Abstract
Plant somatic cells have the remarkable ability to regenerate an entire organism. Many species in the genus Kalanchoë, known as “mother of thousands,” develop plantlets on the leaf margins. Using key regulators of organogenesis (STM) and embryogenesis (LEC1 and FUS3) processes, we analyzed asexual reproduction in Kalanchoë leaves. Suppression of STM abolished the ability to make plantlets. Here, we report that constitutive plantlet-forming species, like Kalanchoë daigremontiana, form plantlets by coopting both organogenesis and embryogenesis programs into leaves. These species have a defective LEC1 gene and produce nonviable seed, whereas species that produce plantlets only upon stress induction have an intact LEC1 gene and produce viable seed. The latter species are basal in the genus, suggesting that induced-plantlet formation and seed viability are ancestral traits. We provide evidence that asexual reproduction likely initiated as a process of organogenesis and then recruited an embryogenesis program into the leaves in response to loss of sexual reproduction within this genus.
Keywords: embryogenesis, LEC1, STM
Unlike animal cells, somatic cells of plants are capable of regenerating the entire adult organism, and this potential for regeneration is called totipotency. In some plants, this ability is used as a mechanism of vegetative reproduction (1) and may represent the only means of reproduction. Species in the genus Kalanchoë (Crassulaceae) reproduce asexually by forming plantlets along their leaf margins. Although some of these species produce plantlets only when placed under stress (induced plantlet-forming species), others spontaneously make plantlets on leaves (constitutive plantlet-forming species). To date, leaf plantlet development in Kalanchoë has been studied extensively at the morphological and anatomical levels (2–10). Although these studies have provided detailed descriptive information, the morphogenic process involved in the origin of these plantlets and the different reproductive strategies undertaken by species of this genus are still not well understood.
Genetic analyses of model species have identified key molecular regulators of organogenesis and embryogenesis. Loss-of-function mutations in Arabidopsis SHOOT MERISTEMLESS (STM), a class 1 KNOTTED1-LIKE HOMEOBOX (KNOX1) gene, result in plants that are unable to form a shoot apical meristem (SAM) and arrest at the seedling stage (11, 12). Transgenic plants constitutively overexpressing KNOX1 genes form ectopic shoots on leaves (13–15). The Arabidopsis LEAFY COTYLEDON1 (LEC1) gene is expressed during embryogenesis, and its expression pattern is similar in both zygotic and somatic embryos (16–18). Loss-of-function mutation of LEC1 results in embryos that do not undergo developmental arrest and are nonviable because they are desiccation-intolerant (19–23). Ectopic expression of LEC1 in transgenic plants induces somatic embryos in vegetative cells (16). Because leaf-plantlet formation resembles aspects of both STM and LEC1 overexpression phenotypes, we investigated the role of these genes in plantlet formation in the genus Kalanchoë. We integrated this information with phylogenetic relationships to draw inferences on the evolution of asexual reproduction within the genus.
Plantlets Share Shoot and Embryo Features.
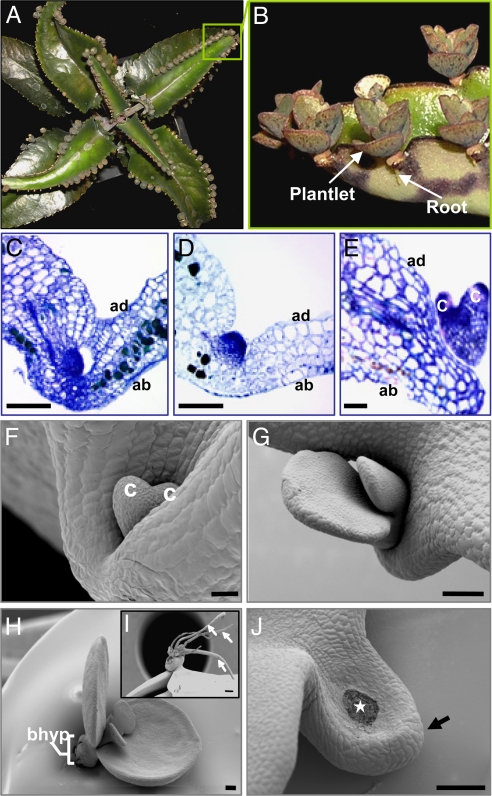
Plantlet development in Kalanchoë daigremontiana (Hamet & Perrier) occurs symmetrically along the leaf margin from leaf tip to base (Fig. 1 A and B). Morphologically, the first stages of leaf plantlet development are dome-like protrusions resembling both globular-stage embryos and shoot meristems (Fig. 1 C and D) (1, 24). Later in development, plantlets proceed through a heart-like embryo stage (Fig. 1 E and F), with cotyledon-like leaves (Fig. 1 G and H), closely resembling embryo development. However, unlike embryos, which form distinct root and shoot apical poles, plantlets resemble shoots in that they produce adventitious roots from the basal “hypocotyl” (Fig. 1 H and I). Once the root system is developed, plantlets detach from the mother leaf, fall to the ground, and grow into new plants. Detachment of the leaf plantlets occurs because of the formation of an abscission zone on the leaf-pedestal (Fig. 1J, arrow) as a consequence of programmed cell death (Fig. 1J, star) (25). Confocal imaging revealed that, like embryos, plantlets have a vascular system independent of the mother tissue at all stages of development (24) [supporting information (SI) Fig. 6 A and B]. On the basis of these morphological similarities to shoots and embryos, we conclude that K. daigremontiana plantlets share features of both organogenesis and embryogenesis.
Fig. 1.
K. daigremontiana leaf plantlet development. (A) K. daigremontiana plant. (B) Plantlets. (C–E) Histology of an early (C), later (D), and heart-like (E) embryo plantlet. (F–H) SEM images of heart-like (F) and cotyledon-like (G) plantlets. (H) Plantlet showing basal “hypocotyl.” (I) Older plantlet showing adventitious roots (arrows). (J) Abscission scar on leaf-pedestal (arrow) after plantlet detachment (star). ad, adaxial leaf; ab, abaxial leaf; bhyp, basal “hypocotyl”; c, cotyledon-like leaves. [Scale bars: 50 μm (C–F); 200 μm (G–J).]
Plantlets Develop Through Recruitment of Organogenic and Embryogenic Programs.
To determine the mechanism by which leaf plantlets arise, we isolated the K. daigremontiana (Kd) STM and LEC1 orthologs. The KdSTM protein shares 75.5% identity with the Arabidopsis STM protein and was placed phylogenetically in a well supported clade of Class 1 KNOX1 genes (SI Fig. 7). The KdLEC1 ortholog shares 72.2% protein sequence identity in the conserved B domain region of the Arabidopsis LEC1-type AHAP3 protein and falls within the well supported LEC1-type clade (SI Fig. 8). Furthermore, KdLEC1 possesses the amino acid residues specific to LEC1-type proteins (SI Fig. 9) (26). Thus, KdSTM and KdLEC1 are STM and LEC1 orthologs. Furthermore, KdSTM and KdLEC1 appear to exist as single copy genes in the K. daigremontiana genome (SI Fig. 10 A and B). Sequence analysis revealed that the KdLEC1 gene has a 20-nucleotide deletion in the C-terminal region of the B domain (SI Fig. 9). This causes the addition of 11 unique amino acids and a premature stop codon in the B domain, resulting in a truncated form of the LEC1 protein.
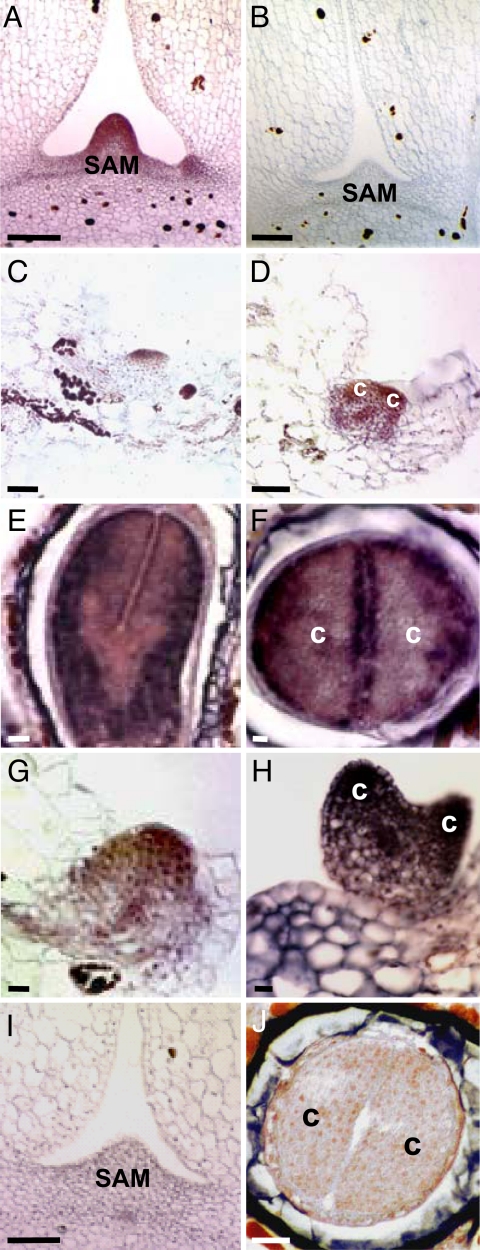
We localized KdSTM mRNA during K. daigremontiana plantlet development by in situ hybridization and RT-PCR analysis. High levels of KdSTM transcript were detected in the SAM and in axillary buds (Fig. 2A). This is consistent with STM expression patterns in most simple-leafed plants (27). In addition, KdSTM mRNA was detected in a small group of cells in leaf margins that were just initiating plantlet formation (Fig. 2C and Fig. 3A, LM2). As the plantlet developed through the heart-like embryo stage, KdSTM transcripts increased in the vascular bundles and in the upper half of the plantlet extending into the cotyledon-like leaves (Figs. 2D and 3A, LM3). This is in contrast to Arabidopsis zygotic embryos, where STM expression is restricted to a few cells in the globular embryo and not seen in cotyledon primordia (18, 28) and is reminiscent of maize somatic embryogenesis, where expression of KNOX1 genes appears to be in a broader domain than seen in maize zygotic embryogenesis (18, 29).
Fig. 2.
In situ distribution of KdSTM and KdLEC1 RNA. (A and B) KdSTM antisense (A) and sense (B) transcripts in the SAM. (C and D) KdSTM expression in an initiating (C) and early heart-like (D) embryo plantlet. (E and F) KdLEC1 expression in longitudinal (E) and transverse (F) sections of a zygotic embryo. (G and H) KdLEC1 expression in an early (G) and heart-like (H) embryo plantlet. (I) KdLEC1 hybridization in the SAM. (J) Sense KdLEC1 transcripts in a zygotic embryo transversal section. c, cotyledon-like leaves; SAM, shoot apical meristem. [Scale bars: 50 μm (A, B, and I); 100 μm (C–H and J).]
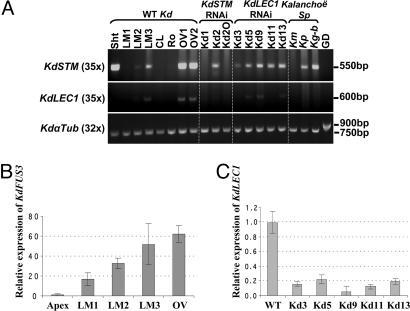
Fig. 3.
RT-PCR analysis of KdSTM, KdLEC1, and KdFUS3 RNA levels in several tissues. (A) RT-PCR in WT K. daigremontiana (Kd) tissues and in LM3 leaf margins of KdSTM and KdLEC1 RNAi plants and other Kalanchoë species. (B) qRT-PCR of KdFUS3 RNA levels in several WT Kd tissues. (C) qRT-PCR of KdLEC1 RNA levels in LM3 of WT and LEC1 RNAi plants. In B and C, RNA levels were normalized to KdGAPDH mRNA and are shown relative to WT apices (B) and LM3 (C). Bars represent SEM over four technical replicates. Km, K. marmorata; Kg-b, K. gastonis-bonnieri; Kp, K. pinnata; Sht, shoot; LM1, LM2, and LM3 are leaf margins from first-stage (0.4–0.7 cm), third-stage (1.5 cm), and fifth-stage (2.5–3.0 cm) long leaves, respectively; CL, central leaf regions; Ro, roots; OV1, young ovaries; OV2, older ovaries; GD, genomic DNA.
Because STM is expressed during both organogenesis and embryogenesis, we analyzed LEC1 expression in developing plantlets. In Arabidopsis and other plants, LEC1 expression is detected only during embryogenesis and not in vegetative development. KdLEC1 transcripts were present in K. daigremontiana zygotic torpedo-stage embryos (Fig. 2 E and F) in a similar pattern to that of LEC1 in Arabidopsis embryos (16) and were not detected in the SAM (Figs. 2I and 3A), demonstrating that KdLEC1 is a marker for embryogenesis in K. daigremontiana as well. We analyzed developing leaf plantlets and showed that KdLEC1 mRNA was also detected in early and heart-like embryo stages in a pattern similar to that of KdSTM (Fig. 2 G and H and Fig. 3A, LM2 and LM3). Together, these results suggest that K. daigremontiana plantlet development proceeds through both organogenesis- and embryogenesis-like stages.
Because the KdLEC1 gene may be defective, we examined another marker of embryogenesis, FUSCA3 (FUS3). FUS3 is a LEC class gene that encodes a B3 domain protein (30, 31). The Arabidopsis fus3 mutants resemble lec1 mutants morphologically (19, 21, 32), suggesting that both proteins regulate a common set of downstream genes (33). The KdFUS3 protein shares 64% identity with Arabidopsis FUS3. Phylogenetic analysis showed that KdFUS3 falls within the monophyletic B3-containing FUS3 protein family (SI Fig. 11). Quantitative (q)RT-PCR results revealed that KdFUS3 is not expressed at a significant level in the shoot apex of K. daigremontiana but, like KdLEC1, is expressed at high levels in pollinated ovaries and in all of the developmental leaf margin stages, being highest in the most advanced plantlet development stage (LM3) analyzed (Fig. 3B). The fact that both embryogenic genes, KdLEC1 and KdFUS3, are present at high levels during plantlet development and in pollinated ovaries but absent or expressed at low levels in the apical meristem suggests that an embryo-like program is also involved in K. daigremontiana plantlet development.
KdSTM Suppression Abolishes Plantlet Formation.
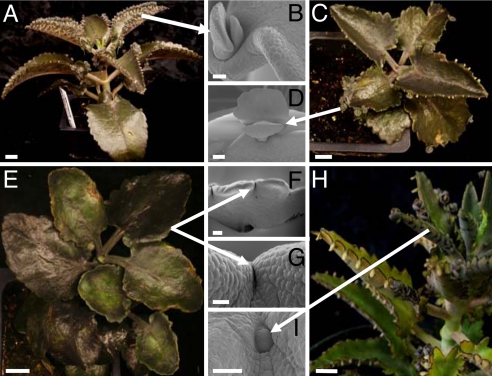
KdSTM is expressed throughout plantlet development in a pattern that differs from that of zygotic and somatic embryos (18, 28). Therefore, we used RNA interference (RNAi) to down-regulate KdSTM RNA levels and determine its function. Plants transformed with the empty vector (Fig. 4 C and D) were morphologically similar to untransformed plants (Fig. 4 A and B). RNAi suppression of KdSTM caused complete inhibition of plantlet formation in seven of eight independent transgenic lines (Fig. 3 E–G). KdSTM mRNA was detected in the leaf margins of one line (Fig. 3A, Kd2) that was able to produce plantlets on leaves (Fig. 4 H and I), suggesting incomplete suppression of KdSTM in this line. Transgenic plants still formed a vegetative SAM because of reduced activity of the Cauliflower Mosaic Virus 35S promoter in the SAM (SI Table 1) and to sequestration of this region from gene-silencing effects (34, 35). The complete suppression of plantlet formation in most of the KdSTM RNAi plants and the high levels of expression of KdSTM at the plantlet-initiation site in WT plants, strongly suggests that KdSTM is required for plantlet formation, likely by initiating and/or maintaining a pool of undifferentiated cells at the leaf notches.
Fig. 4.
KdSTM RNAi transgenic phenotypes. (A) Nontransformed plant. (B) SEM of a plantlet from A. (C) Plant transformed with empty vector. (D) SEM of a plantlet from C. (E) KdSTM RNAi plant showing complete suppression of plantlet formation. (F and G) SEM images of leaf margins from E showing no plantlet formation. (H) Single KdSTM RNAi event (Kd2) showing plantlet formation. (I) SEM image of an early stage Kd2 plantlet. [Scale bars: 2 cm (A); 250 μm (B); 1 cm (C, E, and H); 600 μm (D); 100 μm (F, I); 700 μm (G).]
KdLEC1 Is Unable to Rescue the lec1 Mutation.
Because the KdLEC1 protein is a truncated form of the Arabidopsis LEC1 protein, we asked whether KdLEC1 was functional and able to suppress the lec1 mutation in Arabidopsis. The lec1 mutation causes embryos to become desiccation-intolerant, and, consequently, seeds do not germinate. Three different versions of the KdLEC1 gene were inserted between the LEC1 promoter and terminator (36) (SI Table 2) and used to transform Arabidopsis Ws-O WT and lec1–1 mutant plants (37). Two versions of the KdLEC1 gene encoding defective B domains, either the endogenous (construct 1) or a modified version (construct 2) (SI Table 2), could not rescue the desiccation-intolerance of lec1–1 mutant embryos (16, 19, 21). However, when the deleted nucleotides of the KdLEC1 B domain were replaced by the corresponding nucleotides from the Arabidopsis LEC1-LIKE (L1L) gene (construct 3) to reconstitute a complete B domain, 0.65% of lec1–1 mutant seeds produced viable seedlings (SI Table 2). This percentage is similar to that seen in transformations with the WT Arabidopsis LEC1 gene. These results indicate that this construct was able to suppress the lec1 mutation by conferring desiccation tolerance to lec1–1 mutant seeds.
Although the KdLEC1 gene is unable to confer desiccation tolerance to seeds, it is possible that it could have acquired additional function(s) in leaf plantlet formation. We therefore examined the effect of KdLEC1 down-regulation in plantlet development. In contrast to KdSTM RNAi transgenic plants, all nine KdLEC1 RNAi plants formed plantlets on their leaf margins (SI Fig. 12 A–C) at the same level as nontransformed (Fig. 4 A and B) or empty-vector control-transformed plants (Fig. 4 C and D). qRT-PCR results showed that KdLEC1 mRNA was reduced 5- to 20-fold in the leaf margins of all RNAi lines relative to nontransformed plants (Fig. 3C). These results suggest that the KdLEC1 gene in K. daigremontiana is not required for plantlet formation. KdLEC1 appears incapable of functioning as a normal LEC1 gene during Arabidopsis embryogenesis, and KdLEC1 RNA accumulation during plantlet development may simply reflect activation of promoters responsive to an embryogenic environment. Thus, the inability of K. daigremontiana to produce viable dried seed likely results, at least in part, because KdLEC1 is not functional in conferring desiccation tolerance to zygotic embryos.
Evolution of Asexuality in Kalanchoë.
In an effort to understand the evolution of plantlet formation in the genus Kalanchoë, we examined STM and LEC1 expression in four representative species: a species that does not produce plantlets (Kalanchoë marmorata); a species with constitutive plantlet formation (K. daigremontiana); a species with induced plantlet formation (Kalanchoë pinnata), and a semiconstitutive plantlet-forming species, which produces plantlets constitutively as well as upon stress induction (Kalanchoë gastonis-bonnieri). RT-PCR analysis performed on the leaf margins of these species revealed that STM is expressed in all plantlet-forming species (Fig. 3A, Kd, Kg-b, and Kp), but is absent in species that do not produce plantlets on leaves (Fig. 3A, Km). Plantlets in K. gastonis-bonnieri have a prominent vascular connection to the mother leaf (SI Fig. 6 C and D) and can only detach when the mother leaf dies. These histological characteristics, and the presence of STM (but not LEC1 RNA) in both K. gastonis-bonnieri and K. pinnata (Fig. 3A, Kg-b and Kp), suggests that these species form plantlets by a process resembling organogenic shoot formation in KNOX1-overexpressing plants (13, 14). These results agree with our hypothesis that an organogenic-like program seems to be involved in plantlet formation. However, in addition to STM expression, LEC1 and FUS3 RNA were both detected exclusively in the leaf margins of constitutive plantlet-forming species (Fig. 3 A and B, Kd). Only plantlets in this specific group share embryo-like morphological features. Thus, an embryogenic program seems to have been recruited into the pool of organogenic cells in the leaf notches, suggesting that both organogenesis and embryogenesis programs are involved in plantlet formation in this specific group of species with nonviable seed.
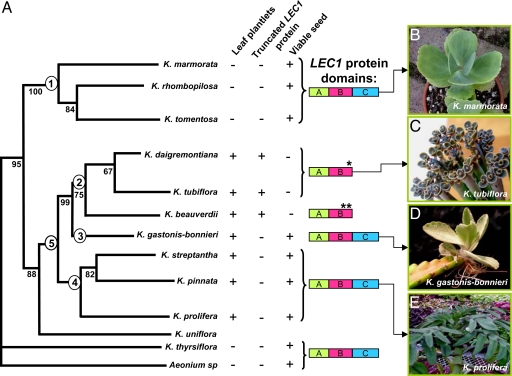
To investigate whether there was a correlation between the presence of a truncated LEC1 protein and seed viability in the Kalanchoë genus, we cloned and sequenced LEC1 orthologs from several groups of species: (i) species that do not produce leaf plantlets (Kalanchoë marmorata, Kalanchoë rhombopilosa, Kalanchoë tomentosa, and Kalanchoë thyrsiflora) (Fig. 5A1 and B); (ii) species with constitutive plantlet formation (Kalanchoë beauverdii, Kalanchoë tubiflora) (Fig. 5 A2 and C); (iii) species with induced plantlet formation (Kalanchoë streptantha, K. pinnata, and Kalanchoë prolifera) (Fig. 5 A4 and E) and; (iv) semiconstitutive plantlet-forming species, which produce plantlets constitutively as well as upon stress induction (K. gastonis-bonnieri) (Fig. 5 A3 and D). Species that do not produce plantlets on leaves have similar protein sequence to the Arabidopsis LEC1-type proteins and have viable seed (Fig. 5A1). Constitutive plantlet-forming species (K. daigremontiana, K. tubiflora and K. beauverdii) all have a truncated LEC1 protein and do not produce viable seed (Fig. 5A2). The LEC1 gene in K. tubiflora is identical to that in K. daigremontiana, whereas in K. beauverdii, two independent mutations have occurred in the LEC1 B domain (SI Fig. 13). These results are consistent with the conclusion that an intact B domain is required for LEC1 activity in seed desiccation tolerance (36). Contrary to constitutive plantlet-forming species, species that require induction to make plantlets have intact LEC1 proteins and produce viable seed (Fig. 5 A3 and A4). According to recent phylogenetic studies (38), K. gastonis-bonnieri is sister (Fig. 5A3) to the clade that contains the constitutive plantlet-forming species (Fig. 5A2). This makes K. gastonis-bonnieri unique among the Kalanchoë species studied here because it may represent a transition step between induced and constitutive plantlet formation. The presence of an intact LEC1-type protein, viable seed production, and induced plantlet formation on this and other inducible plantlet-forming species, suggest that these traits are ancestral (Fig. 5A5). Moreover, the constitutive plantlet-forming species reproduce solely by asexual means and fall in a monophyletic group (Fig. 5A2). Interestingly, within this group, LEC1 is found to have undergone deletions independently in the highly conserved B domain, leading to frame shifts (SI Fig. 13). Truncation of the KdLEC1 protein appears to have negated its activity and adversely affected maturation of zygotic embryos in these species.
Fig. 5.
Kalanchoë LEC1 proteins and their relationship with plantlet formation and seed viability. (A) Phylogenetic tree adapted from Gehrig et al. (38). (B–E) Phenotypes of representative species. (B) Species that do not produce plantlets (1). (C) Constitutive plantlet-forming species (2). (D) Semiconstitutive plantlet-forming species, which produce plantlets constitutively as well as upon stress induction (3). (E) Inducible plantlet-forming species in which plantlets are produced upon stress induction (4). Plantlet-forming species (5) possessing ancestral traits (3 and 4). +, presence; −, absence; *, one deletion; **, two deletions in LEC1 B domain.
Our molecular and genetic data combined with the most recently published phylogenetic relationships in Kalanchoë (38) allow us to generate a model for how leaf plantlet formation evolved in this genus. An inductive signal triggered by the expression of early acting genes, such as STM, may have initiated a developmental switch to meristematic competence and generated a pool of undifferentiated cells in the leaf-notches. This led to the formation of shoot-like-plantlets by organogenesis in species that produce viable seed as is seen in the basal members of the genus Kalanchoë. The loss of LEC1 function only in the clade of species that form plantlets constitutively may have resulted in desiccation-intolerant seed embryos, causing sexual sterility. Mutations resulting in truncated LEC1 proteins appear to be of a selective advantage in creating somatic propagules, because we show that such mutations occurred in parallel at least twice within this clade. In these species, the embryogenic program appears to have been recruited into the pool of organogenic cells in the leaf notches. Survival of these embryos was not affected by the loss of LEC1 function, because they do not go through desiccation. Further experiments are required to determine whether the origin of the truncated LEC1 protein is causal or consequential for constitutive plantlet formation. Analysis of LEC1 function in other spontaneous somatic embryo-producing species outside the genus Kalanchoë may shed some light on the role of the LEC1 gene during the evolution of this unique mode of vegetative propagation.
Materials and Methods
Plant Materials.
Kalanchoë plants were grown in the greenhouse at 29°C and in a 16-h photoperiod. Kalanchoë first whole leaves and margins, counting from the top of the plant, were harvested at different developmental stages: 0.4–0.7 cm length (first stage or LM1); 1.5 cm length (third stage or LM2); and 2.5–3 cm length (fifth stage or LM3) leaves. Leaves and margins at these developmental stages, young shoot apical meristems, and ovaries were used for scanning electron microscopy (SEM), histological analyses, in situ hybridizations, RT-PCR, and quantitative RT-PCR (qRT-PCR) analysis. The Arabidopsis thaliana (L.) Heynh ecotype Wassilewskija (Ws-O) was used as lec1–1 mutant and WT plants and were grown as described (21).
Gene Isolation.
KdSTM cDNA clones were isolated by using degenerate primers based on STM orthologs available in the GenBank database. The KdLEC1 was isolated by using degenerate primers based in LEC1 sequences available (26) and by using an inverse PCR (iPCR) technique (39). LEC1 orthologs from other Kalanchoë species were isolated by using a KdLEC1-specific primer and a primer for the 3′ UTR of the next gene. K. daigremontinana FUSCA3 (KdFUS3), α-TUBULIN (KdαTUB), and GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (KdGAPDH) genes were isolated by using degenerate primers based on sequences available in the GenBank database. For primer sequences and experimental details see SI Materials and Methods.
Histology and in Situ Hybridization.
Tissues for histology and in situ hybridization were fixed and sectioned as described (27, 40). In situ hybridizations were performed according to the method of Long et al. (41), with several modifications. For details, see SI Materials and Methods.
Plasmids and Plant Transformation.
Attempts to reduce expression of KdSTM and KdLEC1 were made by using plasmid pRNA69 (42) for the RNA interference (RNAi) approach. The KdLEC1 complementation constructs are described in SI Table 2 and were driven by the Arabidopsis LEC1 promoter and terminator (43). The Agrobacterium tumefaciens LBA4404 strain, carrying the empty pBIB-KAN vector and knockout constructs was transformed into K. daigremontiana by using a compilation of several Kalanchoë transformation methods (44–46). The Agrobacterium tumefaciens GV3101 strain containing all LEC1 constructs was infiltrated into Arabidopsis plants according to the method of Bechtold et al. (37). For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ernesto Sandoval and Tim Metcalf (Section of Plant Biology Conservatory, University of California, Davis, CA) for plant materials; Julie Kang, Rakefet David-Schwartz, Seisuke Kimura, and Dan Koenig for critical comments; and Minsung Kim for help with cloning and expression analyses. This work was supported by National Science Foundation Grants IBN 0316877 and 0344743 (N.R.S.) and Fundação para a Ciência e Tecnologia, Portugal, Grant PRAXIS XXI 3/3.1/CTAE/1930/95, PhD Fellowship GGPXXI/BD/3377/96, Fundação Luso-Americana para o Desenvolvimento (Portugal), and Centro de Biotecnologia Vegetal (Lisboa, Portugal) (H.M.P.G.).
Abbreviation
- SAM
shoot apical meristem.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genes described in this paper have been deposited in the GenBank database [accession nos. DQ674267 (KdLEC1), DQ674268 (KdSTM), Bankit 811357 (KdαTUB), Bankit 870460 (KdGAPDHc), Bankit 870458 (KdFUSCA3), Bankit 908377 (KmLEC1), Bankit 908591 (KrLEC1), Bankit 908565 (KtLEC1), Bankit 908579 (KbLEC1), Bankit 910314 (Kg-bLEC1), Bankit 908583 (KsLEC1) Bankit 908587 (KpLEC1), Bankit 910326 (KprLEC1), Bankit 910304 (KthLEC1), Bankit 910328 (AsLEC1), and Bankit 910334 (KtomLEC1)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0704105104/DC1.
References
- 1.Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge, UK: Cambridge Univ Press; 1989. [Google Scholar]
- 2.Howe MD. Am J Bot. 1931;18:387–390. [Google Scholar]
- 3.Naylor E. Am J Bot. 1932;19:32–40. [Google Scholar]
- 4.Yarbrough JA. Am J Bot. 1932;19:443–453. [Google Scholar]
- 5.Yarbrough JA. Am J Bot. 1934;21:467–484. [Google Scholar]
- 6.Freeland RO. Am J Bot. 1933;20:467–480. [Google Scholar]
- 7.Warden J. Separata Revista Biologica. 1968;6:357–374. [Google Scholar]
- 8.McVeigh I. Am J Bot. 1938;25:7–11. [Google Scholar]
- 9.Warden J. Separata Portugaliae Acta Biologica. 1970;11:385–394. [Google Scholar]
- 10.Batygina TB, Bragina EA, Titova GE. Acta Soc Bot Poloniae. 1996;65:127–133. [Google Scholar]
- 11.Long J, McConnell J, Fernandez A, Grbic V, Barton MK. Plant Physiol. 1996;111:12. [Google Scholar]
- 12.Vollbrecht E, Reiser L, Hake S. Development (Cambridge, UK) 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- 13.Sinha N, Williams RE, Hake S. Genes Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- 14.Chuck G, Lincoln C, Hake S. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sentoku N, Sato Y, Matsuoka M. Dev Biol. 2000;220:358–364. doi: 10.1006/dbio.2000.9624. [DOI] [PubMed] [Google Scholar]
- 16.Lotan T, Ohto M-A, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda-Iwai M, Satoh S, Kamada H. J Exp Bot. 2002;53:1575–1580. doi: 10.1093/jxb/erf006. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Wong L, Meng L, Lemaux PG. Planta. 2002;215:191–194. doi: 10.1007/s00425-002-0735-3. [DOI] [PubMed] [Google Scholar]
- 19.Meinke DW. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- 20.Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parcy F, Valon C, Kohara A, Misera S, Giraudat J. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vicient CM, Bies-Etheve N, Delseny M. J Exp Bot. 2000;51:995–1003. doi: 10.1093/jexbot/51.347.995. [DOI] [PubMed] [Google Scholar]
- 24.Arnold VS, Sabala I, Bozhkov P, Dyachok J, Filonova L. Plant Cell Tissue Organ Cult. 2002;69:233–249. [Google Scholar]
- 25.Pedroso MC, Durzan DJ. Ann Bot (London) 2000c;86:983–994. doi: 10.1006/anbo.2000.1260. [DOI] [PubMed] [Google Scholar]
- 26.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 28.Long JA, Barton MK. Development (Cambridge, UK) 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- 29.Smith LG. Dev Genet. 1995;16:344–348. [Google Scholar]
- 30.Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luerssen H, Kirik V, Herrmann P, Misera S. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- 32.Keith K, Kraml M, Dengler NG, McCourt P. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. Plant Cell Physiol. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- 34.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 35.Foster TM, Lough TJ, Emerson SJ, Lee RH, Bowman JL, Forster RL, Lucas WJ. Plant Cell. 2002;14:1497–1508. doi: 10.1105/tpc.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bechtold N, Ellis J, Pelletier G. Comptes Rendus Acad Sci Ser III. 1993;316:1194–1199. [Google Scholar]
- 38.Gehrig H, Gaussmann O, Marx H, Schwarzott D, Kluge M. Plant Sci. 2001;160:827–835. doi: 10.1016/s0168-9452(00)00447-7. [DOI] [PubMed] [Google Scholar]
- 39.Britt AB, Earp DJ. In: The Maize Handbook. Freeling M, Walbot V, editors. New York: Springer; 1994. pp. 586–591. [Google Scholar]
- 40.Kessler S, Kim M, Pham T, Weber N, Sinha N. Int J Plant Sci. 2001;162:475–492. [Google Scholar]
- 41.Long JA, Moan EI, Medford JI, Barton MK. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 42.Yesbergenova Z, Yang G, Oron E, Soffer D, Fluhr R, Sagi M. Plant J. 2005;42:862–876. doi: 10.1111/j.1365-313X.2005.02422.x. [DOI] [PubMed] [Google Scholar]
- 43.Gleave AP. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 44.Jia SR, Yang MZ, Ott R, Chua NH. Plant Cell Rep. 1989;8:336–340. doi: 10.1007/BF00716668. [DOI] [PubMed] [Google Scholar]
- 45.Aida R, Shibata M. Plant Sci (Shannon) 1996;121:175–185. [Google Scholar]
- 46.Truesdale MR, Toldi O, Scott P. Plant Physiol (Rockville) 1999;121:957–964. doi: 10.1104/pp.121.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.