Abstract
Antibodies to double-stranded DNA are pathognomonic of systemic lupus erythematosus and deposit in the kidneys of lupus patients to cause glomerulonephritis. Recent data suggest that a significant proportion of anti-DNA antibodies may cross-react with renal antigens and be sequestered in the kidney by virtue of this cross-reactivity. If this is true, antigenic competition for pathogenic antibodies might prevent their deposition in kidneys and the ensuing tissue damage. To generate surrogate antigens that could be used for this purpose, we have used peptide display phage libraries to identify peptides that react with R4A, a pathogenic mouse monoclonal anti-DNA antibody that deposits in glomeruli. We have demonstrated that the peptides bind in or near the double-stranded DNA binding site. Furthermore, the peptides are bound preferentially by the R4A antibody as compared with two closely related antibodies derived from it, one of which deposits in renal tubules and one of which displays no renal pathogenicity. Administration of one of these peptides in a soluble form protects mice from renal deposition of the R4A anti-DNA antibody in vivo. This represents a new therapeutic approach in systemic lupus erythematosus that focuses on protecting target organs from antibody mediated injury.
Keywords: systemic lupus erythematosus, peptide library, cross-reactivity, lupus nephritis
Anti-double-stranded DNA (dsDNA) antibodies are diagnostic of systemic lupus erythematosus, and serum titers correlate with disease activity in both humans and mice (1). The isolation of anti-DNA antibodies from the kidneys of lupus patients first led to the belief that these autoantibodies were involved in renal pathogenesis (2). More recently, the demonstration that a transgene-encoded, secreted form of an anti-DNA antibody induces lupus-like glomerulonephritis in nonautoimmune mice has unequivocally shown that anti-DNA antibodies can cause renal disease (3). However, there is still controversy about whether DNA elicits or is the target of the anti-DNA response, or whether it is merely a marker antigen for antibodies with more physiologically relevant antigenic cross-reactivities (4). While it is clear that complexes of nucleosomes and anti-DNA antibody can deposit in the kidney, many anti-DNA antibodies have been demonstrated to cross-react with some renal tissue antigens and cause damage by virtue of that cross-reactivity (5–8).
The observation that anti-DNA antibodies bind directly to renal tissue makes it possible to consider abrogating renal binding by targeting non-tissue-bound antigens to anti-DNA antibodies. Peptides are small molecules that can occupy an antibody combining site and could compete for binding of anti-DNA antibody to tissue antigens. The development of peptide display phage libraries has expedited the identification of peptides that bind to particular monoclonal antibodies (9–11). Whether antibodies bind protein antigens or non-protein antigens, such as carbohydrates, a peptide library can provide a surrogate antigen or mimotope that will inhibit binding to the original antigen (10, 12, 13).
To see if this is true for anti-dsDNA antibodies, we have screened a peptide library with mouse monoclonal antibodies that bind dsDNA. We have previously described R4A, a mouse monoclonal IgG2b anti-dsDNA antibody that causes glomerulonephritis in nonautoimmune mice (14). We have also described two mutants of R4A generated by site-directed mutagenesis (15). One mutant, 52b3, has three amino acid substitutions in framework 3 of the H chain (arginine to glutamine at position 66, threonine to alanine at position 82b, and arginine to serine at position 83). 52b3 displays a 10-fold increase in its binding of dsDNA and a shift in tissue pathology from the predominantly glomerular deposition of the parental R4A antibody to tubular deposition. The second mutant derived from R4A is 95, which has a single amino acid substitution in the third complementarity determining region of the H chain (aspartic acid to glycine at position 95). 95 shows no detectable dsDNA binding and does not deposit in kidneys. Thus, changes in only a few amino acids result in marked changes in both affinity for dsDNA and pathogenicity (15).
The R4A and 52b3 antibodies were used to select phage from a peptide display library. Each antibody binds distinct, although related, peptides. Furthermore, a synthetic peptide representing one of these motifs blocks the deposition of the R4A antibody in vivo. Thus, peptides can help characterize the binding site of anti-DNA antibodies with differential pathogenic potential and could have utility as therapeutic reagents to protect kidneys from antibody-mediated injury.
MATERIALS AND METHODS
Origin of Library.
The decapeptide library L100 expressed on the minor phage coat protein pIII was constructed using the vector fUSE5 (9) with the amino terminus NH2-ADGSGGX10GAPSGA. Details of the complete construction of the library are given elsewhere (16).
Isolation of Phage Clones Bound by Antibody.
Library screening was performed using modifications of the technique described by Scott and Smith (9). Three or four rounds of affinity selection were performed. Monoclonal antibody was incubated for 4 hr with gentle agitation with an aliquot of the phage library containing 1.2 × 1011 transducing units in 100 μl of buffer [10 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1% (wt/vol) BSA/0.1% (vol/vol) Tween 20/20 mM NaN3). Antibody concentration was 1 μM for the first round and 10 nM for the subsequent rounds of selection.
For the first round of selection, 50 μl of streptavidin-coated magnetic beads (Advanced Magnetics, Cambridge, MA) were incubated with 100 μl of 5 μM biotinylated protein G (Pierce) for 1 hr at room temperature with agitation. In subsequent rounds of selection, 10 μl of streptavidin beads and 0.05 μM of biotinylated protein G were used. To saturate the biotin binding sites on the beads, 1 μl of free biotin 10 mM (Sigma) was added and incubated for 15 min at room temperature with continued agitation. Beads were washed three times with 500 μl of buffer using an Eppendorf magnetic particle concentrator (Dynal, Oslo).
The streptavidin bead/biotinylated protein G complex was combined with the phage/antibody mixture and incubated for 30 min at room temperature with agitation. Beads were collected, and unbound phage were removed by washing. Phage were eluted with 100 μl of 0.1 M glycine·HCl (pH 2.2) for 10 min and neutralized with 10 μl of 2 M Tris Base (pH 9.0).
Eluted phage were amplified in Escherichia coli K91 kan and used as input in the subsequent round of selection. After three or four rounds, individual phage clones were randomly selected for sequencing. Phage clones were grown overnight in terrific broth (per liter of water: 13.33 g of bacto-tryptone, 26.64 g of yeast extract, and 4.4 ml of glycerol) with antibiotic selection (20 μg/ml tetracycline). The phage were precipitated with polyethylene glycol/NaCl (16.7%/3.3 M) for 4 hr on ice and pelleted by a 15-min centrifugation at 32,500 × g at 4°C. The pellet was resuspended in TBS (Tris-buffered saline/10 mM Tris·HCl, pH 7.5/150 mM NaCl). Phage DNA was prepared for sequencing by standard techniques, using serial steps of phenol and sevag [chloroform/isoamyl alcohol 24:1 (vol/vol)] extraction, followed by ethanol precipitation. Phage DNA was sequenced by the dideoxynucleotide termination method (17), using sequenase (Version 2.0; United States Biochemical) and the fUSE 35S sequencing primer (18).
ELISAs.
Binding of antibody to phage was confirmed by ELISA. Microtiter 96-well plates were coated with 50 μl of antibody at 1 μg/ml in TBS overnight at 4°C. Plates were washed, and 2.5 × 1010 purified phage in 50 μl TBS were added to each well, incubated for 2 hr at 37°C, washed, and then blocked with 150 μl of 2% BSA in TBS for 1 hr at 37°C. Plates were incubated with 50 μl of 1:4000 dilution of biotinylated sheep antibody to M13 phage (5 Prime → 3 Prime) for 1 hr at 37°C, followed by a 1:4000 dilution of alkaline phosphatase-conjugated streptavidin (Southern Biotechnology Associates) for 1 hr at 37°C. The assay was developed with alkaline phosphatase substrate tablets (p-nitrophenyl phosphate; Sigma). OD at 405 nm was read with a Titertek ELISA reader (ICN).
The ELISA assay for dsDNA binding was performed as described (15). Briefly, Immulon II microtiter plates (Dynatech) coated overnight with 100 μg/ml dsDNA were blocked and then incubated with 5 μg/ml purified monoclonal antibody for 2 hr at 37°C. The plates were washed, and alkaline phosphatase conjugated goat anti-mouse IgG2b (Southern Biotechnology Associates) was added for 1 hr at 37°C. After addition of substrate, OD was read at 405 nm.
For competition ELISAs, an additional step of incubation of the antibody with varying concentrations of peptide was performed. The combination was then transferred to a dsDNA-coated ELISA plate, and the assay was continued as described above.
Synthesis of Peptide.
The amidated, acetylated peptides ADWADWLDYP (16), DWEYS, and RHEDGDWPRV were synthesized in the Macromolecular Analysis Facility at the Albert Einstein College of Medicine. Purity of the peptides was greater than 90% as assessed by HPLC.
Assay of Anti-dsDNA Antibody Glomerular Deposition.
Glomerular deposition of the nephritogenic R4A antibody was examined by injecting purified antibody into SCID mice. R4A was obtained from cell culture supernatants or from ascites of mice injected with the cell line. Antibody was purified on a protein G column (Gamma Bind, Pharmacia). SCID mice were obtained from the breeding colony maintained at the Albert Einstein College of Medicine. Six- to eight-week-old female mice were used in the experiments. Anti-dsDNA antibodies and IgG2b levels were undetectable in these mice before injection. Individual mice were simultaneously injected i.p. with 75 μg of purified R4A and 150 μg of the appropriate peptide. The mice were killed 16 hr after the injection of antibody.
One kidney from each animal was fixed in 10% formalin and embedded in paraffin. Four-micrometer-thick sections were obtained by microtome, deparaffinized, rehydrated, blocked with 2% BSA in PBS in moist chambers, and stained for 1 hr with biotinylated goat anti-mouse IgG at a 1:800 dilution at room temperature (Vector Laboratories). The sections were then incubated for 45 min with streptavidin-alkaline phosphatase ABC reagent (Vectastain ABC kit) and developed with substrate for alkaline phosphatase (5-bromo-4-chloro-3-indoylphosphate p-toludine salt and nitroblue tetrazolium chloride substrate; GIBCO/BRL). Color development was stopped by the addition of distilled water. The sections were mounted on coverslips with cytosol mounting medium (Aqua polymount; Polysciences), sealed, and viewed with a Zeiss microscope.
RESULTS
Amino Acid Motifs Are Identified by the R4A and 52b3 Anti-dsDNA Antibodies.
The R4A antibody was reacted with a phage library containing random decapeptides to determine if peptide sequences could be recognized by an anti-DNA antibody. Three rounds of selection were performed, and selected phage were randomly chosen for sequencing after the third round. A distinct consensus motif that was present in most of the selected phage could be identified (Table 1). The dominant motif for R4A was D/E-W-D/E-Y-S/G. Interestingly, some of the R4A selected peptides had a cysteine residue near each end of the peptide, permitting the possibility that a disulfide bridge constrained the conformation of the internal sequence (19); some of these sequences also contained parts of the dominant motif (Table 1).
Table 1.
Insert sequences of R4A and 52b3 phage
| Phage | Sequence | Frequency |
|---|---|---|
| R4A selected phage | ||
| Φ1–3 | D W E Y S V W L S N | 23 |
| Φ7–6 | L Y F E D Y R C E L | 3 |
| Φ7–3 | D W D Y G A L M W A | 1 |
| Φ1–10 | Y S D W D Y S E G L | 1 |
| Φ1–2 | S T E H S E A D L W | 8 |
| Consensus | D/E W D/E Y S/G | |
| Φ1–1 | F S D C Y H S G C P | 3 |
| Φ7–21 | W C E A D Y G R C P | 23 |
| Φ7–26 | V P V C D W E L N C | 1 |
| 52b3 selected phage | ||
| ΦB11 | R H E D G D W P R V | 1 |
| ΦB25 | L L D D G F W P R V | 1 |
| ΦB23 | C G V D G R W P R W | 1 |
| ΦB24 | S L I S D E W P R W | 1 |
| ΦB56 | D G E W P R E G W S | 1 |
| ΦB22 | E D L E G E W P M R | 1 |
| ΦB1 | S L D E L D W D S M | 4 |
| Consensus | D/E G D/E W P R | |
Insert sequences of anti-dsDNA antibody-selected phage. Boldface capital letters denote amino acids in the peptide insert that are identical to the derived consensus motif. Cysteine residues are bold and underlined. Frequency denotes the number of phage clones with identical inserts. Beneath the upper consensus motif are phage clones containing peptides with internal cysteine bridges, with or without internal residues identical to the consensus motif.
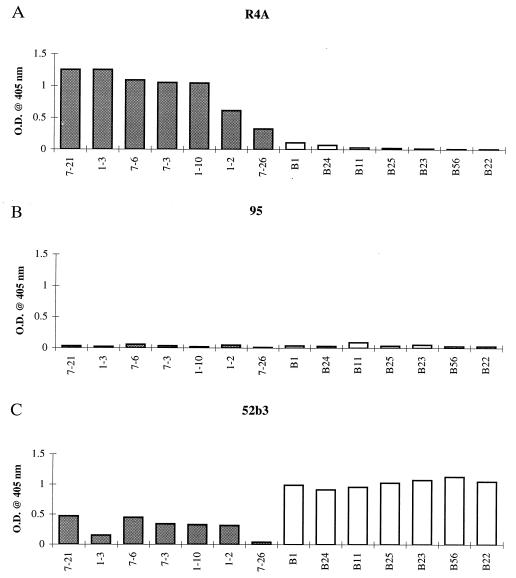
To confirm binding of R4A to these selected phage, ELISAs were performed. All selected phage were bound by the selecting antibody (Fig. 1A). To determine the specificity of these peptides for the glomerular binding R4A antibody, we screened R4A selected phage for binding by two highly related antibodies: 52b3, which has three amino acid substitutions, binds DNA, and deposits in renal tubules, and 95, which has one amino acid substitution, no longer binds DNA and displays no renal deposition (15). 95 did not bind the R4A selected phage (Fig. 1B). 52b3 did bind to R4A selected phage but at a greatly reduced level (Fig. 1C). To further evaluate the ability of peptides to distinguish the binding sites of related antibodies, we then screened the decapeptide library with 52b3. Four rounds of selection were performed before sequencing the phage inserts. A consensus motif D/E-G-D/E-W-P-R was identified (Table 1). 52b3 selected phage were bound strongly by 52b3 (Fig. 1C), while the related antibodies, R4A and 95, did not recognize 52b3 selected phage (Fig. 1 A and B).
Figure 1.
R4A, 95, and 52b3 binding by ELISA to various phage clones. Microtiter 96-well plates were coated with 1 μg/ml purified antibody, and 2.5 × 1010 phage were added to each well, followed by biotinylated antibody to M13 phage, and streptavidin-alkaline phosphatase. OD after addition of substrate was monitored at 405 nm. Shaded boxes (left) are R4A selected clones, and open boxes (right) are 52b3 selected clones. (A) R4A reactivity with R4A and 52b3 selected clones. (B) 95 reactivity with R4A and 52b3 selected clones. (C) 52b3 reactivity with R4A and 52b3 selected clones.
Peptides Are Mimetics for dsDNA.
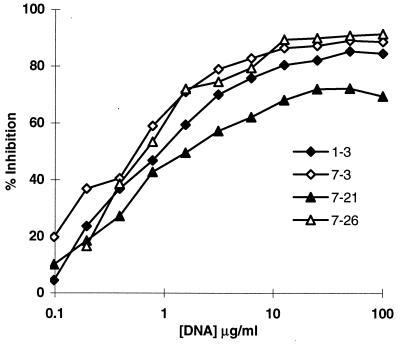
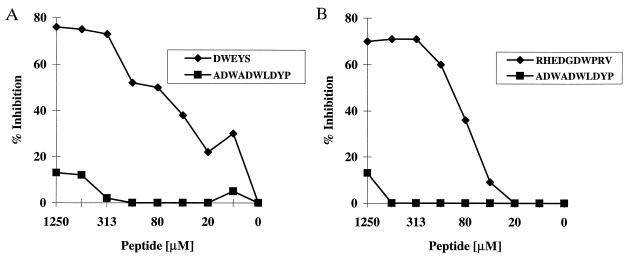
To determine whether there is a single binding site for both peptide and dsDNA, inhibition assays were performed. Calf thymus DNA inhibited the binding of R4A to R4A selected phage (Fig. 2) and the binding of 52b3 to 52b3 selected phage (data not shown). In addition, the synthetic R4A peptide DWEYS and the synthetic 52b3 peptide RHEDGDWPRV were assayed for their ability to inhibit binding of R4A and 52b3, respectively, to dsDNA. DWEYS inhibited the dsDNA binding of R4A, and RHEDGDWPRV inhibited the dsDNA binding of 52b3 (Fig. 3). We also used a larger peptide from phage 1-3 containing the DWEYS motif, and it also inhibited dsDNA binding by R4A (data not shown). The lower maximal inhibition of dsDNA binding by the 52b3 peptide suggests that dsDNA may possess two or more epitopes recognized by 52b3, and the synthetic peptide may inhibit binding to only one epitope. It is also possible that RHEDGDWPRV self-associates at high concentration to obscure the antigenic epitope, in which case increasing the peptide concentration would not result in further inhibition of dsDNA binding.
Figure 2.
Inhibition of R4A binding to selected phage by calf thymus dsDNA. 1–3, 7–3, 7–21, and 7–26 are R4A selected phage clones containing the consensus motif to which R4A showed binding by ELISA (Fig. 1A). Microtiter 96-well plates were coated with 2.5 × 1010 phage from each R4A selected phage clone. R4A at a concentration of 1 μg/ml was incubated with varying concentrations of dsDNA for 1 hr at 37°C and then transferred to the plates precoated with phage. Following a 2-hr incubation at 37°C, the plates were washed, and alkaline phosphatase linked goat anti-mouse IgG2b antibody was added for 1 hr at 37°C. OD after addition of substrate was monitored at 405 nm.
Figure 3.
Peptide inhibition of anti-dsDNA antibody binding to dsDNA. Immulon II plates were coated overnight with 100 μg/ml dsDNA. R4A and 52b3 at a final concentration of 5 μg/ml were incubated with varying concentrations of the specific (DWEYS or RHEDGDWPRV, respectively) or irrelevant (ADWADWLDYP) peptide and were transferred to the plate precoated with dsDNA. After a 2-hr incubation at 37°C, the plates were washed and alkaline phosphatase-linked goat anti-mouse IgG2b antibody was added for 1 hr at 37°C. After addition of substrate, OD was monitored at 405 nm. (A) DWEYS vs. ADWADWLDYP inhibition of R4A binding to dsDNA. (B) RHEDGDWPRV vs. ADWADWLDYP inhibition of 52b3 binding to dsDNA.
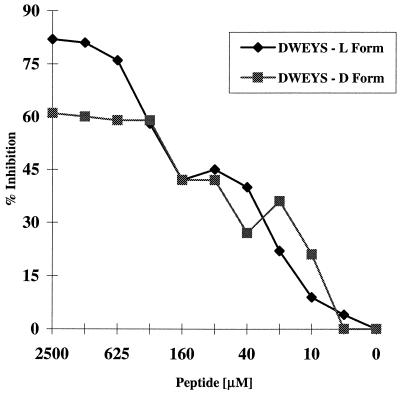
We wished to test whether peptides might block the in vivo binding of an anti-dsDNA antibody to the kidney. We chose to assay the abrogation of binding by R4A, as glomerular binding is what is most commonly found among lupus anti-DNA antibodies. Since peptides synthesized from d amino acids are more resistant to proteolytic digestion and have a longer half life in vivo, we assayed the ability of the R4A-specific peptide DWEYS made from d amino acids to inhibit binding of R4A to dsDNA. The d and l peptides of DWEYS both inhibited dsDNA binding, although the l peptide showed greater maximal inhibition (Fig. 4).
Figure 4.
DWEYS (l and d peptides) inhibition of R4A binding to dsDNA. Immulon II plates were coated overnight with 100 μg/ml salmon sperm dsDNA. R4A at a final concentration of 5 μg/ml was incubated with varying concentrations of l or d DWEYS, and transferred to the plate precoated with dsDNA. After a 2-hr incubation at 37°C, the plates were washed, and alkaline phosphatase-linked goat anti-mouse IgG2b antibody was added for 1 hr at 37°C. OD after addition of substrate was monitored at 405 nm.
R4A-Specific Peptide Inhibits R4A Deposition in Glomeruli.
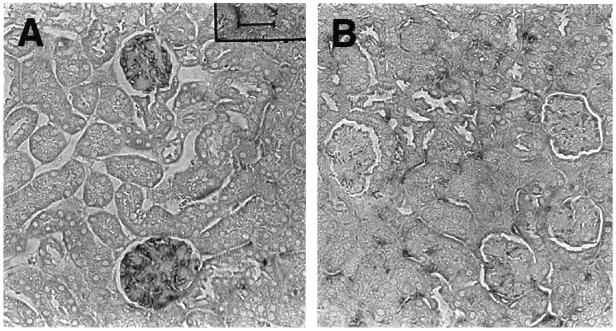
To assess whether peptides that bind to anti-dsDNA antibodies have a potential therapeutic application, we examined whether the DWEYS peptide inhibits R4A deposition in the kidney. We simultaneously injected 75 μg of purified R4A antibody and 150 μg of DWEYS as either the l or d peptide i.p. into SCID mice (five mice in each group) and assessed immunoglobulin deposition in glomeruli 16 hr later. The d peptide DWEYS markedly inhibited the deposition of R4A in the kidney, whereas the l peptide did not (Fig. 5). The SCID mice receiving the d and l DWEYS peptides displayed equivalent titers of IgG2b in their sera (data not shown), demonstrating that in each set of animals a similar amount of antibody had entered the vasculature. This observation suggests that the decreased susceptibility of the d peptide to proteolysis allowed a greater proportion of antibody to remain complexed to the antigen and prevented binding to glomerular tissue in vivo.
Figure 5.
Peptide inhibition of R4A deposition in glomeruli. R4A (75 μg) and peptide (150 μg) were injected i.p. into SCID mice. Mice were killed 16 hr later, and the kidneys immunostained for IgG deposition. (A) A representative section from a mouse injected with R4A, and the R4A-specific l peptide DWEYS. (B) A representative section from a mouse injected with R4A, and the R4A-specific d peptide DWEYS. More anti-dsDNA antibody is present in glomeruli from mice injected with the l peptide.
DISCUSSION
Current therapies for systemic lupus erythematosus are generally based on reducing autoantibody production through administration of relatively nonspecific cytotoxic agents such as cyclophosphamide or reducing inflammation with agents such as corticosteroids (20). Because both of these approaches have significant toxicity, there would be a great advantage if anti-DNA antibodies could be specifically blocked from depositing in the kidney. The approach we have used here is to introduce a small soluble monomeric surrogate antigen to inhibit the binding of an anti-DNA antibody to renal tissue. Peptide libraries provide an unprecedented opportunity to identify the cross-reactive antigens for particular antibody molecules. An analysis of the peptides bound by a given monoclonal antibody demonstrates that different peptide motifs can react with a single binding site (16). Furthermore, through use of peptide libraries to probe antibody binding sites, it has been possible to show that an anti-carbohydrate antibody can also bind a peptide mimotope (12, 13, 16, 21, 22) and that cross-reactive antigens need not have obviously homologous structures (12, 13). Here we have shown that the R4A and 52b3 monoclonal antibodies that bind dsDNA can also bind peptides. The peptide binding site appears, by competitive inhibition studies, to overlap the DNA binding site. Furthermore, the peptides bound by R4A are distinct from those bound by 52b3, even though the antibodies are highly homologous, differing by only three amino acids in the H chain variable region. The sensitivity of peptides as probes for the antigen binding site is further confirmed by the finding that 95, which differs by only a single amino acid from R4A and by four amino acids from 52b3 but has no reactivity with dsDNA, binds neither R4A nor 52b3 selected peptides. Both R4A and 52b3 bind anionic peptides mimicking DNA with its negative charge. However, even though 95 is more cationic than R4A and 52b3, having an aspartic acid to glycine substitution in the third complementarity determining region of the H chain, it does not bind these anionic peptides. Thus, the negative charge is not sufficient to explain the binding of these peptides to antibody. This is confirmed by the lack of binding of both R4A and 52b3 to the anionic peptide ADWADWLDYP.
The peptide motifs selected by both pathogenic antibodies have the D/E-X-D/E sequence. These acidic residues, while apparently important in binding to these anti-DNA antibodies, do not define the fine specificity difference that causes R4A to deposit in glomeruli and 52b3 to deposit in tubules. This apparent difference in specificity is consistent with the finding that they bind distinct peptides. It is also noteworthy that both the R4A and 52b3 consensus motifs have aromatic residues, tryptophan and tyrosine. It is tempting to speculate that the negatively charged aspartate and glutamate residues are analogous to phosphate groups, the aromatic tryptophan residues are analogous to the pentose sugar backbone, and the tyrosine and serine residues are analogous to the hydroxyl groups attached to the pentose moiety in the DNA molecule, thus explaining why these peptides are mimics of DNA. The DWEYS peptide can indeed be designated a dsDNA mimotope, as immunization of nonautoimmune BALB/c mice with peptide in complete Freund’s adjuvant leads to production of anti-dsDNA antibodies (unpublished work).
One potential use of peptide surrogates for dsDNA is to try to block binding of anti-DNA antibodies to renal tissue in vivo. Administration of R4A in vivo with a peptide surrogate for dsDNA composed of d amino acids markedly inhibited anti-dsDNA antibody deposition in the kidney of SCID mice. It has been shown that some anti-peptide antibodies will bind peptides in the d as well as the l form (23, 24), while others do not (25). As the basis for isoform cross-reactivity is not currently clearly understood, it is not possible to predict which antibodies will react with both the d and l form of a given peptide. However, both the d and the l DWEYS peptide inhibited the binding of R4A to dsDNA. It is interesting to note that while the l peptide inhibited dsDNA binding better than the d peptide in vitro, only the d peptide conferred renal protection in vivo, presumably because the d peptide was more resistant to proteolysis in the plasma and had a longer half-life. This likely allowed the d peptide of DWEYS to remain complexed with the glomerulotropic R4A antibody and prevent its deposition in the kidney.
In our in vivo study, peptide was used in a high molar ratio to antibody. Because the actual concentration of nephritogenic antibody is low in serum, a high ratio of peptide to nephritogenic antibody might be achievable in humans. In SCID mice, we could not assess whether the peptides themselves were immunogenic. This is unlikely, however, as monovalent soluble antigens are poorly immunogenic; rather, it is possible that they will be tolerogenic for B cells producing antibodies that react with the peptide.
The finding that peptide can markedly inhibit deposition of R4A in glomeruli in vivo suggests a therapeutic strategy. It will be necessary first, however, to determine the heterogeneity of the binding sites of nephritogenic anti-DNA antibodies. Although it is known that these antibodies are encoded by multiple variable region genes, it is not clear how extensively they differ in fine antigenic specificity. For example, the screening of nephritogenic antibodies with peptide libraries might reveal a limited number of specificities. While the exquisite specificity of the peptides for individual binding sites may suggest that this is unlikely, it is also possible to sequentially screen libraries with different antibodies to seek a peptide that recognizes shared conformations in different though related antibody binding sites (16). If a small panel of peptides could be identified that inhibits the binding of the anti-dsDNA antibodies in the serum of lupus-prone mice, or of patients with systemic lupus erythematosus, then it is possible that peptides can provide a significant therapeutic option.
Acknowledgments
These studies were supported by grants from the National Institutes of Health to B.D. (AI33184 and AR32371), to B.D. and M.D.S. (PO1 AI33184), and to M.D.S. (CA39838). B.D. is partially supported by the Mitrani Chair in Cardiovascular Research. M.D.S. is partially supported by a Chair for Cancer Research from the National Women’s Division of the Albert Einstein College of Medicine. P.V. is partially supported by the Philippe Foundation. C.P. is the recipient of an Arthritis Foundation Physician-Scientist Development Award.
ABBREVIATION
- dsDNA
double-stranded DNA
References
- 1.Pearson L, Lightfoot R W., Jr J Immunol. 1981;126:16–19. [PubMed] [Google Scholar]
- 2.Koffler D. Annu Rev Med. 1974;25:149–164. doi: 10.1146/annurev.me.25.020174.001053. [DOI] [PubMed] [Google Scholar]
- 3.Tsao B P, Ohnishi K, Cheroutre H, Mitchell B, Teitell M, Mixter P, Kronenberg M, Hahn B H. J Immunol. 1992;149:350–358. [PubMed] [Google Scholar]
- 4.Foster M H, Cizman B, Madaio M P. Lab Invest. 1993;69:494–507. [PubMed] [Google Scholar]
- 5.Naparstek Y, Ben-Yehuda A, Madaio M P, Bar-Tana R, Schuger L, Pizov G, Neeman Z, Cohen I R. Arthritis Rheum. 1990;33:1554–1559. doi: 10.1002/art.1780331013. [DOI] [PubMed] [Google Scholar]
- 6.Jacob L, Lety M A, Louvard D, Bach J F. J Clin Invest. 1985;75:315–317. doi: 10.1172/JCI111692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fillit H, Shibata S, Sasaki T, Spiera H, Kerr L D, Blake M. Autoimmunity. 1993;14:243–249. doi: 10.3109/08916939309077372. [DOI] [PubMed] [Google Scholar]
- 8.Madaio M P, Carlson J, Cataldo J, Ucci A, Migliorini P, Pankewycz O. J Immunol. 1987;138:2883–2889. [PubMed] [Google Scholar]
- 9.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 10.Devlin J J, Panganiban L C, Devlin P E. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 11.Cwirla S E, Peters E A, Barrett R W, Dower W J. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oldenburg K R, Loganathan D, Goldstein I J, Schultz P G, Gallop M A. Proc Natl Acad Sci USA. 1992;89:5393–5397. doi: 10.1073/pnas.89.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott J K, Loganathan D, Easley R B, Gong X, Goldstein I J. Proc Natl Acad Sci USA. 1992;89:5398–5402. doi: 10.1073/pnas.89.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shefner R, Kleiner G, Turken A, Papazian L, Diamond B. J Exp Med. 1991;173:287–296. doi: 10.1084/jem.173.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz J B, Limpanasithikul W, Diamond B. J Exp Med. 1994;180:925–932. doi: 10.1084/jem.180.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valadon P, Nussbaum G, Boyd L F, Margulies D H, Scharff M D. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith G P, Scott J K. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 19.O’Neil K T, Hoess R H, Jackson S A, Ramachandran N S, Mousa S A, DeGrado W F. Proteins. 1992;14:509–515. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 20.Fox D A, McCune W J. Rheum Dis Clin N Am. 1994;20:265–299. [PubMed] [Google Scholar]
- 21.Westernik M A, Giardina P C, Apicella M A, Kieber-Emmons T. Proc Natl Acad Sci USA. 1995;92:4021–4025. doi: 10.1073/pnas.92.9.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoess R, Brinkmann U, Handel T, Pastan I. Gene. 1993;128:43–49. doi: 10.1016/0378-1119(93)90151-r. [DOI] [PubMed] [Google Scholar]
- 23.Verdoliva A, Ruvo M, Cassani G, Fassina G. J Biol Chem. 1995;270:30422–30427. doi: 10.1074/jbc.270.51.30422. [DOI] [PubMed] [Google Scholar]
- 24.Nomizu M, Utani A, Shiraishi N, Kibbey M C, Yamada Y, Roller P P. J Biol Chem. 1992;267:4118–14121. [PubMed] [Google Scholar]
- 25.Schumacher T N, Mayr L M, Minor D L, Jr, Milhollen M A, Burgess M W, Kim P S. Science. 1996;271:1854–1857. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]