Abstract
TLX is a transcription factor that is essential for neural stem cell proliferation and self-renewal. However, the molecular mechanism of TLX-mediated neural stem cell proliferation and self-renewal is largely unknown. We show here that TLX recruits histone deacetylases (HDACs) to its downstream target genes to repress their transcription, which in turn regulates neural stem cell proliferation. TLX interacts with HDAC3 and HDAC5 in neural stem cells. The HDAC5-interaction domain was mapped to TLX residues 359–385, which contains a conserved nuclear receptor–coregulator interaction motif IXXLL. Both HDAC3 and HDAC5 have been shown to be recruited to the promoters of TLX target genes along with TLX in neural stem cells. Recruitment of HDACs led to transcriptional repression of TLX target genes, the cyclin-dependent kinase inhibitor, p21CIP1/WAF1(p21), and the tumor suppressor gene, pten. Either inhibition of HDAC activity or knockdown of HDAC expression led to marked induction of p21 and pten gene expression and dramatically reduced neural stem cell proliferation, suggesting that the TLX-interacting HDACs play an important role in neural stem cell proliferation. Moreover, expression of a TLX peptide containing the minimal HDAC5 interaction domain disrupted the TLX–HDAC5 interaction. Disruption of this interaction led to significant induction of p21 and pten gene expression and to dramatic inhibition of neural stem cell proliferation. Taken together, these findings demonstrate a mechanism for neural stem cell proliferation through transcriptional repression of p21 and pten gene expression by TLX–HDAC interactions.
Keywords: coregulators, NR2E1, self-renewal
Nuclear receptors are ligand-dependent transcription factors that regulate the expression of genes critical for a variety of biological processes, including development, growth, and differentiation. TLX is an orphan nuclear receptor that plays an important role in vertebrate brain functions (1–6). We have shown that TLX is an essential regulator of neural stem cell proliferation and self-renewal in the adult brain (3). TLX could act by controlling the expression of a network of downstream target genes to establish the undifferentiated and self-renewable state of neural stem cells. Elucidating the network regulated by TLX in producing these outcomes will be a significant advance in understanding neural stem cell self-renewal and neurogenesis.
Nuclear receptors carry out transcriptional functions through the recruitment of positive and negative regulatory proteins, referred to as coactivators and corepressors (7, 8). At least one mechanism underlying the repression activity of nuclear receptors is through the recruitment of histone deacetylase (HDAC) complexes. For example, HDAC1 and HDAC2 have been found in complexes with Sin 3 (9), nucleosome remodeling and deacetylation (NuRD) (10), nuclear receptor corepressor (N-CoR) (11), silencing mediator of retinoic and thyroid hormone receptors (SMRT) (12), and SMRT and HDAC-associated repressor proteins (SHARP) (13). HDAC4, HDAC5, and HDAC7 have been shown to interact with SMRT, N-CoR (14, 15), and myogenic transcription factor-2 (16). How TLX activity is modulated by transcriptional coregulators remains largely unknown.
We show here that TLX interacts with a set of HDACs in neural stem cells. TLX recruits these HDACs to its target genes to repress their expression. Chemical inhibition of HDAC activity, siRNA knockdown of HDAC expression, or disruption of TLX–HDAC interaction using a TLX peptide containing the minimal HDAC5 interaction site, all led to induction of TLX target genes, p21 and pten, along with marked inhibition of neural stem cell proliferation. These results suggest that transcriptional repression mediated by TLX and its associated HDACs serves as an important mechanism for neural stem cell proliferation.
Results
TLX Interacts with HDACs in Neural Stem Cells.
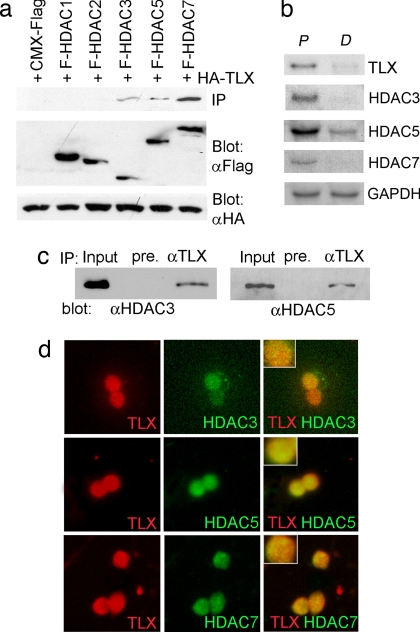
Transcriptional silencing is an essential regulatory mechanism for stem cell maintenance and self-renewal. HDACs play an important role in transcriptional repression by catalyzing histone deacetylation and chromatin condensation. To determine whether TLX recruits HDACs to mediate transcriptional repression, HA-tagged TLX was transfected into HEK293 cells with Flag-tagged HDACs. Cell lysates were immunoprecipitated with a Flag-specific antibody. The immunoprecipitates were examined for the presence of TLX by Western blot analysis with an HA-specific antibody. Among the HDACs examined, TLX was found to be associated with HDAC3, HDAC5, and HDAC7, whereas association with HDAC1 and HDAC2 was not detected (Fig. 1a). This result indicates that TLX interacts with HDAC3, HDAC5, and HDAC7 specifically.
Fig. 1.
The interaction of TLX and HDACs. (a) TLX interacts with HDACs in HEK293 cells. Lysates of TLX and HDAC transfected cells were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-HA antibody. Protein expression was shown by immunoblotting cell lysates with HA- or Flag-specific antibody. (b) Expression of TLX and HDACs in neural stem cells cultured under proliferation (P) or differentiation (D) conditions. GAPDH was included as a loading control. (c) TLX interacts with HDAC3 and HDAC5 in neural stem cells analyzed by immunoprecipitation analysis. pre, preimmune serum. (d) Double immunostaining of neural stem cells with antibodies specific for TLX (red) and antibodies for individual HDACs (green). The merged images are shown in yellow. (Insets) Enlarged images.
Next we asked whether these TLX-interacting HDACs are expressed in neural stem cells, where TLX is expressed. Neural stem cells were isolated from adult mouse brains and cultured as described (3). RNAs were prepared from neural stem cells or differentiated cells and analyzed by Northern blot analysis. The three HDACs that were shown to interact with TLX are all expressed in neural stem cells with significantly reduced expression in differentiated cells, similar to the expression profile of TLX (Fig. 1b). Expression of these HDACs in neural stem cells was further confirmed by immunostaining analysis. Nuclear-specific staining of HDAC3, HDAC5, and HDAC7 was readily detected in neural stem cells but was much lower in differentiated cells. In addition, cytoplasmic localization of HDAC5 and HDAC7 was observed in differentiated cells [supporting information (SI) Fig. 7].
To determine whether endogenous TLX interacts with HDACs in neural stem cells, we developed a TLX-specific rabbit polyclonal antibody (data not shown). Cell lysates of neural stem cells were immunoprecipitated with the TLX-specific antibody. The immunoprecipitates were examined for the presence of HDACs by Western blotting with HDAC-specific antibodies. Both HDAC3 and HDAC5 were detected in the TLX immunocomplex in neural stem cells (Fig. 1c), although we were not able to detect HDAC7 in cell lysates by Western blot analysis. Double staining revealed colocalization of TLX with HDAC3, HDAC5, and HDAC7 in the nucleus of neural stem cells (Fig. 1d), further supporting the notion of TLX–HDAC interaction in neural stem cells.
Mapping the TLX–HDAC5 Interaction Domain.
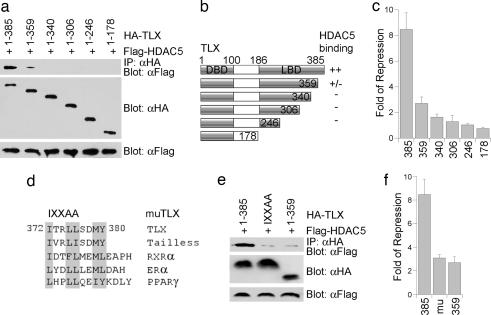
To determine the functional relevance of TLX–HDAC interactions, we mapped the minimal domain of TLX for HDAC interaction. HDAC5 was chosen for this study because it is most abundantly expressed in neural stem cells among the HDACs we have examined (data not shown). The HDAC5-interaction domain was determined by serial deletion analysis of TLX. HA-tagged TLX deletion mutants were transfected into HEK293 cells with Flag-tagged HDAC5. The interaction of TLX with HDAC5 was detected by immunoprecipitation analysis using an HA-specific antibody and subsequent Western blotting with a Flag-specific antibody. This study located the HDAC5 interaction site to TLX residues 359–385, with secondary interaction to TLX residues 340–359 (Fig. 2 a and b). To determine whether deletion of TLX residues 340–385 affects TLX transcriptional repression activity, the same set of deletions was introduced into Gal4-TLX construct and analyzed in reporter assays. Deletion of TLX residues 359–385 led to 3-fold relief of TLX-mediated repression of basal transcription (Fig. 2c). Further deletion to residue 340 resulted in another 1.6-fold loss of repression. These results suggest that the HDAC5 interaction sites are critical for TLX repression activity.
Fig. 2.
Mapping HDAC5-interaction domain in TLX. (a) Immunoprecipitation analysis to determine HDAC5-interaction sites in TLX. HEK cells were transfected with Flag-tagged HDAC5 and HA-tagged full-length (1–385) or deletion mutants of TLX. (b) Schematics of the TLX deletion mutants and summary of the HDAC5 binding results. (c) Deletion of HDAC5 interaction sites relieved TLX repression activity. Fold repression of Gal4-TLX full-length (1–385) or deletion mutants was determined relative to Gal4-DBD activity. (d) The conserved IXXLL motif in mammalian TLX, Drosophila tailless, and other nuclear receptors. (e) Mutation of the IXXLL motif in TLX impaired its ability to interact with HDAC5 in immunoprecipitation analyses. (f) Mutation of the IXXLL motif (mu) relieved TLX repression activity. 385, TLX 1–385; 359, TLX 1–359.
Sequence analysis of TLX residues 359–385 revealed a conserved IXXLL motif (Fig. 2d). This motif was initially identified as the motif in coregulators that mediate nuclear receptor–coregulator interactions (17) and has been shown recently to mediate the recruitment of coregulators in transcription factors (18–20). To determine the role of this motif in TLX–HDAC5 interaction, site-directed mutagenesis was performed to mutate the IXXLL motif to IXXAA. Mutation of the IXXLL motif led to significant loss of TLX–HDAC5 interaction, similar to the loss resulted from deletion of TLX residues 359–385 (Fig. 2e). The IXXAA mutation also led to 2.7-fold relief of TLX transcriptional repression activity (Fig. 2f). These results suggest that the conserved IXXLL motif is important for both TLX–HDAC5 interaction and TLX-mediated transcriptional repression.
HDAC Inhibitor Treatment Led to Marked Induction of TLX Target Genes and Significant Reduction of Neural Stem Cell Proliferation.
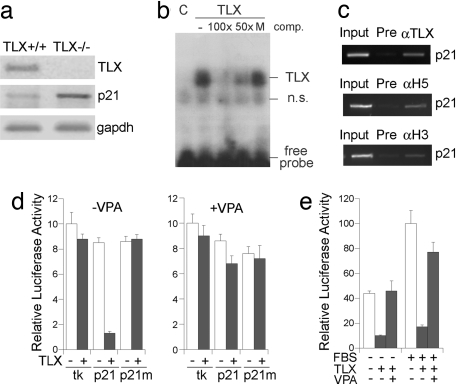
To further determine the contribution of HDACs to TLX-mediated transcriptional repression, we examined whether TLX recruits HDACs to its target genes to repress their expression. Among genes with altered expression in adult brains of WT and TLX-null mice, a significant up-regulation of p21, a cyclin-dependent kinase inhibitor, was detected in adult TLX-null brains (Fig. 3a). Sequence analysis revealed a consensus TLX-binding site, AAGTCA, in the proximal promoter of p21. Gel-shift assays showed that TLX binds to this sequence in vitro (Fig. 3b). The binding specificity was demonstrated by mutating the consensus TLX binding site. WT p21 promoter sequence efficiently competed for TLX binding, but the mutated sequence did not, suggesting that TLX binding is sequence-specific. To confirm the binding of TLX to the p21 promoter, a ChIP analysis was performed in neural stem cells by using a TLX-specific antibody. TLX was detected at the consensus TLX binding site in p21 promoter as expected (Fig. 3c). Interestingly, both HDAC3 and HDAC5 were detected on the same site of p21 promoter along with TLX (Fig. 3c), supporting the idea that TLX recruits HDACs to its target gene.
Fig. 3.
TLX recruits HDACs to repress p21 gene expression. (a) Gene expression in brains of adult WT (TLX+/+) and TLX−/− mice analyzed by RT-PCR analyses. (b) TLX binds to p21 promoter in gel-shift assays. TLX binding was competed by unlabeled WT (100x, 50x), but not by mutant (M) p21 oligoes. C, control lysates. (c) TLX, HDAC3, and HDAC5 were detected on p21 promoter by ChIP assays. Input, DNA input; Pre, preimmune serum. (d) TLX repressed p21 reporter and VPA relieved the repression. Luciferase activity of control tk-luc (tk), p21-tk-luc (p21), and mutant p21-tk-luc (p21m) was measured in the absence (−) or presence (+) of TLX expression in CV-1 cells. The transfected cells were treated with solvent (−VPA) or 4 mM VPA (+VPA). (e) VPA treatment relieved TLX-mediated repression of natural p21 promoter-driven luciferase activity. Neural stem cells were transfected with control vector (−) or TLX (+) and were treated with solvent (−), FBS/forskolin (FBS), 4 mM VPA, or both.
To validate the regulation of p21 gene expression by TLX and HDACs, we placed a luciferase reporter gene downstream of the consensus TLX binding site in the p21 promoter and a minimal thymidine kinase (tk) promoter (p21-tk-luc). Cotransfection of TLX with the reporter gene led to 6-fold repression of the reporter. Mutation of the consensus TLX binding site upstream of the reporter gene (p21m) abolished the repression (Fig. 3d Left). These results suggest that TLX functions as a transcriptional repressor of p21 by binding to a consensus binding site in p21 promoter. Treatment of a HDAC inhibitor, valproic acid (VPA), relieved the TLX-mediated repression from 6-fold to 1.4-fold (Fig. 3d), whereas no effect was detected on the reporter activity in the absence of TLX transfection. Moreover, we cloned a 3-kb natural p21 promoter containing the consensus TLX binding site and placed it upstream of a luciferase reporter gene (p21-luc). Cotransfection of TLX led to 4-fold repression of basal p21-luc activity and 6-fold repression of FBS/forskolin (FSK)-induced p21-luc activity in neural stem cells. Treatment of VPA relieved TLX-mediated repression of both basal and FBS/FSK-induced p21 promoter activity significantly (Fig. 3e). These results together suggest that TLX recruits HDACs to a target gene, p21, to repress its expression.
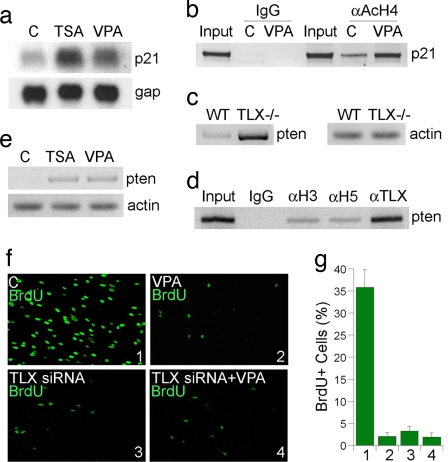
We further tested whether HDACs contribute to repression of endogenous p21 gene expression in neural stem cells. Neural stem cells were treated with solvent or HDAC inhibitors, Trichostatin A (TSA) and VPA. Northern blot analyses revealed increased expression of p21 in TSA- and VPA-treated neural stem cells (Fig. 4a), consistent with the results from the reporter assays (Fig. 3 d and e). ChIP assays revealed significantly increased acetylated histone H4 levels on p21 promoter upon VPA treatment (Fig. 4b), supporting increased p21 expression in VPA-treated cells (Fig. 4a).
Fig. 4.
HDAC inhibitors induce p21 and pten gene expression and inhibit neural stem cell proliferation. (a) Gene expression regulated by HDAC inhibitors, TSA, and VPA. “C” indicates solvent control in all sections. (b) VPA treatment led to increased acetylated histone H4 (AcH4) levels in neural stem cells. (c) Increased expression of pten in TLX−/− adult brains analyzed by RT-PCR analysis. (d) TLX, HDAC3, and HDAC5 were detected on p21 promoter by ChIP assays. (e) Increased expression of pten in TSA- and VPA-treated neural stem cells analyzed by RT-PCR analysis. (f) Cell proliferation as revealed by BrdU labeling (green) in solvent (C) or VPA-treated neural stem cells that were transfected with control GFP siRNA (Upper) or TLX siRNA (Lower). (g) Percent of BrdU+ cells in control (bar 1), VPA- (bar 2), TLX siRNA- (bar 3), and TLX siRNA+VPA- (bar 4) treated neural stem cells. Error bars are standard deviation of the mean; assays were repeated three times.
In addition to p21, a significant increase of pten gene expression was also observed in TLX−/− adult brains (Fig. 4c). This result, along with the observation that TLX binds to a consensus binding site in pten gene promoter and represses pten promoter-driven luciferase reporter activity (21), suggests that TLX functions as a transcriptional repressor of pten gene in adult brains. ChIP analyses revealed the binding of TLX to its consensus binding site in pten gene promoter. Interestingly, HDAC3 and HDAC5 were detected on the same site of pten gene promoter along with TLX (Fig. 4d), suggesting that TLX recruits HDACs to pten promoter. Treatment of neural stem cells with TSA and VPA induced pten gene expression significantly (Fig. 4e), suggesting that HDACs contribute to repression of pten gene expression in neural stem cells, presumably through interaction with TLX.
The interaction of TLX and HDACs in neural stem cells and the resulting repression of p21 and pten gene expression suggest that the TLX-interacting HDACs play an important role in neural stem cell proliferation. To test this hypothesis, we treated neural stem cells with HDAC inhibitor VPA. BrdU treatment was performed to label actively dividing cells. The percentage of BrdU-positive (BrdU+) cells in total living cells was compared between solvent- and VPA-treated cells. A remarkable decrease in the rate of BrdU-positive cells was seen in VPA-treated neural stem cells (Fig. 4 f and g), suggesting that the HDAC inhibitors exert strong inhibitory effect on neural stem cell proliferation. Moreover, neural stem cells were treated with TLX siRNA, which led to marked reduction of cell proliferation (Fig. 4 f and g). The inhibitory effect on cell proliferation by VPA treatment is reduced significantly in TLX siRNA-treated neural stem cells (Fig. 4 f and g). These results together suggest a role of TLX–HDAC interactions in neural stem cell proliferation.
siRNA Knockdown of HDAC Expression Revealed an Important Role of the TLX-Interacting HDACs in Neural Stem Cell Proliferation.
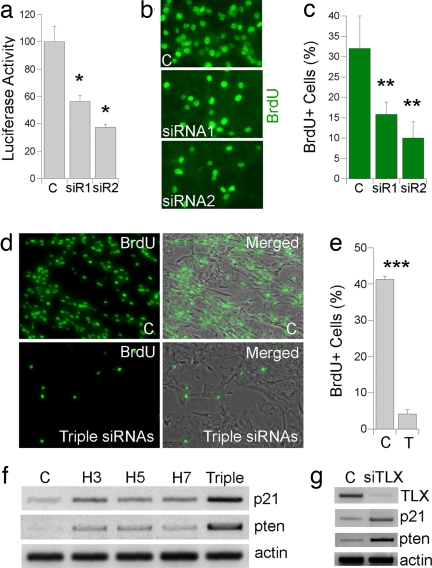
In addition to blocking HDAC activity globally, we attempted to knockdown individual TLX-interacting HDAC expression using siRNAs. We first targeted HDAC5 expression by using its sequence-specific siRNAs. The HDAC5-specific siRNAs were cotransfected with a luciferase reporter gene upstream of HDAC5 cDNA. Among the six siRNAs tested, siRNAs 1 and 2 repressed the luciferase activity significantly (Fig. 5a). These siRNAs were therefore selected to treat neural stem cells. Both siRNAs led to considerable knockdown of HDAC5 expression in neural stem cells by Western blot analysis (data not shown). Cell proliferation was examined by BrdU labeling analyses. The percentage of BrdU+ cells was compared among control GFP siRNA and HDAC5 siRNA-transfected cells. Treatment of HDAC5 siRNAs led to a 50–70% decrease of BrdU labeling (Fig. 5 b and c), suggesting that HDAC5 plays an important role in neural stem cell proliferation.
Fig. 5.
Knockdown of HDAC expression led to reduced cell proliferation and induced p21 and pten gene expression in neural stem cells. (a) HDAC5-specific siRNA1 (SiR1) and siRNA2 (SiR2) inhibit HDAC5-mediated luciferase reporter activity. *, P < 0.01. GFP siRNA was included as a negative control (C) in all sections. (b) HDAC5 siRNAs reduced neural stem cell proliferation as revealed by decreased BrdU labeling (green). (c) Decreased percent of BrdU+ cells in HDAC5 siRNA-treated neural stem cells. **, P < 0.05. (d) A combination of HDAC3-, HDAC5-, and HDAC7-specific siRNAs reduced neural stem cell proliferation as revealed by reduced BrdU labeling (green). Phase-contrast images were shown in gray. (e) Decreased percent of BrdU+ cells in neural stem cells treated with a combination of HDAC3-, HDAC5-, and HDAC7-specific siRNAs (T). ***, P < 0.001. (f) Induction of p21 and pten gene expression in cells treated with single HDAC-specific siRNAs (H3, H5, H7) or a combination of HDAC3, HDAC5, and HDAC7 siRNAs (triple). (g) Induction of p21 and pten gene expression in neural stem cells treated with TLX-specific siRNA (siTLX). Actin was included as a loading control.
To determine whether knockdown of all three TLX-interacting HDACs has more dramatic effect on neural stem cell proliferation, we screened siRNAs for HDAC3 and HDAC7 (data not shown) and selected the ones that have the strongest inhibitory effect. Neural stem cells were transfected with siRNAs for HDAC3, HDAC5, and HDAC7 individually or together. Triple knockdown led to much more dramatic inhibition of cell proliferation (Fig. 5 d and e) compared with individual HDAC knockdown (Fig. 5 b and c and data not shown). RT-PCR analysis revealed that knockdown of all three HDACs led to more dramatic induction of p21 and pten expression (Fig. 5f) compared with the induction by individual HDAC siRNA treatment, providing a molecular basis for the significantly reduced cell proliferation upon triple HDAC siRNA treatment (Fig. 5 d and e). siRNA inhibition of TLX expression also led to significant induction of both p21 and pten gene expression (Fig. 5g), similar to that which resulted from HDAC siRNA treatment (Fig. 5f), strengthening the link of TLX and HDAC in p21 and pten gene regulation.
Disruption of TLX–HDAC5 Interaction Resulted in Dramatic Inhibition of Neural Stem Cell Proliferation.
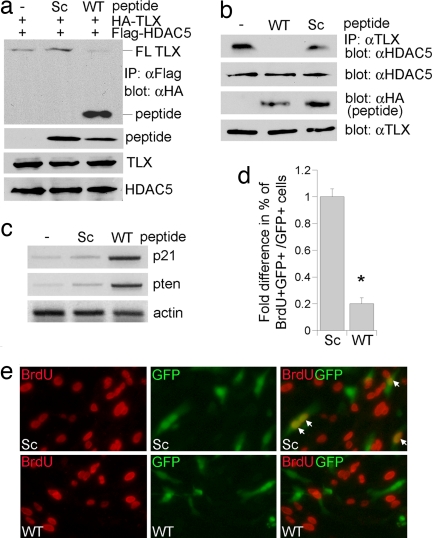
To determine whether the TLX–HDAC interaction is important for TLX-regulated target gene expression and neural stem cell proliferation, we tried to disrupt the interaction by using a TLX peptide containing the minimal HDAC interaction site. We have mapped the primary HDAC5 interaction domain to TLX residues 359–385 with a conserved IXXLL motif located at residues 372–376 (Fig. 2). A peptide spanning TLX residues 362–382 with an HA tag was transfected into HEK293 cells along with Flag-tagged HDAC5 and HA-tagged TLX. The interaction of HDAC5 with the TLX peptide was detected readily (Fig. 6a and SI Fig. 8). The expression of this peptide abolished the interaction of full-length TLX with HDAC5 (Fig. 6a). In contrast, a scrambled peptide had no effect on this interaction. To determine whether the TLX peptide affects endogenous TLX and HDAC5 interaction, we transfected the TLX peptide-expressing vectors into neural stem cells. Expression of the WT TLX peptide led to dramatic loss of endogenous TLX–HDAC5 interaction, whereas the scrambled peptide had minimal effect on this interaction (Fig. 6b). RT-PCR analysis revealed marked induction of both p21 and pten gene expression in the WT peptide-transfected neural stem cells (Fig. 6c). Moreover, expression of the WT peptide led to up to 80% inhibition of neural stem cell proliferation compared with that in the scrambled peptide-transfected cells (Fig. 6 d and e). These results strongly argue that the TLX–HDAC5 interaction is an important component of TLX-mediated transcriptional repression and neural stem cell proliferation.
Fig. 6.
Peptide interference of TLX–HDAC interaction. (a) A TLX peptide competed for full-length (FL) TLX–HDAC5 interaction in immunoprecipitation analyses. Expression of a WT TLX peptide abolished FL TLX and HDAC5 interaction in transfected HEK cells. An empty vector (−) and a scrambled peptide (Sc) were included as negative controls. Expression of the TLX peptides, FL TLX, and HDAC5 was shown by immunoblotting cell lysates with HA- or Flag-specific antibody. (b) A TLX peptide disrupted endogenous TLX and HDAC5 interaction in neural stem cells as revealed by immunoprecipitation analyses. Expression of endogenous TLX, HDAC5, and the transfected peptides was shown by immunoblotting cell lysates with individual antibodies. (c) Induction of p21 and pten gene expression in neural stem cells by expression of WT TLX peptide analyzed by RT-PCR analysis. (d) Decreased percent of BrdU+ cells in the WT TLX peptide-transfected neural stem cells. Percent of BrdU+ cells in the WT peptide-transfected cells (BrdU+GFP+/GFP+) was calculated and normalized with the percent of BrdU+ cells in the Sc peptide-transfected cells. *, P < 0.001. (e) Peptide treatment led to reduced neural stem cell proliferation as revealed by BrdU labeling (red). Cells transfected with the TLX peptide-expressing vectors were green due to the expression of GFP. The BrdU+GFP+ cells were shown in yellow (indicated by arrows).
In summary, we have shown that TLX interacts with HDAC3 and HDAC5 in neural stem cells. TLX recruits the HDACs to the promoters of its target genes, p21 and pten, to repress the expression of these genes. Both chemical inhibition of HDAC activity and siRNA knockdown of HDAC expression led to marked induction of p21 and pten gene expression and significantly reduced neural stem cell proliferation. Moreover, we have mapped the primary HDAC5 interaction site to TLX residues 359–385 and showed that a conserved IXXLL motif in this region is critical for both TLX–HDAC5 interaction and TLX repression activity. A TLX peptide containing this motif disrupted TLX–HDAC5 interaction. Expression of this peptide led to marked induction of p21 and pten gene expression and significant inhibition of neural stem cell proliferation. Taken together, these data strongly support the hypothesis that recruitment of HDACs is an important component in TLX-mediated transcriptional repression and neural stem cell proliferation.
Discussion
We have shown that TLX is an essential regulator of neural stem cell self-renewal (3). In this study, we have identified two TLX target genes, p21 and pten, in neural stem cells, and characterized HDACs as TLX transcriptional corepressors. We have shown that TLX recruits HDACs to p21 and pten genes to repress their expression, which represents an important mechanism for neural stem cell proliferation.
To determine the role of the HDACs in neural stem cells, neural stem cells were treated with HDAC inhibitors, which resulted in significant inhibition of cell proliferation. However, chemical inhibition could have pleiotropic effects. We therefore knocked down individual TLX-interacting HDAC expression by using sequence-specific siRNAs (Fig. 5 and data not shown). Both induction of TLX target gene expression and reduced neural stem cell proliferation were observed upon individual HDAC knockdown. However, the gene regulation and growth arrest effect mediated by individual HDAC siRNA is not as dramatic as that induced by HDAC inhibitors. One possible reason is that the HDAC inhibitors target a panel of HDACs, whereas each HDAC-specific siRNA only reduces one HDAC expression. Indeed, when we treated neural stem cells with a combination of HDAC3, HDAC5, and HDAC7 siRNAs, a much more marked effect was detected upon the induction of p21 and pten gene expression and inhibition of cell proliferation compared with individual HDAC siRNA treatment.
To further determine whether the TLX–HDAC interaction is critical for TLX function, a TLX peptide spanning the minimal HDAC5 interaction site was expressed to disrupt full-length TLX–HDAC5 interaction by competing for HDAC5 binding. Treatment of this peptide led to significant induction of p21 and pten gene expression and marked inhibition of neural stem cell proliferation. Of note, more dramatic effect on cell proliferation (up to 80% reduction) was detected upon TLX peptide treatment compared with the 50–70% reduction in cell proliferation upon siRNA knockdown of HDAC5 expression. This discrepancy could be due to the fact that siRNA only inhibits HDAC5 expression partially (≈60%), whereas the TLX peptide treatment led to almost complete disruption of TLX–HDAC5 interaction.
Cell proliferation was regulated by complex signaling pathways for which the cell cycle machinery holds a central position (22). Cell cycle progression has been shown to be modulated by cyclin-dependent kinase inhibitors. The first of these proteins identified was p21, which has been implicated in growth arrest that precedes terminal differentiation. Results presented here demonstrate that TLX recruits HDACs to repress the expression of its target gene, p21. In addition, we showed that the expression of the tumor suppressor gene pten was up-regulated in adult TLX−/− brains. HDAC3 and HDAC5 were detected along with TLX on the consensus TLX binding site in pten gene promoter, and they repress pten gene expression. pten encodes a lipid phosphotase that regulates cell proliferation by negatively regulating phosphoinositide 3-kinase signaling (23). Conditional pten loss in neural stem cells led to enlarged brains resulted from increased cell proliferation, suggesting that pten negatively regulates neural stem cell proliferation (24). Repression of p21 and pten gene expression by TLX and HDAC interactions provides a mechanism for TLX-mediated neural stem cell proliferation and self-renewal.
Nuclear receptor–HDAC interactions are often mediated by nuclear receptor corepressors SMRT and N-CoR (12, 14, 15, 25, 26). However, we failed to detect the interaction of TLX with SMRT and N-CoR in our assays (data not shown). Others have also reported the lack of interaction between TLX and SMRT/N-CoR (21, 27). Recent studies identified atrophin as a TLX modulator in yeast two-hybrid assays (21, 27). Atrophin has been shown to interact with HDAC1 and HDAC2. However, we showed that TLX interacts with HDAC3 and HDAC5 specifically in neural stem cells. Whether atrophin is in the TLX–HDAC complex in neural stem cells remains to be determined. It is worth noting that the results presented here do not exclude the possibility that TLX recruits HDAC-containing transcriptional corepressor complexes to mediate its cellular function. Exploring the isolation and characterization of TLX corepressor complexes may allow better understanding of the mechanism of TLX-regulated gene expression and its role in neural stem cell proliferation and self-renewal.
Stem cells provide great hope for the treatment of a variety of human diseases that lack efficacious therapies to date. Identifying factors that control stem cell proliferation and self-renewal is an important step in moving stem cell technology from the laboratory to the clinics. One molecule that plays an important role in regulating this process is TLX. Uncovering the regulatory cascade of this nuclear receptor will be critical to implementation of neural stem cell-based cell replacement therapy for the treatment of neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases. The results of this study have provided insights into the TLX signaling pathway and have defined elements that control neural stem cell proliferation. Each component of the TLX signaling network, either downstream target genes or interacting modulators, can be molecular targets for therapeutic intervention of neurodegenerative diseases.
Materials and Methods
Plasmid DNA and Transient Transfections.
GAL4-TLX and HA-TLX were generated by cloning TLX cDNA into CMX-GAL4 DBD or CMX-HA vectors. Flag-HDAC constructs were described in ref. 13. p21-tk-luc was generated by cloning three copies of TLX binding sites in p21 promoter into tk-luc. HDAC5-luc was generated by cloning mouse HDAC5 cDNA into pSicheck 2.2 (Promega, Madison, WI). The WT and scrambled TLX peptides containing mouse TLX residues 362–382 were fused in frame to three copies of nuclear localization signals and HA tags and expressed under CMV promoter. A GFP marker under an independent CMV promoter was included in the vector. CV-1 cells were transfected as described in ref. 13. Neural stem cells were transfected using Nucleofector kit (Amaxa, Gaithersburg, MD). Reporter luciferase activity was normalized by the level of β-galactosidase activity in CV-1 cells and the Renilla luciferase activity in neural stem cells. Each transfection was carried out in triplicate at least three times.
Cell Culture, Immunoprecipitation (IP), and ChIP Assay.
Neural stem cells were cultured in DMEM F12 medium with 1 mM l-glutamine, N2 supplement (GIBCO/BRL, Carlsbad, CA), EGF (20 ng/ml), FGF2 (20 ng/ml), and heparin (50 ng/ml). For differentiation, cells were exposed to 0.5% FBS and 5 μM forskolin for a week. IP of neural stem cells was performed using TLX antibody followed by protein A Sepharose incubation. IP of HEK293 cells was performed as described in ref. 28. ChIP assays were performed using EZ-ChIP kit (Upstate, Lake Placid, NY) and 5 μg of antibody per reaction.
Northern Blot, RT-PCR, Gel Shift, and RNA Interference Assays.
Northern blotting was performed as described in ref. 13. Reverse transcription in RT-PCR analysis was performed using Omniscript RT kit (Qiagen, Valencia, CA). Gel-shift assay was performed using in vitro translated TLX and γ-32P-labeled DNA probes. HDAC siRNA duplexes were transfected into neural stem cells by using SilentFect (Bio-Rad, Hercules, CA) and transfected into HEK293 cells by using Fugene (Roche, Indianapolis, IN).
BrdU Treatment and Immunofluorescence Analysis.
Neural stem cells were treated with BrdU for 2 h. Cells were fixed and acid-treated, followed by rat anti-BrdU antibody (diluted 1:250; Accurate, Westbury, NY) incubation. Quantitative studies were based on four or more replicates. Immunostaining of neural stem cells with antibodies for TLX (1:500), HDAC3, HDAC5, and HDAC7 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) was performed as described in ref. 3.
Supplementary Material
Acknowledgments
We thank Drs. Kamil Alzayady, Barry Forman, Wendong Huang, and Rama Natarajan for critical reading of the manuscript and Dr. Judith Singer-Sam (Beckman Research Institute of City of Hope) for providing the p21 DNA. G.S. is a Herbert Horvitz Postdoctoral Fellow. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. Y.S. is a Kimmel Scholar. This work was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke Grant R01 NS059546 (to Y.S.).
Abbreviations
- HDAC
histone deacetylase
- N-Cor
nuclear receptor corepressor
- SMRT
silencing mediator of retinoic and thyroid hormone receptors
- tk
thymidine kinase
- TSA
trichostatin A
- VPA
valproic acid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704089104/DC1.
References
- 1.Yu RT, McKeown M, Evans RM, Umesono K. Nature. 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 2.Monaghan AP, Bock D, Gass P, Schwager A, Wolfer DP, Lipp HP, Schutz G. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 4.Roy K, Kuznicki K, Wu Q, Sun Z, Bock D, Schutz G, Vranich N, Monaghan AP. J Neurosci. 2004;24:8333–8345. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenman JM, Wang B, Campbell K. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie BR, Li AM, Redila VA, Booth H, Wong BK, Eadie BD, Ernst C, Simpson EM. Neuroscience. 2006;137:1031–1037. doi: 10.1016/j.neuroscience.2005.08.091. [DOI] [PubMed] [Google Scholar]
- 7.McKenna NJ, Lanz RB, O'Malley BW. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 8.Perissi V, Rosenfeld MG. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 11.Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 12.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao HY, Downes M, Ordentlich P, Evans RM. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 15.Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 16.McKinsey TA, Zhang CL, Olson EN. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 17.Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JW. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litterst CM, Pfitzner E. J Biol Chem. 2002;277:36052–36060. doi: 10.1074/jbc.M203556200. [DOI] [PubMed] [Google Scholar]
- 19.Zor T, De Guzman RN, Dyson HJ, Wright PE. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Razeto A, Ramakrishnan V, Litterst CM, Giller K, Griesinger C, Carlomagno T, Lakomek N, Heimburg T, Lodrini M, Pfitzner E, et al. J Mol Biol. 2004;336:319–329. doi: 10.1016/j.jmb.2003.12.057. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Genes Dev. 2006;20:1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherr CJ. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 25.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Genes Dev. 2006;20:525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Mosser DD, Morimoto RI. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.