitor cells located at the colon crypt base. Perturbation of the pathways regulating stem cell renewal contributes significantly to neoplastic transformation. The current basis of our understanding of intestinal stem cells is primarily derived from studying the small intestine, which shares major regulatory pathways with the colon. Specifically, several key regulatory signals are involved in intestinal stem cell renewal and differentiation, including the Wnt, bone morphogenetic protein (BMP), and Notch pathways (1–3).
Among these pathways, canonical Wnt signaling plays a major role in maintaining intestinal stem cell fate and progenitor cell proliferation (4). BMP signaling, in contrast, has been reported to inhibit intestinal stem cell activation and promote intestinal differentiation (5). Cell fate decisions in the intestine have been shown to involve Notch signaling, which specifically directs cells toward a secretory lineage in the gut (6). All of the evidence suggests there is a close interaction of several key pathways in directing intestinal epithelial stem cell renewal and differentiation. Yet how these different pathways coordinate in the specific anatomical compartment of the intestine remains mostly unknown.
Intestinal epithelial stem cells are supported by underlying myofibroblasts known as intestinal subepithelial myofibroblasts (ISEMFs), which are in close proximity to the smooth muscle cells of the muscularis mucosae layer. These cells at the base of intestinal crypts may contribute to the stem cell niche and act as regulators of intestinal stem cell self-renewal and differentiation.
Several genomic studies have been applied to study mouse intestinal epithelial stem cells and their differentiation program by using either expression array technology or cDNA library sequencing (7–9). These gene expression analyses have provided valuable information and candidate markers for mouse gastrointestinal stem/progenitor cells, as well as revealing the differentiation program of these cells. However, no information regarding the stem cell niche environment, specifically for the supporting cells, is known because previous experiments used microdissected or isolated epithelial cells. Furthermore, no data are available with regard to the human intestine, especially for the human colon. Data on the proliferation program governing the stem/progenitor cell compartment and the differentiation program of colon epithelial cells are of particular importance because colon cancer is one of the most common cancer types, whereas small intestinal cancer is exceedingly rare in humans.
In this article, we characterized the gene expression profiles of the human colon by comparing the gene expression pattern between the top and basal crypt compartments. We identified a comprehensive list of differentially expressed genes encompassing major pathways regulating intestinal epithelial stem cell renewal. Among these pathways, we identified elements that contribute to the stem cell niche, which were then validated by cellular localization and in vitro functional studies. Our data set provides a comprehensive picture of the human colonic epithelial cell differentiation program and helps identify elements that contribute to the maintenance of the intestinal stem cell niche.
Results
Gene Expression Signatures of Human Colon Top and Bottom Crypt Compartments.
Using cDNA microarrays containing 44,500 cDNA clones representing ≈30,000 unique genes, we generated gene expression profiles from nine paired horizontally dissected human colon top versus bottom crypt tissue compartments. We next applied significance analysis of microarrays (SAM) to the array data set and identified 969 cDNA clones representing ≈736 unique genes that are differentially expressed in colon top versus bottom crypts, with a false discovery rate of <0.1%. Among these genes, 367 cDNA clones (299 unique genes) were highly expressed in colon bottom crypts, and 602 cDNA clones (437 unique genes) were expressed in colon tops [see supporting information (SI) Table 1 for the corresponding list of genes].
Careful examination of the genes that are highly expressed at colon basal crypts revealed that, apart from previously well known genes such as the c-myc and the EphB family (EPHB2, EPHB3, and EPHB4), two major clusters exist (clusters I and II in Fig. 1). Cluster I includes many genes involved in cell proliferation and cell cycle regulation, as well as candidate oncogenes (e.g., CDC20, Cyclin B2, PTTG1, and FYN). These genes are cell cycle-regulated and are highly expressed in tumor cells, compared with normal tissues in a variety of tumor types (10). As such, these genes are most likely to be expressed by proliferating cryptic progenitor cells. Cluster II includes many genes that encode secretory proteins and genes involved in cell matrix or matrix modeling (e.g., Fibronectin, TIMP3, ADAMTS1, and TAGLIN). Some of these genes (including Fibronectin and TAGLIN) have been found to be expressed by myofibroblasts as well as smooth muscle cells (11, 12). Therefore, we suspect that genes in this cluster most likely represent genes that are expressed by cryptic stromal cells. Strikingly, there are three BMP antagonists expressed in this cluster: gremlin 1 (GREM1), gremlin 2 (GREM2), and chordin-like 1 (CHRDL1), whose expression and role in the normal human colon are mostly unknown. The genes expressed in the colon top include genes that inhibit cell proliferation (p21 and MAD), cell adhesion molecules (CDH1 and TJP3), and genes encoding functional proteins of gut epithelial cells (membrane transporters ABCB1, ABCG2, or enzymes like CA4). Together the data support that our microarray analysis accurately captures the global gene expression patterns of colon top versus basal crypts.
Fig. 1.
Hierarchical clustering of genes differentially expressed in colon top and basal crypt as identified by SAM. Cluster I is enriched in genes associated with cell proliferation, and cluster II is enriched in genes expressed in pericryptal mesenchymal cells.
To further characterize the functional significance of genes expressed in colon basal crypts and tops, we performed gene ontology (GO) term analysis and identified GO terms, which are enriched in each gene list with a cutoff P value of <0.05 (SI Table 2). GO term analysis facilitates the interpretation of data by providing biological, physiological, and functional descriptions of gene products. The GO terms that are enriched and unique in the basal crypt gene list include “M phase,” “cell cycle,” “protein biosynthesis,” “macromolecular biosynthesis,” and “DNA replication.” These terms are clearly related to the cell proliferation and cell renewal at basal crypts. In contrast, GO terms that are enriched and unique in the colon top gene list include “cell communication,” “digestion,” “establishment of localization,” “transport,” “ion transport,” etc. These GO terms are consistent with the expression of genes required for digestive function and transport in mature intestinal epithelial cells.
Expression Profiling in Different Molecular Pathways.
To gain a broader picture of gene expression changes and to elucidate the molecular and biological pathways involved in colon crypt maturation, we examined the global expression profile data set by using paired t test. Of the 25,132 cDNA clones, 6,087 were found to be significantly altered between the two compartments with the cutoff value at P < 0.01 (approximate false discovery rate of 4%) (SI Table 3). These 6,087 transcripts were then visualized by using GenMapp software to examine their relationship in various biological pathways. Expression data of genes in key signal transduction pathways regulating stem cell renewal also were extracted by using a threshold of P < 0.05 in paired t test.
Cell Cycle and Apoptosis.
A significant increased gene expression signature enriched in the cell cycle pathway was observed in bottom crypts, consistent with the findings that proliferative activity is located within this compartment (SI Fig. 6A). In particular, 85% of the differentially expressed genes within this pathway were significantly up-regulated in the bottom compartments. By contrast, inhibitors of cell cycle, including CDKN1A and CDKN2A, were down-regulated in the bottom compartment. Genes involved in RNA and protein processing, including ribosomal proteins and translation factors, also were up-regulated in the bottom crypts (SI Fig. 7). We next examined genes involved in the apoptosis pathway and noted that most of these genes, including TNF, its receptor TNFRSF1B, CRADD, CASP10, and BAK1, are significantly down-regulated in the colon bottoms (SI Fig. 6B). Our array data are consistent with the occurrence of cell maturation and elimination of epithelial cells through apoptosis at the colon top compartment.
We next examined the expression of an essential group of genes that control cell growth: the Myc/Mad/Max network (SI Fig. 8A). As expected, oncogenic MYC was highly expressed in the proliferative bottom crypt, whereas its dimerization partner MAX and its antagonist MAD were restricted to the upper crypt. In addition, the MXI1 gene that functions to antagonize MYC by competing for MAX also was highly expressed at colon tops. Our findings suggest that proliferation is prohibited in the upper mature colon compartment by expression of multiple MYC antagonists.
Wnt Signaling Pathway.
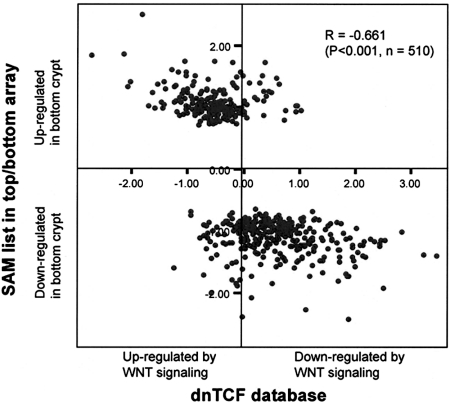
To verify the key contribution of the Wnt signaling pathway in controlling colon crypt development, we correlated the 969 cDNA clones that were differentially expressed as identified by SAM with the previously published Wnt target gene data set obtained by using inducible dnTCF-4 in CRC cell lines by van de Watering et al. (13). Interestingly, we observed an exceedingly high concordance of expression between the two data sets (Pearson correlation coefficient, −0.661; P < 0.001) (Fig. 2): Genes highly expressed in colon tops are mostly induced by interruption of Wnt signaling through dnTCF4 (e.g., p21, BMP2, MAD, and CDH18), whereas genes highly expressed in colon crypts are mostly repressed by dnTCF4 (e.g., MYC, CDCA7, EPHB2, and EPHB3) (SI Fig. 9). These results provide direct evidence that Wnt/β-catenin signaling pathways are a major determinant of gene expression patterns along the colon crypt axis.
Fig. 2.
Significant correlation between genes differentially expressed in colon top and basal crypt and Wnt/β-catenin signaling targets. Microarray data of inducible expression of dnTCF4 in Ls174 cells were retrieved from van de Wetering et al. (13), and overlapping clones with colon top–bottom crypt gene list as identified by SAM were selected and calculated for correlation. The x axis measures mean gene expression change (log2) 23 h after dnTCF4 induction, and the y axis measures mean fold change (log2) of bottom versus top colon crypt compartments.
BMP Signaling Pathway.
We noted differential expression of multiple BMP components along the colon crypt axis (SI Fig. 8B). BMP1, BMP2, BMP5, BMP7, SMAD7, and BMPR2 were highly expressed in colon tops, whereas BMP antagonists CHRDL1, GREM1, and GREM2 were enriched in basal colon crypts. This observation suggests that BMP signaling is activated in the upper crypt, whereas secretory inhibitors CHRDL1, GREM1, and GREM2 located at the bottom antagonize BMP signaling in the intestinal epithelial stem cell niche.
NOTCH Signaling Pathway.
It is known that the transmembrane NOTCH receptor is cleaved upon activation by its ligand (Delta/JAG), releasing the intracellular domain of Notch (NICD). NICD then migrates to the nucleus and activates the transcriptional regulator RBPSUH/RBP-Jk by binding to it. We observed an expression profile consistent with the activation of NOTCH signaling in the bottom crypt, where NOTCH1, NOTCH2, NOTCH3, RBPSUH, and TLE2 were highly expressed at the basal crypt and the NOTCH ligand JAG1 was expressed at the top (SI Fig. 8C).
The EPH Family.
We noted a distinct expression gradient of multiple members of the EPHA and EPHB family of tyrosine kinase receptors as well as their ligands in the colon crypt axis (SI Fig. 8D). Expression of EPHB receptors and their ligands are implicated in maintaining the correct positioning as well as driving proliferation of the progenitor compartment in the crypt–villus axis of the mouse intestine (14, 15). Consistent with the published data on the EPHB families, we noted expression of EPHB1, EPHB2, EPHB3, EPHB4, and EPHB6 in the crypt base, whereas the ligand EFNB2 was expressed at colon tops. Interestingly, we also noted differential expression of the EPHA receptor family in the colon crypt axis, with high expression of EPHA1, EPHA4, and EPHA7 at the crypt base and high expression of EPHA2, EPHA5, and the ligand EFNA1 in colon tops. Our results call for further study of the role of the EPHA family in controlling colon crypt maturation and its possible involvement in the oncogenic process.
Quantitative RT-PCR Validation of Differentially Expressed Genes.
To verify our bottom–top array data, several genes belonging to different key pathways were selected for validation by using quantitative RT-PCR in four pairs of samples, including MXI1 (Myc/Mad/Max family); APC and SFRP1 (WNT signaling); GREM1, GREM2, and CHRDL1 (BMP signaling); JAG1 (Notch pathway); EFNA1 (Eph family); DUSP5 (MAPK pathway); and GPC4 (candidate stem cell marker). All of the selected genes were confirmed to be differentially expressed between colon bottom–top compartments by quantitative RT-PCR (SI Table 4).
BMP Antagonists Are Expressed by Subepithelial Myofibroblasts and Smooth Muscle Cells at Colon Crypts.
One of the most intriguing observations is the distribution of BMP signaling pathway molecules along the colon crypt axis, including BMP ligands and receptor and signaling molecules. In the colon top, BMP1, BMP2, BMP5, BMP7, SMAD7, and BMPR2 are highly expressed, whereas the basal crypt exhibits high expression of three BMP antagonists, GREM1, GREM2, and CHRDL1 (Fig. 1 and SI Fig. 8B). The latter information, which was previously unknown, suggests the unusual requirement to block BMP signaling at the colon basal crypt region. Furthermore, we discovered that the three BMP antagonists are part of a cocluster of genes that are enriched for those putatively expressed by intestinal cryptic stromal cells, such as the fibronectin gene (Fig. 1), which prompted us to suspect that these BMP antagonists also may be expressed by stromal cells.
To investigate the cellular origin of BMP antagonists, we performed in situ hybridization for GREM1 and GREM2 on human colon tissues. In situ hybridization revealed strong expression of GREM1 and GREM2 in the mesenchymal cells in the basal part of the lamina propria and muscularis mucosae of the colon (Fig. 3 A and B), whereas sense probes showed no staining (data not shown). Expression corresponded to cells on serial sections that stained for fibronectin (Fig. 3C) and α-smooth muscle actin (Fig. 3D), markers expressed in both myofibroblasts and smooth muscle cells. Transcripts of GREM1 and GREM2 were not detected in epithelial cells, mesenchymal cells in the top part of the lamina propria and submucosa, or smooth muscle cells in the muscularis propria (data not shown).
Fig. 3.
Expression and localization of GREM1 and GREM2 by myofibroblast cells and smooth muscle cells at colon crypt. (A and B) In situ hybridization (ISH) for GREM1 (A) and GREM2 (B). Dark brown dots indicate positive staining. (C and D) Immunohistochemical staining of fibronectin (C) and α-smooth muscle actin (D) as markers for intestinal myofibroblasts as well as smooth muscle cells. Dark brown staining indicates positive staining. (E and F) Double labeling for GREM1 mRNA (E, red) and myofibroblast marker vimentin (F, green) at colon basal crypt region. (G) Combined image showing coexpression of GREM1 and vimentin (yellow dots indicated by white arrows) at scattered pericryptal mesenchymal cells corresponding to myofibroblasts. See SI Fig. 10 for the enlarged version of fluorescent ISH/immunostaining. (H) RT-PCR analysis of BMP antagonists expression in four intestinal myofibroblast isolates (CMF11, CMF7B, IMF11B, and 18Co) as well as three colon cancer cell lines (Caco-2, DLD-1, and HT29).
In the colon, it has been shown that myofibroblasts are vimentin-positive, whereas smooth muscle cells are vimentin-negative (16). Although expression of GREM1 and GREM2 in α-smooth muscle actin-positive smooth muscle cells of the muscularis mucosae is unequivocal, specific expression of GREM1 and GREM2 by cryptal myofibroblasts remained unclear because of their inconspicuous morphology. Thus, we performed coimmunofluorescence with vimentin to further define the cellular origin. Consistently, we observed no GREM1 signal in colon tops. In addition, we found GREM1 mRNA (Fig. 3E) and vimentin staining (Fig. 3F) colocalized (Fig. 3G, white arrows; also see SI Fig. 10 for the enlarged version of the coimmunostaining) in certain mesenchymal cells surrounding the basal crypts, suggesting that gremlin 1 also is secreted by myofibroblasts.
To further validate our findings, we isolated primary colonic myofibroblasts from histologically normal human colonic tissue and assayed for gene expression by RT-PCR. The myofibroblast features of isolated cell lines were confirmed by immunofluorescent staining for fibronectin, vimentin, and α-smooth muscle actin (SI Fig. 11). The mRNA for BMP antagonists GREM1, GREM2, and CHRDL1 were detected in human colonic and ileal myofibroblasts, but never or weakly in colon tumor epithelial cells (Fig. 3H).
Taken together, the data demonstrate that gremlin 1, gremlin 2, and chordin-like 1 in the gastrointestinal tract likely originate from myofibroblasts and smooth muscle cells, both located at the crypt base in proximity to the stem cell niche. Thus, we hypothesized that, through inhibiting BMP signaling locally, these BMP antagonists may function to maintain Wnt signaling and inhibit differentiation at the crypt base.
Gremlin 1 Partially Inhibits Caco-2 Cell Differentiation.
To determine whether gremlin 1 interferes with differentiation in intestinal epithelial cells, Caco-2 cells were treated with recombinant gremlin 1, and gene expression of intestinal differentiation markers was assayed by quantitative RT-PCR. Caco-2 cells have been shown to spontaneously differentiate into an enterocyte phenotype in 21 days upon reaching confluence and form a polarized monolayer resembling the intestine (17). In a microarray study of Caco-2 cell differentiation, it was found that expression levels of mature differentiation marker genes reach a plateau at 4 to 7 days postconfluence, and the expression levels do not significantly go up during the rest of the 21 days in culture (A. Saaf, personal communication). We have further validated these results by quantitative RT-PCR (data not shown). Therefore, we chose 7 days postconfluence to study the effect of gremlin 1 on Caco-2 cell differentiation. We assayed the expression of two genes: p21/CDKN1A, a marker for cell cycle inhibition; and ANPEP, a brush border enzyme. We found that 7 days of gremlin 1 treatment consistently decreased p21 gene expression by 20–30% in Caco-2 cells compared with control cells (Fig. 4A). Similarly, 7 days of gremlin 1 treatment consistently decreased ANPEP gene expression by 40–50% in Caco-2 cells compared with control cells (Fig. 4A). These findings suggest that gremlin 1 partially inhibits intestinal differentiation, and thus gremlin 1 may play a crucial role in inhibiting differentiation near the crypt base.
Fig. 4.
Gremlin 1 partially inhibits Caco-2 cell differentiation and activates Wnt/β-catenin signaling in normal intestinal cells. (A) Quantitative RT-PCR analysis revealed a statistically significant decrease in expression of intestinal epithelial differentiation markers ANPEP and p21 at day 7 when Caco-2 cells were cultured in growth media supplemented with gremlin 1. The analysis detected a significant up-regulation of the AXIN2 transcript in Caco-2 cells after a 4-h treatment with gremlin 1 (*, P < 0.05). (B) Quantitative RT-PCR analysis demonstrated a statistically significant increase in AXIN2 expression in normal rat intestinal cells IEC-6 and IEC-18 after 48-h treatment with gremlin 1 (*, P < 0.01). (C and D) Gremlin 1 induces nuclear/cytoplasm localization of β-catenin in IEC-18 cells.
Gremlin 1 Activates Wnt Signaling in Intestinal Cells.
In a previous study, overexpressing the BMP antagonist noggin in the intestine promoted Wnt activity and the development of ectopic crypts (18). Consistent with the hypothesis that BMP antagonists may activate Wnt signaling, we noticed that, in Caco-2 cell differentiation assays, gremlin 1 is able to transiently induce expression of the known Wnt target gene AXIN2 (19, 20) in Caco-2 cells at 4 h (Fig. 4A). To test our hypothesis that gremlin 1 assists in maintaining Wnt signaling in normal intestine, we treated two normal rat intestinal epithelial cell lines, IEC-6 and IEC-18, with gremlin 1 for 48 h and examined the expression of AXIN2. Quantitative RT-PCR analysis revealed that the expression of AXIN2 was significantly up-regulated by gremlin 1 treatment in both tested cell lines (Fig. 4B). We next examined whether gremlin 1 affects β-catenin activity by assaying the subcellular localization of β-catenin in IEC-18 cells. We found that, in untreated IEC-18 cells, none of the cells displayed nuclear β-catenin staining. After incubating with gremlin 1, nuclear β-catenin was observed in a small number of IEC-18 cells (Fig. 4 C and D). All these data support that gremlin 1 is able to activate Wnt signaling in intestinal epithelial cells.
In summary, our data support that the BMP antagonists gremlin 1, gremlin 2, and chordin-like 1 are expressed by colon crypt myofibroblasts and smooth muscle cells and contribute to the stem cell niche by activating Wnt signaling and inhibiting differentiation of basal crypt epithelial cells.
Discussion
In this manuscript, we provide a comprehensive genomic analysis of genes differentially expressed at human colon top and basal crypt compartments. Our results reveal alteration in a diverse spectrum of genes reflecting not only a difference in cell proliferation versus differentiation/apoptosis along the colon crypt axis but also changes in various components of key signaling pathways regulating colon stem cell renewal. Although many similarities were noted in comparison with an expression profiling database derived from mouse small intestine (8), our data extend the findings to humans and provide unique information about the colon, including elements highly relevant to colon carcinogenesis. Specifically, our data captured information not only from the epithelial cells, but also the supporting tissue microenvironment, which may contribute critical elements for creating and maintaining the stem cell niche.
The identification of genes highly expressed in colon crypts provides us with a unique opportunity to search for markers of intestinal stem/progenitor cells. We compared the crypt gene list with genes that are highly expressed in human ES and embryonic carcinoma (EC) cells (21) and identified 31 genes, including GAB1, PTTG1, EBAF, GPC4, and MYBL, which are highly expressed in ES and EC cells as well as in colon crypts (SI Fig. 12 and SI Table 5). These genes mutually expressed in basal crypts and ES and EC cells represent potential markers for intestinal stem or progenitor cells. Some potential cell surface proteins (e.g., GPC4) might be useful markers for the purification of intestinal stem/progenitor cells. One has to be cautious, however, because some of these genes may simply represent proliferating cell signatures in ES, EC, and cryptic progenitor cells. Further studies to address the cellular localization of these genes in the intestinal compartment and their function in intestinal stem/progenitor cell differentiation will improve our understanding of intestinal stem/progenitor cells.
Although we observed gene expression profiles reflecting activated Wnt signaling in colon crypts (Fig. 2), the exact mechanism leading to Wnt activation remains unclear. We have observed differential expression of several members involved in transduction or regulation of Wnt signaling along the colon crypt axis. Specifically, APC, WNT5B, and TCF4 were localized at the crypt top, whereas AXIN2, DKK3, TCF3, SFRP1, SFRP2, FZD2, FZD3, FZD7, and FZDB were all restricted to the bottom (SI Table 3). Many of these observed differential expression patterns were consistent with an expression study based on in situ hybridization in mouse intestine (22). The reason for the expression of several Wnt secretory inhibitors in the colon crypt base is unclear. It may suggest either a negative-feedback regulation or the need to fine-tune Wnt activity through an intricate balance of positive and negative regulators in this specific anatomical location.
Altogether, based on our expression profiling data, we generated a model depicting the components and signaling molecules featured in this study and their differential expression along the colon crypt axis (Fig. 5), including activated Wnt and Notch signaling at the crypts and BMP signaling at the tops. The differential distribution of Eph receptors and their ligands, as well as MYC and MYC antagonists, helps maintain crypt polarity through regulating cell positioning and proliferation.
Fig. 5.
Graphical view of human colon intestinal epithelial cell development and stem cell niche maintenance. Only genes with significant differential expression in paired t test (P < 0.05) are listed. ISEMF, intestinal subepithelial myofibroblast; SMC, smooth muscle cell.
The discovery of genes localized to the colonic stem cell niche provides further understanding of the signaling pathways important in this region. However, the detailed mechanisms of how the different signaling pathways coordinate to create this niche remains largely unknown. It has been hypothesized that Wnt signaling is required but not sufficient for intestinal stem cell activation and self-renewal. A second signal to antagonize BMP is required so as to release its inhibitory effect on nuclear translocation of β-catenin. In mouse intestine, transient expression of the BMP antagonist noggin has been observed in pericryptal mesenchymal cells and intestinal epithelial stem cells, which may contribute to this required second signal (23). Although a cDNA clone corresponding to noggin was not present in our array, we found expression of three different BMP antagonists, including GREM1, GREM2, and CHRDL1, in colon basal crypts by pericryptal mesenchymal cells. Moreover, we found a similar effect of gremlin 1 in promoting nuclear translocation of β-catenin and activating Wnt signaling. These findings led us to propose a model of how BMPs and their antagonists, including gremlin 1, contribute to create the colonic epithelial stem cell niche through modulation of Wnt activity (Fig. 5). In this model, BMPs act to restrict stem cell expansion and are expressed at colon tops with a decreasing gradient toward the crypt. BMP antagonists, including gremlin 1, expressed by ISEMFs and smooth muscle cells in turn create an opposite gradient to antagonize BMPs, thus maintaining Wnt activity at the crypt base. This gradient then creates an environment that promotes stem cell self-renewal and expansion at the crypt base region. Indeed, the expression of multiple BMP antagonists by ISEMFs and smooth muscle cells has provided an optimal anatomical setup for the creation and maintenance of the stem cell niche in the basal crypt region of the colon.
Both BMPs and their antagonists play essential roles in stem cell biology, although their functions may vary in different stem cell systems (24). In a recent study, GREM1 was reported to be expressed in stromal cells of basal cell carcinoma of the skin, and gremlin 1 was shown to inhibit differentiation and promote proliferation in basal cell carcinoma cells in vitro (25). Expression of GREM1 also was noted in stromal cells in diverse types of human cancer, including colon cancer. Consistently, we observed GREM1 expression by stromal cells in a subset of human colon cancer samples (SI Fig. 13). The staining of GREM1 in tumor stromal cells tends to be stronger than that in normal myofibroblast and smooth muscle cells at the colon crypt. The data suggest that GREM1 expression is up-regulated during the development of a subset of colon tumors, and thus BMP antagonists may represent important stem cell niche factors in both normal and neoplastic conditions. It would be of great interest to further investigate and clarify the role of BMP antagonists in the colon cancer stem cell niche. Such studies may provide new opportunities for therapeutic strategy through the modulation of BMP activity.
Materials and Methods
Tissue Samples, Microarrays, and Data Analysis.
Colectomy specimens were received fresh from the operating theater immediately upon resection. Morphologically normal colon mucosae were laid completely flat on a metal surface and frozen in liquid nitrogen. Ten-microgram-thick serial horizontal sections were cut such that the early sections contained the top compartment, whereas the deeper sections contained the basal crypt compartment (SI Fig. 14). Based on interval sections stained for H&E, tissues from top and basal crypt compartments were selected for expression profiling, skipping tissue from the mid-crypt region. Total RNA was isolated from nine pairs of colon top and crypt compartments, amplified together with universal human reference RNA (Stratagene, La Jolla, CA) and hybridized to cDNA microarrays produced by Stanford Functional Genomics Facility. The raw data were deposited in Stanford Microarray Database at http://smd.stanford.edu. The raw data also were submitted to Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo, accession no. GSE6894). Paired SAM (26) was performed to identify genes differentially expressed in colon top versus crypt. The GO Term Finder program (27) was used to analyze the list of differentially expressed genes for enrichment of specific functional groups.
Quantitative RT-PCR, Immunohistochemistry, and in Situ Hybridization.
The procedure for quantitative RT-PCR was performed by using TaqMan Gene Expression Assay (Applied Biosystems, Foster City, CA) for validation of expression array data and SYBR Green Master mix for cell culture experiments. Primer sequences are listed in SI Table 6. In situ hybridization and double staining by using immunofluorescence and in situ hybridization were carried out as described previously (28). Immunohistochemical staining was performed by using the standard method.
Cell Culture and Cell Treatment.
Primary intestinal myofibroblast cultures were established from histologically normal margins of surgically resected colonic or ileal tissue by using the outgrowth method described by Mahida et al. (29). All other cells were obtained from American Type Culture Collection (Manassas, VA). Recombinant gremlin (R&D Systems, Minneapolis, MN) was added to the cells for various times as described in the text.
See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank the Stanford Functional Genomic Facility and Stanford Microarray Database for support on microarray experiments and Chris Haqq, Julie Sneddon, Patrick Brown, and Linheng Li for experimental advice. This work was supported in part by National Institutes of Health Grants R21DK-069309 (to X.C.) and R01DK-055783 (to D.W.P.), Research Grants Council of the Hong Kong Special Administrative Region Grant HKU7524/06M (to S.Y.L., S.T.Y., T.L.C., and X.C.), and a California Institute for Regenerative Medicine stem cell fellowship (to C.K.).
Abbreviations
- BMP
bone morphogenetic protein
- EC
embryonic carcinoma
- GO
gene ontology
- ISEMF
intestinal subepithelial myofibroblast
- SAM
significance analysis of microarrays.
Footnotes
The authors declare no conflict of interest.
Data deposition: The array data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/projects/geo (accession no. GSE6894).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707210104/DC1.
References
- 1.Rubin DC. Curr Opin Gastroenterol. 2007;23:111–114. doi: 10.1097/MOG.0b013e3280145082. [DOI] [PubMed] [Google Scholar]
- 2.Crosnier C, Stamataki D, Lewis J. Nat Rev Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- 3.Leedham SJ, Brittan M, McDonald SA, Wright NA. J Cell Mol Med. 2005;9:11–24. doi: 10.1111/j.1582-4934.2005.tb00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 5.He XC, Zhang J, Li L. Ann NY Acad Sci. 2005;1049:28–38. doi: 10.1196/annals.1334.005. [DOI] [PubMed] [Google Scholar]
- 6.van Es JH, Clevers H. Trends Mol Med. 2005;11:496–502. doi: 10.1016/j.molmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Stappenbeck TS, Mills JC, Gordon JI. Proc Natl Acad Sci USA. 2003;100:1004–1009. doi: 10.1073/pnas.242735899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mariadason JM, Nicholas C, L'Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, et al. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, Gordon JI. J Biol Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield ML, George LK, Grant GD, Perou CM. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 11.Pourreyron C, Dumortier J, Ratineau C, Nejjari M, Beatrix O, Jacquier MF, Remy L, Chayvialle JA, Scoazec JY. Int J Cancer. 2003;104:28–35. doi: 10.1002/ijc.10898. [DOI] [PubMed] [Google Scholar]
- 12.Lawson D, Harrison M, Shapland C. Cell Motil Cytoskeleton. 1997;38:250–257. doi: 10.1002/(SICI)1097-0169(1997)38:3<250::AID-CM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 14.Batlle E, Henderson JT, Beghtel H, van de Born MMW, Sancho E, Huls G, Pawson T, Clevers H. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 15.Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisen J. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Adegboyega PA, Mifflin RC, DiMari JF, Saada JI, Powell DW. Arch Pathol Lab Med. 2002;126:829–836. doi: 10.5858/2002-126-0829-ISOMIN. [DOI] [PubMed] [Google Scholar]
- 17.Chantret I, Barbat A, Dussaulx E, Brattain MG, Zweibaum A. Cancer Res. 1988;48:1936–1942. [PubMed] [Google Scholar]
- 18.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 19.Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER. J Biol Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 20.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, Clevers H. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 23.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Li L. Dev Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Sneddon JB, Zhen HH, Montgomery K, van de Rijn M, Tward AD, West R, Gladstone H, Chang HY, Morganroth GS, Oro AE, Brown PO. Proc Natl Acad Sci USA. 2006;103:14842–14847. doi: 10.1073/pnas.0606857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West RB, Rubin BP, Miller MA, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli RJ, De Luca A, Downs-Kelly E, et al. Proc Natl Acad Sci USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahida YR, Galvin AM, Gray T, Makh S, McAlindon ME, Sewell HF, Podolsky DK. Clin Exp Immunol. 1997;109:377–386. doi: 10.1046/j.1365-2249.1997.4481346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.