Abstract
Drosophila Polycomb group (PcG) and Trithorax group (TrxG) proteins are responsible for the maintenance of stable transcription patterns of many developmental regulators, such as the homeotic genes. We have used ChIP-on-chip to compare the distribution of several PcG/TrxG proteins, as well as histone modifications in active and repressed genes across the two homeotic complexes ANT-C and BX-C. Our data indicate the colocalization of the Polycomb repressive complex 1 (PRC1) with Trx and the DNA binding protein Pleiohomeotic (Pho) at discrete sequence elements as well as significant chromatin assembly differences in active and inactive regions. Trx binds to the promoters of active genes and noncoding transcripts. Most strikingly, in the active state, Pho covers extended chromatin domains over many kilobases. This feature of Pho, observed on many polytene chromosome puffs, reflects a previously undescribed function. At the hsp70 gene, we demonstrate in mutants that Pho is required for transcriptional recovery after heat shock. Besides its presumptive function in recruiting PcG complexes to their site of action, our results now uncover that Pho plays an additional role in the repression of already induced genes.
Keywords: Antennapedia, Bithorax, chromatin, gene regulation, HOX
The Polycomb (PcG) and the Trithorax (TrxG) group proteins form the basis of a cellular memory system maintaining the transcriptional state of their target genes heritable during development. Initially, identified in Drosophila as regulators of HOX genes, the chromatin-associated control by the PcG/TrxG proteins has been found to regulate a variety of target genes in many different organisms, up to epigenetic processes such as X chromosome inactivation in mammals. In Drosophila, the genes controlled by the PcG/TrxG system have PcG response elements (PREs) to which these proteins bind and either keep the gene permanently repressed (PcG) or active (TrxG). PREs are composed of a complex set of binding sites for proteins that seem to render a general target gene specificity to the PcG/TrxG-complex as well as a tissue and temporal specificity (1).
Three biochemically distinct PcG complexes have been identified from embryonic nuclear extracts (2, 3). The Polycomb repressive complex 1 (PRC1) contains the four PcG proteins Polycomb (Pc), Polyhomeotic (Ph), Posterior sex combs (Psc), and dRing/Sex combs extra (Sce). PRC1 in vitro is able to repress transcription and inhibit the ATP-dependent chromatin remodelling mediated by the human SWI/SNF complex, a complex related to the Drosophila TrxG Brahma (BRM) complex (4–6). The PRC2 complex contains the SET domain protein E(Z), which preferentially methylates histone H3 at lysine 27 (H3K27me3) (7–10). Because the chromodomain of Pc binds to H3 histone tails trimethylated at lysine 27, this mark has been proposed to be specific for the recruitment of the PRC1 complex, leading to a subsequent silencing of the target gene (11). Most recently, the Pho repressive complex (PhoRC) has been identified that consists of the two PcG proteins Pleiohomeotic (Pho) and dSfmbt (12). Both proteins are crucial for HOX gene silencing with Pho sequence specifically binding to DNA and dSfmbt recognizing the tails of histones H3 and H4 mono- or dimethylated at H3K9 and H4K20 (12).
Maintenance of the active gene expression state utilizes an antisilencing function of the TrxG proteins, requiring at least two enzymatic activities. A nucleosome remodelling activity is provided by the BRM complex that, however, besides counteracting PcG-silencing is involved in more general aspects of gene transcription (6). Additionally, covalent modifications of histones, mediated by TrxG factors such as Trx, Ash1, and their interaction partners, were found to be of importance for a stable maintenance of the active state (10, 13–16). Trx is present in the TAC1 complex that contains the histone acetyltransferase dCBP (13). The role of histone acetylation in PcG/TrxG-dependent processes is supported by the observation that disruption of PRE-mediated silencing in Drosophila transgenes is accompanied by local accumulation of hyperacetylated histone H4 (17). Currently it is unclear how the interplay between DNA elements, histones and their modifications, and the PcG/TrxG-chromatin-associated proteins results in a stable gene expression state, which is heritable through DNA replication and mitosis. A key to the understanding of the molecular mechanisms is to uncover the chromatin sites of action of the PcG/TrxG proteins and to correlate their binding patterns with the presence of histone modifications as well as with the expression level of their target genes. The advent of ChIP maps demonstrated an accumulation of PcG/TrxG proteins at PREs and some of the associated promoters (18, 19). Interestingly, PcG proteins were found to be bound at PREs, even if the associated gene was active (20, 21). In addition, a recent study that concentrated on the HOX gene Ultrabithorax (Ubx) in Drosophila imaginal discs showed that members of the PRC1/2 complexes and Trx are constitutively bound at the regulatory sites of Ubx regardless of its activity state (16). Because there is strong evidence that the function and composition of PcG complexes is modulated in different tissues (22) and in different chromatin environments (21, 23), we wanted to generate high-resolution protein distribution data to uncover possible regulatory hallmarks for the different expression states.
To address this intention we generated a DNA tiling array that comprises the ANT-C and the BX-C as a series of overlapping 1-kb PCR fragments and thus covers several HOX genes. On the basis of this tiling array, we investigated the distribution of three core components of the PRC1 (Pc, Ph, and Psc), the DNA binding protein Pho, Trx, hyperacetylated H4, and H3K27me3 in two different Drosophila embryonic cell lines that show different expression states for the AbdB and Dfd genes. We observe that in particular the Pho protein depicts a distribution profile that reflects the expression state of the target gene.
Results
To investigate the distribution of PcG and TrxG proteins in the ANT-C and the BX-C we chose the two Drosophila melanogaster embryonic cell lines Kc and SF4 as starting material. The diploid SF4 cells show a higher degree of homogeneity regarding their karyotype compared with the heteroploid SL2 cells from which they originated (D. Arndt-Jovin, personal communication). To correlate protein distributions with gene expression we characterized the HOX gene expression profile in the two cell lines [supporting information (SI) Fig. 5]. The AbdB domain and the Dfd gene are highly active in SF4 and in Kc cells, respectively, whereas the other HOX genes are silent. A detailed description of the HOX gene expression profile is given in the SI Text, and all significant binding sites we identified are summarized in SI Tables 1 and 2.
Distribution of Pho, PRC1, and Trx in the Silent ANT-C and BX-C.
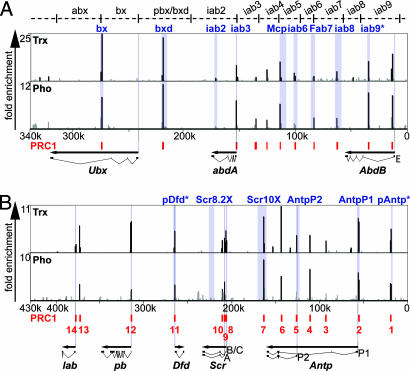
Several studies suggest the DNA binding protein Pho as a targeting factor for PRC1 (24–26), whereas others have shown, by immunostaining of polytene chromosomes, that the binding of Pc, Ph, and Psc to most sites is not altered in pho, pho-like double mutants (pho-like encodes a Pho-related protein, which binds to the same DNA motif) (27). So far our knowledge about Pho binding sites on chromatin has been limited to low-resolution polytene chromosome stainings and the PCR analysis of Ubx sequences and the iab-7 PRE after ChIPs (12, 16, 26). To gain a better insight into the interdependency of Pho and PRC1, we investigated the distribution of Pho in the homeotic complexes. As indicated in Fig. 1, Pho binds at many discrete sites and nearly all known regulatory regions in the silent BX-C and ANT-C.
Fig. 1.
At silent HOX genes, Trx and Pho colocalize at most elements with PRC1. Normalized ChIP/input ratios >1, indicating enriched fragments, are plotted across the BX-C (A) and ANT-C (B). The various regulatory regions of the BX-C (abx to iab9) are indicated at the top. Published PREs and promoters are indicated in blue; iab9* may be the PRE of the iab9 region. pDfd* and pAntp* have been identified in an algorithmic approach as potential PREs (30) but had not been experimentally confirmed previously. The coordinates based on the complete sequences of the BX-C (U31961, 0–340,000) and ANT-C (AE001572, 0–430,000) are shown on the x axis with proximal to the left. The HOX genes are drawn with exon structure. For AbdB, only the longest transcription unit (AbdB-RE) is indicated. Scr contains three alternative promoters, RA, RB, and RC; the latter two are not resolvable on our array. Antp contains two alternative transcription units, AntpP1 and AntpP2. PRC1-bound fragments identified in these cells are indicated in red at the bottom of the profiles (for more details, see SI Fig. 6). The numbers of PRC1 sites in B correspond to the numbering in SI Table 1 in which the coordinates of binding sites are listed. (B) Pho is missing at the lab and Scr-RB/C promoters. ChIP/input ratios with an FDR <5% are drawn in black.
For a direct comparison, we also mapped the binding sites for PRC1 by chromatin immunoprecipitations of three core subunits of the complex, assuming that the simultaneous binding of Pc, Ph, and Psc at the same site occurs as part of a functional PRC1. The identified PRC1 sites are indicated in Fig. 1, and the detailed profiles are shown in SI Figs. 6 and 7. The distribution of PRC1 in SF4 cells resembles the results of Schwartz et al. (28) who also used a Schneider cell derivative to map Pc and Psc sites. Remarkably, all promoter regions of the silent ANT-C genes are occupied by PRC1, including all alternative promoters of Antp and Scr (Fig. 1B, PRC1 sites 2, 5, 8, and 9). PRC1 site 13 is very closely located to lab, making it a candidate for a lab PRE. Scr8.2X is devoid of PRC1, unlike Scr10X, which is bound in the distal part (PRC1 site 7). The lack of PRC1 binding at Scr8.2X might be an indication for the tissue-specific requirement of PREs. In these embryonic cell lines, the inactive state of Scr is maintained without using Scr8.2, whereas other Scr regulatory regions are occupied by PRC1/Pho (Fig. 1, Scr10X and PRC1 site 10). Additional binding sites can be found in intronic regions of Antp (PRC1 site 3, 4, and 6). Also in the BX-C PRC1 binds to discrete sequence elements and shows a nearly complete overlap in both cell lines (Fig. 1A and SI Figs. 6 and 7).
We found a strong correlation of Pho and PRC1 binding at most regulatory sequences of silent HOX genes (Fig. 1). We never observed Pho enrichment outside a PRC1 site in the silent condition. However, PRC1 binding might not exclusively depend on Pho as demonstrated at the lab and AntpP2 promoters.
An important function for the maintenance of HOX gene activity is the action of the Trx protein. In both cell lines we detected a complete coverage of PRC1/Pho sites with Trx at silent genes (Fig. 1). This data set provides additional evidence against the simple model that PRC1 and Trx compete for binding regulatory sequences, at least not in the target genes' inactive state.
Active Versus Silent Transcription States.
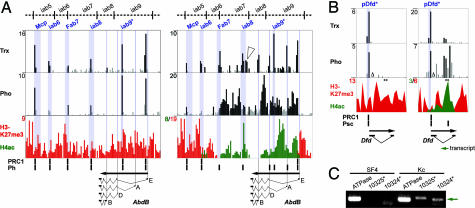
The AbdB gene contains several alternative promoters that are embedded in a complex regulated domain. The two different cell lines allowed us to compare this domain in the active and silent state (Fig. 2). Previous studies revealed a central role of trimethylated H3K27 at PcG regulated genes (16, 28, 29). Supporting this data, we detected H3K27me3 in broad regions encompassing entire inactive transcription units such as the AbdB domain in Kc cells (Fig. 2A Left and SI Figs. 6 and 7). PRC1, Pho, and Trx are bound to all regulatory elements. The promoter regions are free of PRC1, Pho, and Trx. In the active AbdB domain we observed a rather different protein-binding pattern (Fig. 2A Right). Here, H3K27me3 is absent in the region from the RE promoter to the Fab7 PRE; instead, we found hyperacetylated histone H4 (H4ac), which is a mark for ongoing transcription. PRC1 is still present at Mcp, iab6, and a fragment near the RE promoter and colocalizes with Pho and Trx. In contrast Fab7, iab8, and iab9* are devoid of a functional PRC1 complex, whereas Trx stays bound. Interestingly, Trx also occupies the promoter regions of the active AbdB transcription units RB, RD, and RA and of a noncoding transcript that is only present in the active AbdB domain (Fig. 2A; D. Enderle and R.P., unpublished results). However, the most striking difference we observed was for the distribution of Pho. In contrast to PRC1, it is not absent from the PREs but seems to spread over the complete active chromatin domain.
Fig. 2.
Comparison of the active and repressed transcription states of AbdB and Dfd. (A) Pho spreading, promoter binding by Trx, and absence of PRC1 determine the active state of the AbdB domain. Shown is the protein distribution on the inactive (Left) and the active (Right) AbdB domain of Kc and SF4 cells, respectively. PREs and promoters are indicated as in Fig. 1. The arrowhead in the Trx profile shows the start of a ncRNA that is only present in the active state (D. Enderle and R.P., unpublished results). Fragments bound by PRC1 or Ph alone are indicated in black below the histone-modification profile. Red indicates H3K27me3, and green indicates H4ac. (B) An alternative chromatin composition maintains the active state of Dfd. Inactive (Left) and active (Right) Dfd gene of SF4 and Kc cells, respectively. The predicted Dfd PRE (pDfd*), which is located at the Dfd promoter, is indicated in blue. Fragments bound by PRC1 or Psc alone, are indicated in black below the histone modification profile. Δ indicates a fragment that is missing on the array. The green arrow indicates the antisense transcription we detected in two fragments located in the intron region of the active Dfd gene (**, fragment 10325 and 10324; 10325 is located to the proximal end). At fragment 10324, Psc and H4ac show peak enrichments. (C) RT-PCR analysis for the detection of antisense transcription in fragments 10325 and 10324 using total RNA isolated from Kc and SF4 cells. ATPase cf6 was used as positive control. In the absence of reverse transcriptase, no PCR product was detected in any of the reactions (data not shown).
The Dfd gene, which is active in Kc cells and inactive in SF4 cells, is much smaller than the AbdB gene and its regulatory region is less complex. For Dfd, only one PRE (pDfd*), located in the promoter region, was identified by an algorithmic PRE prediction approach (30), which was confirmed as a binding site for PRC1 in our study. There were no other PRC1-bound sequence elements detectable surrounding Dfd, either in SF4 or in Kc cells.
Consistent with the situation of AbdB, in the inactive state, pDfd* is occupied by PRC1, Pho, and Trx, and the gene is covered by H3K27me3 (Fig. 2B Left). In the active state, we observed several differences compared with AbdB (Fig. 2B Right). All proteins remain enriched in the pDfd*, PRC1 is present, and Trx and Psc are enriched at two independent sites in the intron. The occurrence of Psc and Trx prompted us to check for an intronic transcript in this region that would provide evidence for an active PRE. Indeed, we detected an antisense transcript covering the Psc/H4ac peak in the active, but not in the inactive state (Fig. 2C). Despite the small size of the Dfd gene, a spreading of Pho over at least five fragments covering a region of 4 kb is visible, which confirmed our observation in the active AbdB domain.
Pho Plays a Role in Actively Transcribed Genes.
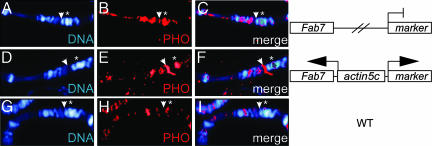
We previously used a transgenic reporter system to study the function of noncoding transcription through the Fab7 PRE (31). In this reporter system transcription through the Fab7 element driven by a constitutively active actin5c promoter leads to the activation of the coupled miniwhite marker, whereas in the absence of the actin5c promoter Fab7 is not transcribed and acts as a silencer for miniwhite. Here we used immunostaining of polytene chromosomes to investigate whether transcription through the Fab7 PRE on the pFAs transgene also leads to a spreading of Pho. In the absence of the actin5C promoter, the repressed Fab7 PRE generated a new binding site for Pho on polytene chromosomes, which was absent in wild type (Fig. 3 A–C and G–I). When the silencing function of Fab7 was abolished by transcription from the actin5C promoter, we observed more intense Pho staining at the transgenic construct (Fig. 3 D–F).
Fig. 3.
Pho is found at sites of active gene transcription. The binding of Pho at the actively transcribed Fab7 PRE was investigated with a previously published transgenic reporter system (31). The transgene constructs are indicated on the right side. (A–C) The repressed Fab7 PRE on the transgene created an ectopic Pho binding site (arrow). (D–F) Transcription of Fab7 through the actin5C promoter leads to stronger Pho staining. (G–I) Wild-type control. There is no endogenous Pho binding at the transgene insertion site. Asterisks denote endogenous Pho binding at 32F.
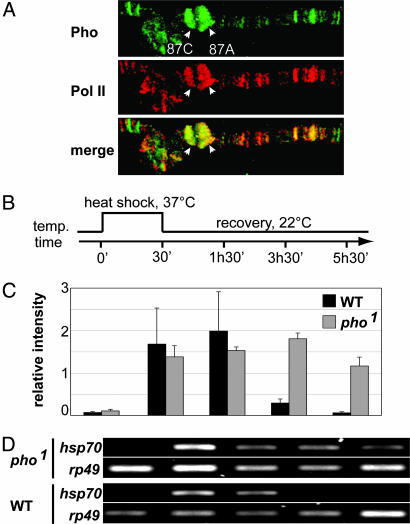
We also observed strong Pho signals at many puffs, which are regions with high transcription rates (data not shown). This finding suggested that Pho may also be involved in more general aspects of gene regulation. To test this hypothesis we prepared polytenes from heat-treated larvae and stained them with antibodies for Pho and serine-5-phosphorylated RNA polymerase II (Pol II). Indeed, chromosomes showed a strong Pho staining covering several heat-shock loci, where “puffing” is initiated in response to induction (Fig. 4A). Altogether, ≈50% of all Pho bands colocalized with predominantly strong signals of active Pol II, suggesting that Pho primarily acts at highly induced genes. These results supported the spreading of Pho in active chromatin domains observed in the ChIP-on-chip approach and furthermore implicate a general role for Pho in actively transcribed genes.
Fig. 4.
Pho is present at highly induced genes on polytene chromosomes and is required for hsp70 recovery after heat shock. (A) After heat induction, Pho covers heat-shock loci completely (arrows at 87A and 87C) and additionally colocalizes with active, serine-5-phosphorylated RNA Pol II at many sites. A partial spread of chromosome 3R is shown. Pho is shown in green, and Pol II is shown in red; yellow indicates colocalization of both proteins. (B–D) Transcriptional recovery after heat-shock induction of hsp70 is slower in homozygous pho1 mutant larvae. (B) Experimental scheme. Larvae were heat shocked at 37°C for 30 min and afterward were transferred to 22°C for recovery. RNA was isolated at the indicated times and assayed by RT-PCR with primers specific for hsp70 and rp49 as standard. (C) Normalized values of quantified hsp70 RT-PCR products (hsp70/rp49-ratio). The standard deviation was calculated for two independent PCRs. (D) For one experiment, ethidium-bromide-stained gels are shown.
Next we asked for the functional relevance of the observed Pho distribution by investigating the heat-shock-induced expression of the hsp70 gene in pho mutant larvae. We monitored the abundance of the hsp70 transcript during and after heat shock by RT-PCR of RNA isolated from larvae that were either wild type or homozygous for pho1, a protein-negative allele of pho (12). As shown in Fig. 4, we observed no significant difference at 30 min of heat shock and 1 h of recovery after heat-shock treatment. However after 3 and 5 h of recovery, the level of hsp70 transcript was still significantly higher in pho1 mutant larvae compared with wild type. This result suggests that the spreading of Pho on transcribed regions is not correlated to a function in activation but pinpoints a possible functional role in the rerepression of a gene after down-regulation of the transcriptional stimulus.
Discussion
In this work we used two Drosophila tissue culture lines to map the distribution of chromatin proteins required for the transcriptional maintenance of the HOX genes. Although compromising on the precise developmental identity, the tissue culture cells provided us with biochemically tractable homogeneous material, which currently would be difficult to obtain from whole animals. We believe that this choice was important to obtain the sharply delineated ChIP profiles, which show a highly significant correlation to mapped genetic elements in the two homeotic complexes. As such, the protein patterns obtained seem to reflect a valid situation as found in material from whole animals. In addition, the ChIP profiles uncovered a new function of Pho, which could be confirmed in whole animals.
Our results for SF4 cells are consistent with the data obtained by Schwartz et al. (28) who also used a Schneider cell derivative for ChIP studies (29). PRC1 binds to discrete sequence elements, whereas H3K27me3 covers large genomic domains, including genic and intergenic regions. These observations indicate that H3K27me3 cannot be solely responsible for PRC1 targeting. How these H3K27 methylated domains influence HOX gene expression and whether the broad methylation pattern is the cause or consequence of gene silencing remains unclear. H3K27me3 may prevent the binding of activating protein factors as e.g., chromatin remodeling complexes and/or prevent the establishment of activating histone modifications. To this regard, we detect a complementary pattern of H3K27me3 and H4ac, which is present in active gene regions.
Several lines of evidence suggest that PcG proteins propagate their silencing effect by the direct interaction with the promoter region, which results in the inhibition of transcription initiation (16, 32–34). In agreement with that, all promoter regions of the silent ANT-C HOX genes are occupied by PRC1. However, the Ubx promoter, which is silent in both cell lines, as well as the silent AbdB transcription units in Kc cells, are devoid of PRC1. Here, probably the numerous PREs, which are occupied by PRC1 in the Ubx and AbdB domains, build up a special chromatin structure that maintains the silent transcription state.
In agreement with the observed H3K27me3 pattern in Drosophila cells, in mammalian Hox clusters inactive domains are covered by H3K27 and active domains are found entirely covered by H3K4 methylation (35, 36). In contrast, the distribution of the enzymes setting the histone marks are completely different. In Drosophila E(Z), Trx, and Ash1 are bound at discrete sequence elements (this study and refs. 16, 28, and 29), whereas the mammalian homologues EZH2 and MLL1 localize to extended regions coincident with the methylation signals (36, 37). MLL1 acts as a functional human equivalent of yeast Set1 (36). Both proteins colocalize with RNA Pol II at the transcription start site of highly expressed genes and catalyze the trimethylation of H3K4 at this location (36, 38–41). Only at active Hox genes MLL1 reveals a different binding behavior covering entire active chromatin domains. In contrast, our data shows that Trx also localizes to promoter regions of silent HOX genes and does not show the spreading behavior of MLL1 but appears at additional discrete sites. We observe a complete colocalization of Trx with PRC1 sites at silent genes, i.e., in this expression state no obvious competition is taking place with regard to binding sites.
What Defines the Active State?.
The comparison of the AbdB gene with the Dfd gene shows that the maintenance of the active state can be performed in alternative ways. The absence of PcG complexes does not seem to be a prerequisite of the active state as observed at the promoter of Dfd in this study and at regulatory regions of Ubx in imaginal discs (16).
In the active AbdB domain Ph stays bound in a minor but significant amount, and Psc is present in the active Dfd intron. In this regard, Ph and Psc could serve as recruiting platforms for other PRC1 subunits in case of the gene switching to the off state. However, both proteins have been reported to be associated with active genes (21). Consistent with this, we also observe Ph in the proximal part of both homeotic complexes binding actively transcribed non-HOX genes (SI Fig. 8). The function of this binding behavior remains elusive.
The transcription of noncoding RNAs (ncRNAs) seem to play an important, although diverse, role in the regulation of the BX-C. Petruk et al. (42, 43) found that noncoding transcription through the bxd PRE is crucial for Ubx repression and that transcription through Mcp overlaps with AbdB transcription in the embryo. Work from others and our laboratory showing that ncRNA transcription in the AbdB domain coincides with an active AbdB gene indicates a nonuniversal, gene specific function for ncRNAs in the BX-C (31, 43, 44).
In the silent state PRC1 is bound to all PREs in the AbdB domain and might be recruited by the action of sequence-specific factors like Pho and the E(Z) histone methyltransferase activity, which may also mark the entire domain as being inactive (29, 44). In the active AbdB domain, ncRNA transcription may directly influence the binding of of PRC1 and E(Z) or may trigger the enzymatic activity of Trx. Consistent with this scenario, Trx has recently been shown to bind single-stranded DNA and RNA in vitro (45). The switch of Trx into an activating mode could lead to the methylation of histones and/or other proteins setting positive transcriptional marks and modulate their activity, respectively. In this case, the displacement of PcG proteins could be directly caused by the Trx action. The binding of Trx to the promoter regions of the active AbdB transcription units could either be caused by (transient) chromatin looping events bridging Trx-bound PREs with the promoters, or Trx could be recruited independently to the active HOX promoters by interaction with RNA Pol II, similar to MLL1, which is recruited to actively transcribed genes in mammalian cells. Trx- and TAC1-interacting histone acetyltransferases may then be responsible for setting epigenetic marks that maintain the active transcription state. Petruk et al. (42) showed that Trx is required for transcription elongation and that it is localized in the gene body of active Ubx, caused by the interaction with elongation factors. In contrast, Papp and Muller (16), who investigated the distribution of PcG and TrxG proteins at the active and repressed Ubx gene in imaginal discs, found the same restricted Trx profile as we did, namely Trx binding at discrete sites. These differences may be explained by the different Trx antibodies used. Trx is most probably proteolytically processed like human MLL which results in two fragments that form a heterodimeric complex (46, 47). Whereas the antibody used by Petruk et al. (42) recognizes the N-terminal fragment of the protein, ours, as well as the antibody used by Papp et al. (16), recognizes the C-terminal fragment. This raises the intriguing question whether the complete heterodimeric Trx complex might get recruited to the promoter and upon gene induction the N-terminal fragment tracked along the gene body together with elongation factors, whereas the C-terminal fragment stayed at the promoter.
Pho Is Involved in the Rerepression of Induced Genes.
We generated Pho maps to investigate its role in the recruitment of PRC1. However, the distribution of Pho suggests that the protein also functions in the gene body of actively transcribed genes. The immunostaining of polytene chromosomes revealed that Pho seems not only to be limited to HOX gene control but plays a general role in gene regulation. The colocalization of Pho with strong signals of active Pol II on polytenes together with the effect of a pho-null mutation on the recovery of induced hsp70 indicates that Pho may be directly involved in the rerepression of highly active genes.
It is difficult to imagine that the spreading of Pho is the result of the ability of this protein to bind sequence specifically to DNA. Instead, we propose a model in which Pho either acts directly at the Pol II elongation complex or it interacts with a remodeling complex, carrying it along the chromatin fiber. In this line, Pho has been shown to interact with BRM and dINO80, two nucleosome remodeling complexes (12, 25). Interestingly, heat-shock gene transcription is independent of BRM but involves the recruitment of the TAC1 complex, possibly through multiple interactions with the elongating Pol II complex (48, 49). The simultaneous action of Trx and Pho at heat-shock genes is striking and might resemble their antagonistic functions at HOX genes. Further studies are necessary to unravel the exact molecular mechanism of Pho in this process.
Concluding Remarks.
Compared with previously published data, our results provide evidence for the notion that the PcG/trxG system is highly dynamic, acting tissue and probably even gene specific. Papp and Muller (16) reported no difference for the binding of Pho, Trx, and PRC1 at the active respectively inactive Ubx gene in larval imaginal discs. This is similar to the situation observed at the Dfd gene but rather different to our data for the AbdB domain. Instead of additional Trx binding sites, the HMT Ash1 is present at active Ubx, which might resemble the function of Trx. Remarkably, Pho has not been detected in the gene body of active Ubx. Assuming that Pho plays a role in the repression of highly induced genes, this function might not be necessary in larval cells, which are already determined to build up a wing. These cells are probably less plastic and more fixed regarding their gene expression state than the embryonic tissue culture cells used in our study. PRC1 absence at the active AbdB domain and the Ubx promoter in embryonic cells on the one hand and its constitutive binding at the PREs and promoter of Ubx in imaginal discs on the other hand is consistent with the observation of Negre et al. (50). They performed a developmental ChIP study with Pc and Ph and show that the association of PRC1 to its target chromatin is dynamic in many cases.
In summary we can conclude that the ChIP studies published so far have been a first step to gain insights into the complex function of PcG/TrxG proteins but more comprehensive high resolution data sets that correlate gene expression state with chromatin composition will be required to describe and understand the cellular memory system in a predictable manner.
Materials and Methods
ChIP.
Drosophila melanogaster Kc and SF4 (obtained from D. Arndt-Jovin, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) cells were cultured in Schneider's Drosophila medium (Invitrogen, Carlsbad, CA) supplemented with 10% FCS. Chromatin fixation and immunoprecipitation were performed essentially as described in ref. 51. Cells (1 × 109) were fixed in 200-ml medium with 1% formaldehyde for 10 min at room temperature. Cross-linked cells were sonicated to produce chromatin fragments of an average size of 500–1,000 bp. Soluble chromatin was separated from insoluble material by centrifugation. The supernatant containing chromatin of 5 × 107 cells was taken for immunoprecipitation. Psc and Ph antibodies were described in ref. 21. Pc and Trx antibodies recognizing the C-terminal 199 aa of Pc and residues 2388–2674 of Trx, respectively, were produced in rabbits and were affinity purified. Antibodies against Pho were obtained from J. Müller (European Molecular Bliology Laboratory, Heidelberg, Germany), and antibodies against H3K27me3 were obtained from T. Jenuwein (Research Institute of Molecular Pathology, Vienna, Austria). Anti-acetyl-histone H4 was purchased from Upstate, Lake Placid, NY (catalog no. 06-866). DNA amplification, array hybridization, construction, and analysis of the microarrays are described in SI Text.
Analysis of Polytene Chromosomes.
Immunostaining of polytene chromosomes was largely performed as described in ref. 52. Details can be found in SI Text.
Collection of pho1 Mutant Larvae, Heat Treatment, and RT-PCR.
pho1 homozygous larvae were collected from a stock that was ey-GAL4/ey-GAL4;+;pho1/GS15194. Larvae homozygous for pho1 do not express GFP. GS15194 flies were obtained from DGRC (Kyoto, Japan). It contains a UAS-GFP transgene insert close to the pho gene that can be used as a marker for the fourth chromosome. ey-GAL4 flies were obtained from W. Gehring (University of Basel, Basel, Switzerland). pho1/pho1 and wild- type larvae were incubated at 37°C for 30 min. The recovery followed at 22°C. At the indicated time points (Fig. 4), two to three larvae were snap-frozen in liquid nitrogen. Larvae were homogenized in TRIzol reagent (Invitrogen), and the RNA was isolated according to the manufacturer's instructions. Two nanograms of RNA was taken to detect the transcripts for hsp70 and rp49 (as standard) by RT-PCR using the FideliTaq PCR Master Mix (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. Primer sequences hsp70, 5′-CGATCTGGGCACCACCTACTC and 5′-CGTGGGCTCATTGATGATGCG; rp49, 5′-CGGATCGATATGCTAAGCTG and 5′-GAACGCAGGCGACCGTTGGGG. The RT-PCR program was as follows: 20 min at 50°C, 3 min at 94°C, 33×(20 sec at 94°C, 45 sec at 55°C, 1 min at 68°C), 5 min at 68°C. The ethidiume bromide-stained agarose gel was photographed, and the bands were quantified with AIDA (Raytest, Straubenhardt, Germany). ey-GAL4 and the insertion of GS15194 do not show an effect on the heat-shock response (data not shown).
Supplementary Material
Acknowledgments
We thank Donna Arndt-Jovin, Thomas Jenuwein, and Jürg Müller for reagents and Peter Angel for providing the microarray scanner. S.S. was supported by a Boehringer Ingelheim Fonds fellowship. This research was supported by Deutsche Forschungsgemeinschaft Grant SFB/Transregio 5.
Abbreviations
- BRM
Brahma
- ncRNA
noncoding RNA
- PRC
Polycomb repressive complex
- Pol II
polymerase II
- PRE
PcG response element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE 7031).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701538104/DC1.
References
- 1.Ringrose L, Paro R. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 2.Levine SS, King IF, Kingston RE. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Muller J, Kassis JA. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 5.King IF, Francis NJ, Kingston RE. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. Development (Cambridge, UK) 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 7.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 8.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 9.Cao R, Tsukada YI, Zhang Y. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 11.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 14.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 15.Klymenko T, Muller J. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papp B, Muller J. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalli G, Paro R. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 18.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strutt H, Cavalli G, Paro R. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringrose L, Ehret H, Paro R. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Strutt H, Paro R. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otte AP, Kwaks TH. Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 23.Rastelli L, Chan CS, Pirrotta V. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Brown JL, Fritsch C, Mueller J, Kassis JA. Development (Cambridge, UK) 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 29.Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. J Biol Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- 30.Ringrose L, Rehmsmeier M, Dura JM, Paro R. Dev Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt S, Prestel M, Paro R. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breiling A, O'Neill LP, D'Eliseo D, Turner BM, Orlando V. EMBO Rep. 2004;5:976–982. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiling A, Turner BM, Bianchi ME, Orlando V. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 34.Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 39.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petruk S, Sedkov Y, Brock HW, Mazo A. RNA Biol. 2007;4:1–6. doi: 10.4161/rna.4.1.4300. [DOI] [PubMed] [Google Scholar]
- 44.Hogga I, Karch F. Development (Cambridge, UK) 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- 45.Krajewski WA, Nakamura T, Mazo A, Canaani E. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh JJ, Cheng EH, Korsmeyer SJ. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 48.Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW. EMBO J. 2002;21:5245–5254. doi: 10.1093/emboj/cdf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 50.Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlando V, Strutt H, Paro R. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 52.Lavrov S, Dejardin J, Cavalli G. Methods Mol Biol. 2004;247:289–303. doi: 10.1385/1-59259-665-7:289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.