Abstract
Recent studies revealed an immunoregulatory role of natural IgG-anti-F(ab′)2 antibodies in both healthy individuals and patients with certain diseases. The implication of anti-F(ab′)2 antibodies in the pathogenesis of diseases prompted us to study the gene segment structure of their antigen-binding domains and their binding characteristics. cDNA was prepared from the lymphocytes of a patient with a high IgG-anti-F(ab′)2 serum titer. Variable heavy and light gene segments were amplified by PCR and inserted into a phagemid surface expression vector. Single-chain antibodies displayed on the phage surface were screened for binding to F(ab′)2 fragments. The subsequent analysis of 95 single clones demonstrated that they all bound specifically to F(ab′)2. Sequence analyses of 12 clones showed that 11 were identical and 1 contained a silent point mutation in the heavy chain and three amino acid exchanges in the light chain. The heavy chains belonged to the VH3 and the light chains to the Vκ2 gene family. The 11 identical light-chain genes were completely homologous to a germ-line sequence (DPK-15). Binding assays showed that the single-chain antibodies bind to F(ab′)2, but not to Fab, Fc, or intact IgG. This binding pattern was confirmed by surface plasmon resonance studies, which revealed a relatively high affinity (Ka = 2.8 × 107 M−1). The strong binding capacity was further demonstrated by competitive inhibition of the serum anti-IgG antibody’s interaction with antigen. The present study defines for the first time to our knowledge the gene segment structure of the antigen-binding domain of two human IgG-anti-F(ab′)2 autoantibody clones and describes the binding kinetics of the purified monomeric fragments.
Keywords: anti-immunoglobulin antibodies, phage display, antibody library
The immune response is tightly regulated by stimulatory and suppressive mechanisms. Excessive or deficient suppression affects the efficiency of an immune response against foreign antigens and may contribute to the initiation of autoimmune reactions. Much has been learned about the immunoregulatory role of antiidiotypic antibodies (1–3). We have focused on the immunosuppressive role of natural IgG-anti-F(ab′)2 autoantibodies of idiotype-independent nature in various physiological and pathological conditions (4–9). In cold agglutination, an autoimmune disease caused by anti-erythrocyte autoantibodies, we found a striking inverse correlation between IgG-anti-F(ab′)2 and autoantibody production (4). Patients with high anti-F(ab′)2 titers had low titers of the deleterious anti-erythrocyte autoantibody and vice versa. Cell culture experiments showed that natural anti-F(ab′)2 antibodies are able to suppress anti-erythrocyte autoantibody-producing B cells (5), possibly by crosslinking the membrane Ig with the Fc receptor on the B cell membrane (10–12). It has been demonstrated that anti-Ig antibodies are able to induce a state of dormancy in malignant B cells (13–15). In patients with cold agglutination, it appears likely that very low IgG-anti-F(ab′)2 antibody titers lead to an increase of anti-erythrocyte autoantibody production. IgG-anti-F(ab′)2 antibodies also were shown to play a role in kidney graft rejection (6), the pathogenesis of AIDS (7), and systemic lupus erythematosus (16, 17). The apparent involvement of these antibodies in the pathogenesis of diseases prompted us to study their genetic structure and binding characteristics.
The advent of new molecular biological techniques made it possible to construct and engineer Ig molecules (18–22). In the current series of experiments, the heavy and light chain variable regions (VH and VL) genes of anti-F(ab′)2 antibodies were cloned, sequenced, and expressed in a bacterial system. After purification of the single-chain products, their binding properties were analyzed by ELISA, surface plasmon resonance, and competitive inhibition.
MATERIALS AND METHODS
Construction of the Phagemid Display Vector pSEX81.
A new vector for generating a phage display library of single-chain Fv (scFv) antibodies was constructed. Briefly, pBluescript II SK(+) (Stratagene) was digested with PvuII, and the resulting 2513-bp fragment was purified by gel electrophoresis. This fragment then was ligated with a PCR product of 2247 bp obtained after amplifying a part of the phagemid pSEX20 (23) with the oligonucleotides 1204 (New England Biolabs) and the pSEX primer (5′-GTCGACGTTAACCGACAAACAACAGATAA-3′). This resulted in the vector pSEX61. Finally, the promoter PA1/04/03 (24) of pSEX61 was exchanged for the lac wild-type promoter of the vector pBluescript II SK(+). The correct sequence of the resulting phagemid vector pSEX81 was confirmed by sequence analysis.
Generation of a scFv Expression Library.
Peripheral blood mononuclear cells were separated from 25 ml of blood of a kidney graft recipient with a high anti-F(ab′)2-antibody titer. After mRNA extraction and first-strand cDNA synthesis, the Fv-encoding regions of γ, κ, and λ chains were amplified by PCR as previously described (25). The amplified light chains were purified by agarose gel electrophoresis, digested with MluI and NotI, and ligated into pSEX81. Electrocompetent Escherichia coli XL-1blue cells (Stratagene) were transformed according to the supplier’s protocol. After overnight incubation on SOB agar plates (SOB medium contains, per liter: 20 g of tryptone, 5 g of yeast extract, and 0.5 g of NaCl; after autoclaving add 10 ml of 1 M MgCl2 and 10 ml of 1 M MgSO4 before use) containing 100 mM glucose and 50 μg/ml ampicillin, the bacteria were harvested, and phagemid DNA containing the light chains was prepared.
Subsequently, the heavy chains were cloned into the κ and λ sublibraries by NcoI/HindIII digestion and transduced into E. coli XL-1blue. The bacteria were plated on SOB agar plates (100 mM glucose, 50 μg/ml ampicillin, and 30 μg/ml tetracycline) and incubated overnight. The harvested bacteria were used to inoculate 2× YT medium (2× yT medium contains per liter: 17 g of tryptone, 10 g of yeast extract, and 5 g of NaCl) containing 100 mM glucose and 50 μg/ml ampicillin. After reaching an optical density of OD600 = 1, M13KO7 helper phages were added (multiplicity of infection = 20).
One hour later, the bacteria were pelleted, resuspended, and cultured in 400 ml of 2× YT (50 μg/ml ampicillin and 50 μg/ml kanamycin) for 5–8 h. Finally, the phages were precipitated and resuspended in 4 ml of phage dilution buffer (10 mM Tris·HCl, pH 7.5/20 mM NaCl/2 mM EDTA). For the analysis of single clones, the phage rescue was performed on a smaller scale.
Selection of Antigen-Binding Phages.
A Maxisorb Immunotube (Nunc) was coated overnight with human F(ab′)2 γ fragments (500 μg per tube) (Jackson ImmunoResearch), washed, and blocked with 2% (wt/vol) skimmed milk in PBS for 3 h. After washing, 1011 recombinant phages were added to the tube and incubated for 2 h. The tube then was extensively washed with 0.1% PBS/Tween 20 and PBS. Finally, the phages were eluted with 100 mM triethylamine. E. coli XL-1blue cells were infected with the eluted phages, plated on SOB agar plates (100 mM glucose, 50 μg/ml ampicillin, 30 μg/ml tetracycline), and incubated overnight at 30°C. The selection procedure was repeated five times.
Phage-ELISA.
Microtiter plates were coated overnight with 1 μg per well of F(ab′)2 γ fragments or 2.5 μg per well of BSA. After blocking with 2% (wt/vol) skimmed milk in PBS, 108 phages per well were added and incubated for 2 h at room temperature. After 5 washings with PBS/Tween 20, 100 μl of rabbit-anti-M13 antibody (dilution 1:1000; Stratagene) was added and incubated for 2 h at room temperature. ELISA was developed with a monoclonal horseradish peroxidase-conjugated goat-anti-rabbit IgG antibody (Jackson ImmunoResearch) and the substrate 3,3′,5,5′- tetramethylbenzidine (Kierkegaard & Perry Laboratories). The A was measured at 620 nm.
Sequence Analysis.
Nucleotide sequencing was performed using the dideoxy termination method with the vector pBluescript II SK(+) and specific primers. Subsequent sequence analyses were performed with the husar sequence analysis program kindly provided by the German Cancer Research Center, Heidelberg, Germany.
Production of Soluble Anti-F(ab′)2 scFv Antibodies.
The scFv2 and scFv6 antibody genes were cloned into the vector pOPE51 (26). E. coli XL-1blueMRF′ bacteria, transformed with either the plasmid pOPE51scFv2 or the plasmid pOPE51scFv6, were propagated as previously described (27). Antibody production was induced by the addition of isopropyl-β-d-thiogalactopyranoside at a concentration of 20 μM. scFv antibodies were isolated from solubilized periplasmic inclusion bodies by immobilized metal affinity chromatography under denaturing conditions, and then refolded as previously described (26). Monomeric scFv antibodies were separated from aggregates and dimers by size-exclusion chromatography on a Superdex 75 HL26/60 column (Pharmacia) (26). The monomeric peak was concentrated using Centriplus10 (Amicon) and dialyzed against Hepes-buffered saline (10 mM Hepes/0.15 M NaCl/3.4 mM EDTA/0.001% Tween 20, pH 7.4). For ELISA analysis, the scFv antibodies were obtained from soluble periplasmic extracts and culture supernatants using the pHOG21 vector under the conditions recently described by Kipriyanov et al. (28).
ELISA Analysis of Anti-F(ab′)2 scFv Antibodies.
Microtiter plates were coated with human IgG (1.5 μg per well), F(ab′)2 (1 μg per well), Fab (1 μg per well), and Fc (0.5 μg per well) fragments (Jackson ImmunoResearch) or BSA (2.5 μg per well) as described above.
Various dilutions of scFv antibodies (in 100 μl) were added followed by the stepwise addition of anti-c-myc mouse monoclonal antibody 9E10 (100 μl, dilution 1:4000) (Cambridge Research Biochemicals), horseradish peroxidase-conjugated goat-anti-mouse IgG antibody (Jackson ImmunoResearch), and 3,3′,5,5′-tetramethylbenzidine substrate.
Surface Plasmon Resonance Analysis.
Binding properties of scFv were characterized by surface plasmon resonance using a BIAcore 2000 instrument with multichannel Integrated μ-Fluidic Cartridge (Pharmacia). Antigens diluted in 10 mM sodium acetate buffer, pH 4.5 at a concentration of 100 μg/ml F(ab′)2, 200 μg/ml Fab, 67 μg/ml IgG, and 200 μg/ml Fc were immobilized with a contact time of 7 min and a flow rate of 10 μl/min. The following resonance units (1000 resonance units correspond to a surface concentration of 1 ng/mm2) were obtained: 4194 for immobilized F(ab′)2, 1671 for Fab, 7034 for IgG, and 4050 for Fc. The negative control consisted of BSA (154 μg/ml; 3792 resonance units). One channel of each sensor chip was used after activation for injection of 10 mM sodium acetate buffer, pH 4.5, followed by deactivation with ethanolamine. This channel was used to control the nonspecific binding of scFv to the carboxymethylated dextran surface. All surface resonance plasmon analysis measurements were carried out at a flow rate of 5 μl/min in HBS at 25.5°C. After each cycle, the surface was regenerated by a single 5-ml injection of 10 mM HCl. Analyses of monomeric scFv2 and scFv6 antibodies were performed at a concentration of 6.25–400 nM. Each injected sample of recombinant antibody fragment (25 μl) was in contact with the antigen for 5 min. The dissociation was followed for 10 min. Kinetic constants were calculated using BIAevaluation 2.1 software (Pharmacia).
Competitive Inhibition of Serum IgG-Anti-F(ab′)2 Antibodies.
Microtiter plates were coated with human F(ab′)2 γ-fragments (Jackson ImmunoResearch) as described. The scFv antibodies were applied in various dilutions (1:4 to 1:64) followed by addition of the patient’s serum (dilution: 1:512). The scFv antibodies were detected as mentioned above. Horseradish peroxidase-conjugated goat-anti-human IgG antibody (Jackson ImmunoResearch) was applied for detection of serum IgG-anti-F(ab′)2 antibodies. 3,3′,5,5′-tetramethylbenzidine substrate was added, and the absorption was measured at 620 nm.
RESULTS
Generation of Antibody Gene Library.
Antibody genes were derived from the peripheral mononuclear cells of a kidney graft recipient with a high IgG-anti-F(ab′)2 antibody titer. The mRNA was extracted and reverse-transcribed into cDNA. Immunoglobulin VH as well as variable light-chain κ and λ regions were amplified by PCR with a specific set of primers (25). Subsequently, the PCR products were subcloned into the surface expression vector pSEX81.
Selection of Anti-F(ab′)2 scFv Antibody-Presenting Phages.
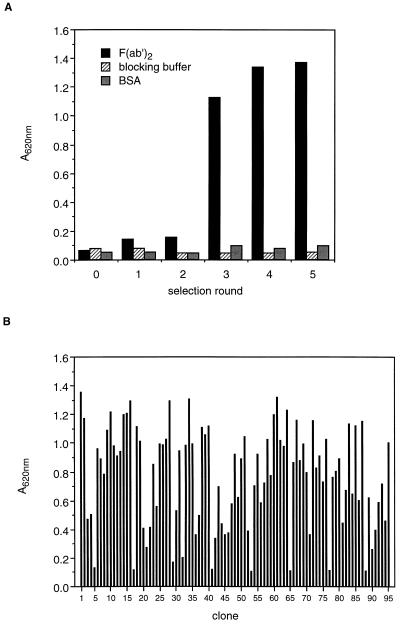
Specific phages were obtained by five consecutive selection rounds performed in F(ab′)2-coated tubes. A marked increase in the number of anti-F(ab′)2-binding phages occurred after the third round (Fig. 1A). After the fifth round, 95 anti-F(ab′)2 single clones were selected and tested for binding activity (Fig. 1B).
Figure 1.
Binding of scFv anti-F(ab′)2-expressing phages to F(ab′)2 fragments. (A) The polyclonal phage population obtained after each selection round was tested in ELISA for binding to F(ab′)2. Negative controls consisted of microtiter plates coated with blocking buffer or BSA. (B) Single clones (n = 95) with anti-F(ab′)2 activity were generated and tested in ELISA (negative controls: A620 of blocking buffer = 0.079 and that of BSA = 0.074).
VH and VL Gene Sequences.
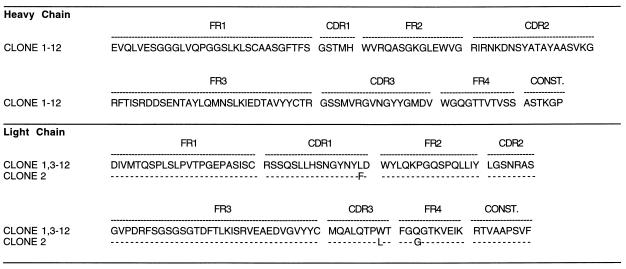
Twelve of the 95 single clones with relatively high binding activity were sequenced (Fig. 2). Eleven of these clones were found to have identical VH and VL genes. One clone (scFv2) differed by a silent point mutation in the FR1 region of the heavy chain and by three amino acid exchanges in the VL region.
Figure 2.
Sequence analysis of VH and VL chain genes of scFv anti-F(ab′)2 antibodies. Amino acid sequences are shown. FR, framework region; CDR, complementarity-determining region; CONST., constant region.
A germ-line sequence comparison (Table 1) revealed that the light chains were almost identical to the DPK-15 germ-line gene. The heavy chains were 88% homologous with the nearest germ-line gene. A comparison with mutated and unmutated Ig sequences from the European Molecular Biology Laboratory databank and the GenBank database showed that the heavy-chain genes have the highest homology to a B cell tumor gene (29) and the light-chain genes to a gene isolated from a human spleen cDNA library (30). The heavy chains belong to the VH3 gene family and the light chains to the Vκ2 gene family.
Table 1.
V gene usage of anti-F(ab′)2 antibodies
| Clone | Closest germ-line V gene | Homology, % | Gene family | Closest rearranged V gene | Homology, % | Accession no. |
|---|---|---|---|---|---|---|
| HC 1,3-12 | DP-29 | 88.7 | VH3 | Ig H chain from a B cell tumor | 89.2 | Z31394Z31394 |
| HC 2 | DP-29 | 88.3 | VH3 | Ig H chain from a B cell tumor | 89.4 | Z31394Z31394 |
| LC 1,3-12 | DPK-15 | 100 | Vκ2 | Ig κ L chain from a spleen library | 99.2 | X72452X72452 |
| LC 2 | DPK-15 | 99.3 | Vκ2 | Ig κ L chain from a spleen library | 97.5 | X72452X72452 |
Comparison of the anti-F(ab′)2 scFv antibodies’ variable heavy (H) and light (L) chains with the V BASE (Medical Research Council, Cambridge, U.K. CB2 2QH) the GenBank and EMBL databases.
Binding Properties of Soluble scFv Anti-F(ab′)2 Antibodies.
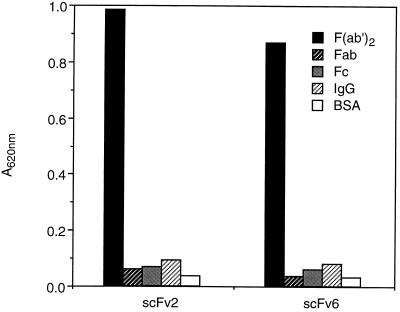
Binding of the monomeric antibody clone scFv6, representative for the 11 identical clones, and clone scFv2 to F(ab′)2, Fab, Fc, and IgG was tested in ELISA (Fig. 3). The antibodies bound to F(ab′)2 fragments, but no significant binding to Fab, Fc, or intact IgG was detected.
Figure 3.
Binding of scFv anti-F(ab′)2 antibodies to different antigens [F(ab′)2, Fab, Fc, IgG, and BSA] is analyzed by ELISA.
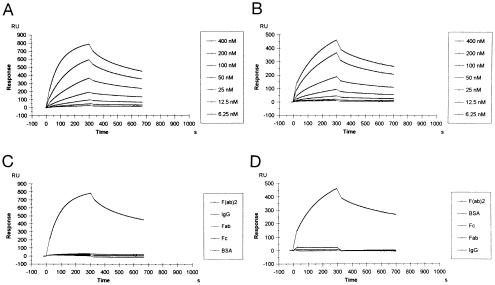
To investigate the kinetics of antigen binding, surface plasmon resonance analyses were performed. The BIAcore sensorgrams for interaction of scFv6 and scFv2 at different concentrations with immobilized F(ab′)2 are shown in Fig. 4A and B. The shape of the curves reveals a slower association of the scFv6 than of the scFv2 antibody, but similar dissociation kinetics. The antibodies bound to F(ab′)2 but no measurable interaction occurred with IgG, Fab, or Fc molecules (Fig. 4 C and D). The calculated values of the association (kon) and dissociation (koff) rate constants as well as the affinity constants (Ka) are shown in Table 2. The association kinetics of both scFv antibodies fit perfectly to the homogeneous kinetic model provided by BIAevaluation software, indicating a single-site interaction between the antibody and the immobilized antigen. As already indicated by the shape of the sensorgram curves, the scFv2 antibody binds 3 times faster to F(ab′)2 than scFv6. In contrast, the dissociation constants for both antibodies were similar. The affinity constant (Ka) of scFv2 (2.78 × 107 M−1) was about 4 times higher than that of scFv6 (0.79 × 107 M−1). These values are relatively high compared with previously described intact anti-IgG autoantibodies of rheumatoid patients (31, 32). To verify whether the cloned antibody fragments are able to inhibit the natural anti-F(ab′)2 activity, a competitive inhibition ELISA was performed. Both anti-F(ab′)2 scFv antibody clones inhibited the serum anti-F(ab′)2 activity, whereas an unrelated human scFv antibody did not (Fig. 5).
Figure 4.
Comparison of BIAcore sensorgrams for the interaction between recombinant antibody fragments and immobilized antigens. Overlap plots showing interaction of scFv2 (A) and scFv6 (B) with immobilized F(ab′)2 at different scFv concentrations. Interaction of scFv2 (C) and scFv6 (D) at 400 nM with different antigens. The association and dissociation phases of the sensorgrams are shown. The bulk effect is subtracted from the relative response. Legends in the right of each plot show the list of scFv concentrations (A and B) or immobilized antigens (C and D) in the order of decreasing resonance signals. RU, response units.
Table 2.
Kinetic parameters for binding of scFv fragments to F(ab′)2 measured by the BIAcore system
| Antibody fragment | kon, M−1·s−1/104 | koff, s−1/10−4 | Ka = kon/koff, M−1/107 | Keq, M−1/107 |
|---|---|---|---|---|
| scFv2 | 2.47 ± 0.11 | 8.86 ± 0.24 | 2.78 ± 0.80 | 2.29 ± 0.63 |
| scFv6 | 0.74 ± 0.09 | 9.26 ± 0.81 | 0.79 ± 0.07 | 0.60 ± 0.18 |
Association and dissociation rate constants were calculated using the homogeneous (monophasic) kinetic model. Affinity (equilibrium association) constants were obtained directly from the ratio kon/koff as well as from steady-state measurements (Keq).
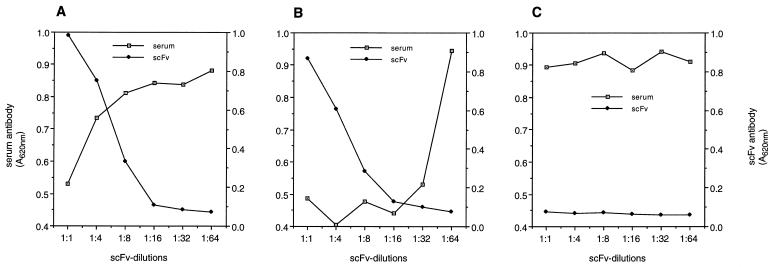
Figure 5.
Competitive inhibition of serum IgG-anti-F(ab′)2 antibodies with scFv anti-F(ab′)2 antibodies. ELISA plates coated with F(ab′)2 γ-fragments were incubated with increasing dilutions of scFv2 (A), scFv6 (B), and a human anti-digoxigenin scFv antibody (C). Serum IgG-anti-F(ab′)2 antibodies were added at a dilution of 1:512. Negative controls consisted of PBS. Maximum binding activity of scFv2 and scFv6 was A620 = 0.983 and 0.869, respectively.
DISCUSSION
The IgG-anti-F(ab′)2 antibodies analyzed in this study belong to the large family of anti-IgG antibodies generally termed rheumatoid factors (RFs). However, whereas classical RFs bind to the Fc region and are an IgM isotype (33), the antibodies described herein bind to the F(ab′)2 region and belong to the IgG isotype. The classical RF was first described in patients with rheumatoid arthritis (34). Later it became evident that the same type of antibody is associated with many other diseases, including various autoimmune conditions, viral or parasitic infections, chronic inflammations, neoplasms, and monoclonal gammopathies (33, 35–37). Increased anti-IgG antibody titers also were detected during normal immune responses to foreign antigens, in both humans and animals (38–42). Although there is circumstantial evidence for an immunoregulatory function of anti-IgG antibodies, their precise role in the physiological immune response or in the pathogenesis of diseases remains to be determined.
A series of recent clinical studies performed in our laboratory revealed striking associations of IgG-anti-F(ab′)2 antibodies with several pathological conditions (4, 6, 7). To better understand the antibodies’ function and to define their relationship with other members of the anti-IgG antibody family, we studied the genetic structure and binding characteristics of recombinant IgG-anti-F(ab′)2 antibodies.
Genes encoding anti-F(ab′)2 were obtained from the cDNA of B cells from a patient with a high anti-F(ab′)2 titer by amplifying the variable immunoglobulin gene segments by PCR (25). The primers were homologous to the 5′ end of the variable domain DNA and to the 5′ end of the IgG CH1 constant region DNA, respectively. The amplified VH genes therefore are derived from an IgG isotype similar to that analyzed in our previous clinical and experimental studies.
The VL chains of the isolated recombinant antibodies were almost identical with a germ-line gene sequence. The light chain, therefore, had not been generated by antigen-driven affinity maturation. In the case of the heavy chain, however, there was a homology of only 88% with the closest germ-line gene. Because of the germ-line gene polymorphism known to exist in humans, an exact analysis of the heavy-chain mutational pattern would have required sequencing of the patient’s own germ-line genes. A computer search for homologous mutated and unmutated immunoglobulin sequences of the GenBank and European Molecular Biology Laboratory databases revealed that the closest related VH gene is from a B cell tumor (29). Of course, this homology has no pathophysiological significance. Although we were not able to ascertain their germ-line origin, our data show that all VH amino acid sequences of the 12 selected clones are identical. Evidently, they are derived from the same paternal gene. The homology of the VL chains indicates that they also originate from one common gene.
It is an interesting question whether the genes encoding these IgG-anti-F(ab′)2 antibody fragments are related to those of IgM anti-IgG antibodies described in patients with monoclonal gammopathies or rheumatoid arthritis. The VH3 gene family, to which the antibodies described herein belong, frequently encodes paraproteins with RF activity (43–47) as well as RF of rheumatoid arthritis patients (32, 48–50) and antibodies against foreign antigens (51). Indeed the VH3 family is the largest and most heterogenous of all gene families (52). Therefore, the simple assignment of a heavy-chain gene to this family does not necessarily demonstrate a functional relationship of the encoded antibodies with other members of the same family. A comprehensive computer search revealed no homology to known RF heavy-chain genes (33).
The analysis of light-chain gene sequences led us to a similar conclusion. Most variable light-chain genes of RF belong to the Vκ3 or λ family (32, 49, 53–55). The light chains of our antibodies, however, belong to the Vκ2 family, thus differing from the classical RF. Furthermore, the antibodies demonstrate different binding patterns. Classical RFs bind to Fc fragments, whereas the antibodies analyzed in this study bind with relatively high affinity to F(ab′)2, but not to Fc fragments. The lack of relationship with other members of the anti-IgG family is not surprising, because previous clinical studies indicated functions of IgG anti-F(ab′)2 antibodies distinct from those of IgM- or IgG-anti-Fc antibodies (6).
Our previous work suggested that human serum antibodies that specifically recognize F(ab′)2 represent a group of antibodies rather than a single antibody (8, 9). One antibody of this group binds to an epitope located in the hinge region of IgG1 (8, 9). Although the recombinant antibodies described herein do not interact with this hinge-region epitope (data not shown), they reduce the binding of natural IgG anti-F(ab′)2 serum antibodies in competitive inhibition experiments. This incomplete inhibition of serum antibodies supports our observation that IgG anti-F(ab′)2 antibodies represent an antibody family, of which the antibodies described herein are members.
The binding properties of the anti-F(ab′)2 antibody fragments clearly differ from those of anti-idiotypic antibodies. Idiotypes are common to F(ab′)2, Fab, and IgG molecules. Consequently, an anti-idiotypic antibody reacts with all three molecules. The anti-F(ab′)2 antibodies studied by us, however, react strongly with F(ab′)2, but not with Fab or intact IgG. The lack of binding to intact IgG has important functional implications. Anti-IgG antibodies, including natural anti-F(ab′)2 antibodies, suppress B cells through membrane immunoglobulin-mediated signaling (5, 10–12, 42). This is possible only if the antibodies do not bind to soluble IgG of the serum, but to membrane IgG expressed on B cells.
Alternatively, the antibodies may bind selectively to antigen-bound IgG. This possibility is supported by previous experiments in animals, demonstrating that some natural anti-IgG antibodies preferentially bind to antigen-complexed IgG or selectively suppress B cells whose membrane Ig is occupied by its ligand (42, 56).
In conclusion, the present study defines the gene segment structure of the antigen-binding domain of a human IgG anti-F(ab′)2 autoantibody and describes the binding kinetics of the recombinant Fv fragments. It thereby contributes to a better understanding of the anti-F(ab′)2 antibody’s role in physiological and pathological conditions.
Acknowledgments
We thank Roland Kontermann and Ian Tomlinson from Greg Winter’s Laboratory (Medical Research Council, Cambridge, United Kingdom) for kindly providing us with germ-line V gene sequences. The oligonucleotide synthesis performed by Wolfgang Weinig and the BIAcore measurements performed by Anne Vogt (German Cancer Research Center) are gratefully acknowledged. We thank Christiane Hain and Martina Finger (Institute of Immunology) for excellent technical assistance and Monika Zewe-Welschof (German Cancer Research Center) for support and very useful advice. M.W. was supported by a grant of the Heidelberg Transplantation Program of Baden Württemberg, S.M.K. by a grant from the German Cancer Aid Foundation (Deutsche Krebshilfe), and F.B. by a grant from the German Science Council (DFG).
ABBREVIATIONS
- scFv
single-chain Fv
- VH
variable region of heavy chains
- VL
variable region of light chains
- RF
rheumatoid factors.
Footnotes
Data deposition: The VH and VL sequences of the scFv antibodies reported in this paper have been deposited in the GenBank database (accession nos. X89053–X89056X89053X89054X89055X89056 and X98749–X98750X98749X98750).
References
- 1.Bona C, Cazenave P A. Lymphocyte Regulation by Antibodies. New York: Wiley; 1981. pp. 205–213. [Google Scholar]
- 2.Bona C, Siminovitch K, Zanetti M, Theofilopoulos A. The Molecular Pathology of Autoimmune Diseases. London: Harwood; 1993. [Google Scholar]
- 3.Zaghouani H, Goldstein D, Shah H, Anderson S, Lacroix M, Dionne G, Kennedy R, Bona C. Proc Natl Acad Sci USA. 1991;88:5645–5649. doi: 10.1073/pnas.88.13.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terness P, Kirschfink M, Navolan D, Dufter C, Kohl I, Opelz G, Roelcke D. Blood. 1995;85:548–551. [PubMed] [Google Scholar]
- 5.Terness P, Marx U, Sandilands G, Roelcke D, Welschof M, Opelz G. Clin Exp Immunol. 1993;93:253–258. doi: 10.1111/j.1365-2249.1993.tb07975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Süsal C, Groth J, Oberg H H, Terness P, May G, Opelz G. Transplantation. 1992;54:632–635. doi: 10.1097/00007890-199210000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Süsal C, Daniel V, Oberg H H, Terness P, Huth-Kühne A, Zimmermann R, Opelz G. Blood. 1992;79:954–957. [PubMed] [Google Scholar]
- 8.Terness P, Kohl I, Hübener G, Battistutta R, Moroder L, Welschof M, Dufter C, Finger M, Hain C, Jung M, Opelz G. J Immunol. 1995;154:6446–6501. [PubMed] [Google Scholar]
- 9.Terness P, Navolan D, Moroder L, Siedler F, Weyher E, Kohl I, Dufter C, Welschof M, Drugarin D, Schneider F, Opelz G. J Immunol. 1996;157:4251–4257. [PubMed] [Google Scholar]
- 10.Sinclair N R, Panoskaltsis A. Contr Microbiol Immunol. 1989;11:96–123. [PubMed] [Google Scholar]
- 11.Phillips N, Parker D C. J Immunol. 1984;132:627–632. [PubMed] [Google Scholar]
- 12.Bijsterbosch M K, Klaus G G B. J Exp Med. 1985;162:1825–1836. doi: 10.1084/jem.162.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Racila E, Scheuermann R H, Picker L J, Yefenof E, Tucker T, Chang W, Marches R, Street N E, Vitetta E S, Uhr J W. J Exp Med. 1995;181:1539–1550. doi: 10.1084/jem.181.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheuermann R H, Racila E, Tucker T, Yefenof E, Street N E, Vitetta E S, Picker L J, Uhr J W. Proc Natl Acad Sci USA. 1994;91:4048–4052. doi: 10.1073/pnas.91.9.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yefenof E, Picker L J, Scheuermann R H, Tucker T F, Vitetta E S, Uhr J W. Proc Natl Acad Sci USA. 1993;90:1829–1833. doi: 10.1073/pnas.90.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zouali M, Eyguem A. Cell Immunol. 1983;76:137–147. doi: 10.1016/0008-8749(83)90356-8. [DOI] [PubMed] [Google Scholar]
- 17.Silvestris F, Bankhurst A D, Searles R P, Williams R C. Arthritis Rheum. 1984;27:1387–1396. doi: 10.1002/art.1780271209. [DOI] [PubMed] [Google Scholar]
- 18.Huston J S, Levinson D, Mudgett-Hunter M, Tai M S, Novotny J, Margolies M N, Ridge R J, Bruccoleri R E, Haber E, Crea R, Oppermann H. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoogenboom H R, Griffiths A D, Johnson K S, Chiswell D J, Hudson P, Winter G. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaghouani H, Steiman R, Nonacs R, Shah H, Gerhard W, Bona C. Science. 1993;259:224–227. doi: 10.1126/science.7678469. [DOI] [PubMed] [Google Scholar]
- 21.Kurucz I, Jost C R, George A J, Andrew S M, Segal D M. Proc Natl Acad Sci USA. 1993;90:3830–3834. doi: 10.1073/pnas.90.9.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen P S, Stryhn A, Hansen B E, Fugger L, Engberg J, Buus S. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dübel S, Breitling F, Fuchs P, Braunagel M, Klewinghaus I, Little M. Gene. 1993;128:97–101. doi: 10.1016/0378-1119(93)90159-z. [DOI] [PubMed] [Google Scholar]
- 24.Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. Gene. 1991;104:147–153. doi: 10.1016/0378-1119(91)90244-6. [DOI] [PubMed] [Google Scholar]
- 25.Welschof M, Terness P, Kolbinger F, Zewe M, Dübel S, Dörsam H, Hain C, Finger M, Jung M, Moldenhauer G, Hayashi N, Little M, Opelz G. J Immunol Methods. 1995;179:203–214. doi: 10.1016/0022-1759(94)00286-6. [DOI] [PubMed] [Google Scholar]
- 26.Kipriyanov S M, Dübel S, Breitling F, Kontermann R E, Little M. Mol Immunol. 1994;31:1047–1058. doi: 10.1016/0161-5890(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 27.Dübel S, Breitling F, Klewinghaus I, Little M. Cell Biophys. 1992;21:69–79. doi: 10.1007/BF02789479. [DOI] [PubMed] [Google Scholar]
- 28.Kipriyanov S M, Kupriyanova O A, Little M, Moldenhauer G. J Immunol Methods. 1996;196:51–62. doi: 10.1016/0022-1759(96)00115-9. [DOI] [PubMed] [Google Scholar]
- 29.Hawkins R E, Zhu D, Ovecka M, Winter G, Hamblin T J, Long A, Stevenson F K. Blood. 1994;83:3279–3288. [PubMed] [Google Scholar]
- 30.Klein R, Jaenichen R, Zachau H G. Eur J Immunol. 1993;23:3248–3271. doi: 10.1002/eji.1830231231. [DOI] [PubMed] [Google Scholar]
- 31.Olee T, Lu E W, Huang D F, Soto-Gil R W, Deftos M, Kozin F, Carson D A, Chen P P. J Exp Med. 1992;175:831–842. doi: 10.1084/jem.175.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantovani L, Wilder R L, Casali P. J Immunol. 1993;151:473–488. [PMC free article] [PubMed] [Google Scholar]
- 33.Carson D A, Chen P P, Fox R I, Kipps T J, Jirik F, Goldfien R D, Silverman G, Radoux V, Fong S. Annu Rev Immunol. 1987;5:109–126. doi: 10.1146/annurev.iy.05.040187.000545. [DOI] [PubMed] [Google Scholar]
- 34.Rose H M, Ragan C, Pearce E, Lipman M O. Proc Soc Exp Biol Med. 1949;68:1–5. doi: 10.3181/00379727-68-16375. [DOI] [PubMed] [Google Scholar]
- 35.Williams R C, Kunkel H G. J Clin Invest. 1962;41:666–672. doi: 10.1172/JCI104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levo Y, Gorevic P P, Kassab H J, Zucker-Franklin D, Franklin E C. N Engl J Med. 1977;296:1501–1504. doi: 10.1056/NEJM197706302962605. [DOI] [PubMed] [Google Scholar]
- 37.Carson D A, Bayer A S, Eisenberg R A, Lawrence S, Theofilopoulos A. Clin Exp Immunol. 1978;31:100–103. [PMC free article] [PubMed] [Google Scholar]
- 38.Bokisch V A, Bernstein D, Krause R M. J Exp Med. 1972;136:799–815. doi: 10.1084/jem.136.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasu H, Chia D S, Knutson D W, Barnett E V. Clin Exp Immunol. 1980;42:378–386. [PMC free article] [PubMed] [Google Scholar]
- 40.Welch M J, Fong S, Vaughan J, Carson D. Clin Exp Immunol. 1983;51:299–304. [PMC free article] [PubMed] [Google Scholar]
- 41.Coulie P G, van Snick J. J Exp Med. 1985;161:88–97. doi: 10.1084/jem.161.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terness P, Opelz G. Transplantation. 1992;54:88–91. doi: 10.1097/00007890-199207000-00016. [DOI] [PubMed] [Google Scholar]
- 43.Capra J D, Kehoe J M. Proc Natl Acad Sci USA. 1974;71:4032–4036. doi: 10.1073/pnas.71.10.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews D W, Capra J D. Proc Natl Acad Sci USA. 1981;78:3799–3803. doi: 10.1073/pnas.78.6.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mageed R A, Dearlove M, Goodall D M, Jefferis R. Rheumatol Int. 1986;6:179–183. doi: 10.1007/BF00541285. [DOI] [PubMed] [Google Scholar]
- 46.Newkirk M M, Mageed R A, Jefferis R, Chen P P, Capra J D. J Exp Med. 1987;166:550–564. doi: 10.1084/jem.166.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman G J, Goni F, Fernendez J, Chen P P, Frangione B, Carson D A. J Exp Med. 1988;168:2361–2366. doi: 10.1084/jem.168.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natvig J B, Thompson K, Randen I, Steinitz M, Taussig M, Beale D, Barker P, Sletten K, Waalen K, Forre O. Scand J Immunol. 1988;75:127–132. doi: 10.3109/03009748809096753. [DOI] [PubMed] [Google Scholar]
- 49.Pascual V, Randen I, Thompson K, Sioud M, Forre O, Natvig J, Capra J D. J Clin Invest. 1990;86:1320–1328. doi: 10.1172/JCI114841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harindranath N, Goldfarb I S, Ikematsu H, Burastero S E, Wilder R L, Notkins A L, Casali P. Int Immunol. 1991;3:865–875. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabat, E. A., Wu, T. T., Perry, H. M., Gottesman, K. S. & Foeller, C. (1991) Sequences of Proteins of Immunological Interest, NIH Publ. No. 91–3242. (Natl. Inst. Health, Bethesda), 5th Ed.
- 52.Berman J E, Mellis S J, Pollock R, Smith C L, Suh H, Heinke B, Kowal C, Surti U, Chess L, Cantor R C, Alt F W. EMBO J. 1988;3:727–738. doi: 10.1002/j.1460-2075.1988.tb02869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radoux V, Chen P P, Sorge J A, Carson D A. J Exp Med. 1986;164:2119–2124. doi: 10.1084/jem.164.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrohenloher R E, Koopman W J. Arthritis Rheum. 1986;29:263–268. doi: 10.1002/art.1780291003. [DOI] [PubMed] [Google Scholar]
- 55.Chen P P, Robbins D L, Jirik F R, Kipps T J, Carson D A. J Exp Med. 1987;166:1900–1905. doi: 10.1084/jem.166.6.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fazekas G, Rajnavölgyi E, Kurucz I, Sintar E, Kiss K, Laszlo G, Gergely J. Eur J Immunol. 1990;20:2719–2729. doi: 10.1002/eji.1830201229. [DOI] [PubMed] [Google Scholar]