Abstract
Sugars repress α-amylase expression in germinating embryos and cell cultures of rice (Oryza sativa) through a sugar response complex (SRC) in α-amylase gene promoters and its interacting transcription factor MYBS1. The Snf1 protein kinase is required for the derepression of glucose-repressible genes in yeast. In this study, we explored the role of the yeast Snf1 ortholog in rice, SnRK1, in sugar signaling and plant growth. Rice embryo transient expression assays indicated that SnRK1A and SnRK1B act upstream and relieve glucose repression of MYBS1 and αAmy3 SRC promoters. Both SnRK1s contain N-terminal kinase domains serving as activators and C-terminal regulatory domains as dominant negative regulators of SRC. The accumulation and activity of SnRK1A was regulated by sugars posttranscriptionally, and SnRK1A relieved glucose repression specifically through the TA box in SRC. A transgenic RNA interference approach indicated that SnRK1A is also necessary for the activation of MYBS1 and αAmy3 expression under glucose starvation. Two mutants of SnRK1s, snrk1a and snrk1b, were obtained, and the functions of both SnRK1s were further studied. Our studies demonstrated that SnRK1A is an important intermediate in the sugar signaling cascade, functioning upstream from the interaction between MYBS1 and αAmy3 SRC and playing a key role in regulating seed germination and seedling growth in rice.
INTRODUCTION
In plants, sugars not only serve as metabolic resources and structural constituents of cells but also have hormone-like regulatory activities. Sugars modulate nearly all fundamental processes throughout the entire life cycle of plants, including embryogenesis, germination, growth, development, reproduction, senescence, and responses to diseases and environmental stimuli (Smeekens, 2000; Halford and Paul, 2003). In general, sugars upregulate genes involved in biosynthesis, transport, and storage of reserves and cell growth and downregulate those associated with photosynthesis, reserve mobilization, and response to stresses (Graham, 1996; Koch, 1996; Ho et al., 2001). Studies with Arabidopsis thaliana and rice (Oryza sativa) have revealed intimate interaction between sugar and plant hormones, environmental stimuli, and metabolic signaling pathways (Sheen et al., 1999; Gibson, 2000; Smeekens, 2000; Coruzzi and Zhou, 2001; Gazzarrini and McCourt, 2001; Rolland et al., 2002, 2006; Hong et al., 2003; Chen et al., 2006). Despite extensive efforts in the identification of mutants defective in sugar responses (Sheen et al., 1999; Rolland et al., 2002, 2006), the identity of essential components in the sugar signaling pathway, the detailed mechanism underlying sugar-regulated transcription, and how they regulate plant growth and development remain mostly unclear.
Yeast is a convenient model for studying the mechanisms of sugar sensing and signal transduction in eukaryotic cells. In yeast, genes required for growth on carbon sources other than glucose are repressed by glucose present in the culture medium and can be derepressed when glucose is removed, a phenomenon known as glucose repression (Carlson, 1987; Gancedo, 1998). The yeast SNF1 (for sucrose nonfermenting1) kinase complex and its mammalian counterpart, the AMP-activated protein kinase (AMPK) complex, are considered to be metabolic sensors that monitor cellular glucose and/or AMP and ATP levels (Hardie et al., 1998). The structures and functions of SNF1 and AMPK are highly conserved between mammals and yeast, serving as central components of kinase cascades in the sugar signaling pathway, and are important for metabolic stress responses (Hardie et al., 1998). In yeast, SNF1 is required for the derepression of nearly all glucose-repressible genes for the utilization of alternative carbon sources, and snf1 mutant strains fail to grow on nonglucose carbon sources (Celenza and Carlson, 1984, 1986).
Both SNF1 and AMPK are Ser/Thr protein kinases and are heterotrimeric protein complexes, consisting of a catalytic activating subunit (α) and two regulatory subunits (β and γ). In yeast, a single gene encodes the α subunit (Snf1) and the γ subunit (Snf4), whereas there are three isoforms of the β subunit (Sip1, Sip2, and Gal83). Snf1 and AMPKα can be divided into two separate functional domains: an N-terminal kinase domain and a C-terminal regulatory domain (Dyck et al., 1996; Jiang and Carlson, 1996, 1997; Crute et al., 1998). In glucose-provided yeast cells, the SNF1 complex exists in an inactive autoinhibited conformation in which the Snf1 kinase domain binds to the Snf1 regulatory domain (Jiang and Carlson, 1996). In glucose-starved yeast cells, Snf4 binds to the Snf1 regulatory domain and the Snf1 kinase domain is released, leading to an active open conformation of the SNF1 complex (Jiang and Carlson, 1996). Sip1/Sip2/Gal83 acts as a scaffold protein binding to both Snf1 and Snf4, and this binding is also regulated by glucose present in the growth medium (Jiang and Carlson, 1996, 1997).
Genes encoding Snf1-related protein kinases (SnRK1s) and orthologs of other subunits of the yeast SNF1 heterotrimeric complex have been identified and characterized in several plant species (Halford and Hardie, 1998; Halford et al., 2003). Functional assays in yeast showed glucose-regulated interaction between subunits of plant SnRK1 and yeast SNF1 complexes, and yeast two-hybrid assays showed physical interactions among these subunits (Lakatos et al., 1999; Kleinow et al., 2000). Interactions among subunits of the SnRK1 heterotrimeric complex have been observed in Arabidopsis (Polge and Thomas, 2007). SnRK1 homologs from various plant species can complement the yeast snf1 mutant phenotype (Alderson et al., 1991; Muranaka et al., 1994; Takano et al., 1998; Bhalerao et al., 1999; Lovas et al., 2003). These studies suggest that similar structural, functional, and regulatory interactions among subunits in the SnRK1 complex to those in yeast might exist in plants.
The yeast Snf1 contains a conserved Thr residue (Thr-210) in its activation loop that is essential for Snf1 activity in vitro and in vivo (Estruch et al., 1992). Glucose limitation leads to the rapid phosphorylation of Snf1 on the activation loop Thr-210 by three upstream kinases, Pak1/Sak1, Tos3, and Elm1 (Hong et al., 2003), and the activated SNF1 protein kinase in turn phosphorylates and regulates downstream transcription factors. Similar upstream protein kinases, LKB1 and CaMK2, which regulate AMPK kinase activity, have been identified in mammals (Hardie and Sakamoto, 2006). The mammalian kinase LKB1 can function heterologously in yeast as well as phosphorylate and activate SnRK1 in vitro (Hong et al., 2003, 2005), indicating some similarity in the regulation of SnRK1 activity in plants. Recently, upstream kinases of SnRK1 were also identified in plants. Two Arabidopsis kinases, At SnAk1 and At SnAk2, were found to complement the yeast sak1 tos3 elm1 triple mutant and phosphorylate the Thr residue in the T-loop of SnRK1 (Shen and Hanley-Bowdoin, 2006; Hey et al., 2007). Plant SnRK1s, yeast SNF1, and mammalian AMPKs appear to be activated by different mechanisms: SnRK1s and SNF1 by sugars, and AMPKs by AMP (Halford et al., 2003). The exact nature of the signal regulating SnRK1 expression or activation in plants is not known.
In addition to SnRK1, plants contain two other SnRK subfamilies, SnRK2 and SnRK3 (Halford et al., 2003). Members of the SnRK2 and SnRK3 families have been shown to be involved in osmotic stress and abscisic acid signaling (Boudsocq et al., 2004; Kobayashi et al., 2004). However, the SnRK1 protein family is most closely related to Snf1 and AMPKα (Halford et al., 2003). Compared with the information available for yeast and mammals, little is known about the physiological role of SnRK1 in plants. SnRK1s are proposed to be directly or indirectly involved in the control of carbohydrate metabolism, starch biosynthesis, fertility, organogenesis, senescence, stress responses, and interactions with pathogens (Polge and Thomas, 2007). A potato (Solanum tuberosum) SnRK1 was shown to be involved in starch accumulation (Purcell et al., 1998; Halford et al., 2003), and SnRK1 silencing caused abnormal pollen development and male sterility in transgenic barley (Hordeum vulgare) (Zhang et al., 2001). These studies suggest that plant SnRK1 may play roles in the regulation of global metabolism in plants; however, the detailed mechanism by which SnRK1 regulates its downstream target gene expression, and the precise physiological functions of SnRK1 in plant growth and development, remain elusive.
In cereals, α-amylases are essential enzymes for the hydrolysis of starch to sugars to support germination and seedling development, with the expression of α-amylase genes activated by sugar depletion in embryos and induced by gibberellins in endosperms (Yu et al., 1992, 1996; Perata et al., 1997; Chen et al., 2006). The expression of α-amylase in cultured rice suspension cells and rice and barley embryos is repressed by almost all metabolizable sugars (Yu et al., 1991; Perata et al., 1997; Umemura et al., 1998). αAmy3 and αAmy8 constitute two major rice α-amylases expressed under sugar-depleted conditions and are important for the catabolism of starch as a carbon source when sugar levels become too low to sustain respiration and growth in rice suspension cells and germinating embryos (Yu et al., 1996; Yu, 1999; Chen et al., 2006). Sugar repression of α-amylase gene expression involves the control of transcription rate and mRNA stability (Chan et al., 1994; Sheu et al., 1994, 1996; Chan and Yu, 1998a, 1998b), and common regulatory elements in αAmy3 and αAmy8 sugar response complexes (SRCs) have been identified (Hwang et al., 1998; Lu et al., 1998; Toyofuku et al., 1998; Chen et al., 2006). The 100-bp αAmy3 SRC contains three essential motifs, the GC box, the G box, and the TA box, for high-level promoter activity under sugar-depleted conditions (Lu et al., 1998). Transcription factor MYBS1, which contains a single DNA binding repeat, binds specifically to the TA box both in vivo and in vitro and functions as a transcriptional activator of the αAmy3 SRC under sugar-depleted conditions (Lu et al., 2002). A previous study with wheat (Triticum aestivum) embryo transient expression assays demonstrated that glucose repression of a wheat α-amylase gene (α-Amy2) promoter could be relieved by an SnRK1 (Laurie et al., 2003).
In this study, the physiological role of SnRK1 in rice was further studied. We showed that SnRK1s are structurally and functionally analogous to their yeast Snf1 ortholog. By both gain- and loss-of-function analyses, and investigations of mutants with two SnRK1 genes tagged by T-DNA, we showed that SnRK1A acts upstream and plays a central role in the sugar signaling pathway regulating both MYBS1 and αAmy3 expression in rice. Additionally, SnRK1A regulates the germination and seedling growth of rice.
RESULTS
SnRK1A Expression in Rice Is Regulated by Sugars at the Posttranscriptional Level
SnRK1A/OSK1 and SnRK1B/OSK24 were previously identified in rice (Takano et al., 1998), but their functions were not clear. The deduced amino acid sequences encoded by these two cDNAs are similar to those of yeast Snf1 in their N-terminal halves (being 67% identical in the protein kinase domain but lacking the His track near the N terminus of yeast Snf1), whereas parts of the C-terminal half have no significant homology. The amino acid sequence identity between SnRK1A/OSK1 and SnRK1B/OSK24 is 74% (Takano et al., 1998). To elucidate the role of SnRK1 in sugar signaling, the full-length cDNAs of rice SnRK1A and SnRK1B were isolated (see Supplemental Figure 1 online) and subcloned into a yeast expression vector. Like SnRK1s from other plant species, the rice SnRK1s were able to complement the yeast snf1 mutant for the utilization of sucrose as a carbon source (see Supplemental Figure 2 online), indicating that rice SnRK1s possess the same function as yeast Snf1 for the expression of glucose-repressible genes.
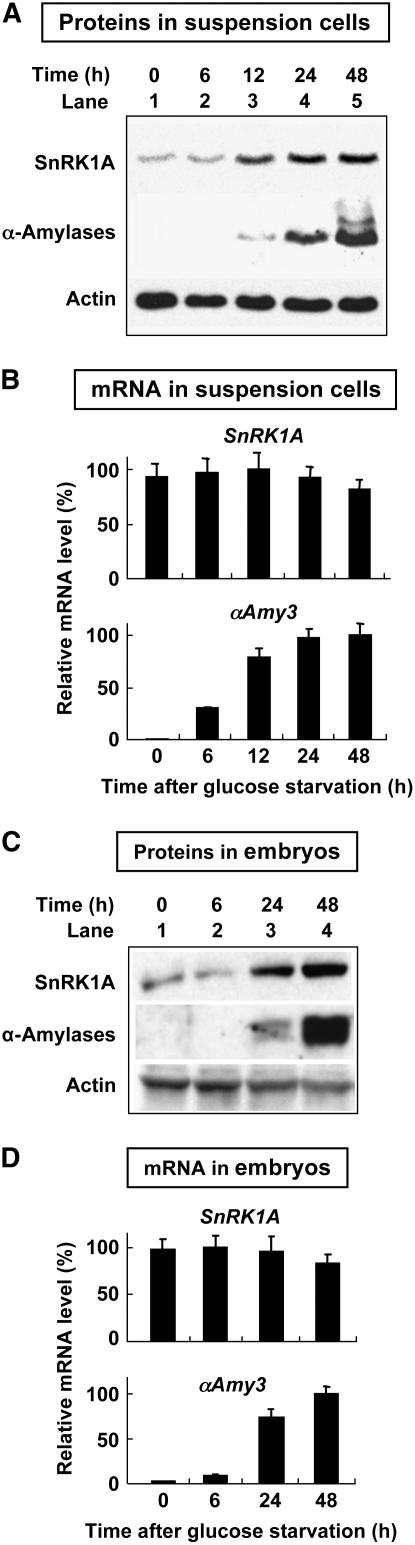
We generated polyclonal antibodies against two SnRK1s using two polypeptides specific to each protein (see Supplemental Figure 1 online). However, only the anti-SnRK1A polyclonal antibodies were obtained. We first determined the accumulation profile of SnRK1A in rice cultured suspension cells and embryos, in which sugar regulation of α-amylase expression is easily detected (Yu et al., 1991, 1996; Perata et al., 1997; Umemura et al., 1998). Rice suspension cells were cultured in 100 mM glucose-containing (+G) medium for 24 h and shifted to glucose-free (–G) medium for various lengths of time. Total soluble proteins of these cells were subjected to gel blot analyses using antibodies against SnRK1A and α-amylase. A low level of SnRK1A was detected, while no α-amylase was detected, within 6 h after cells were shifted from +G to –G medium (Figure 1A, lanes 1 and 2). Levels of both SnRK1A and α-amylase started to increase at 12 h after glucose starvation (Figure 1A, lanes 3 to 5). Rice embryos were also shifted from +G to –G medium for various lengths of time. A low level of SnRK1A was detectable within 6 h after embryos were shifted from +G to –G medium (Figure 1C, lanes 1 and 2), and both SnRK1A and α-amylase increased at 24 h after glucose starvation (Figure 1C, lanes 3 and 4).
Figure 1.
SnRK1A Expression in Rice Is Regulated by Sugars at the Posttranscriptional Level.
(A) and (C) Rice suspension cells (A) and rice embryos (C) prepared for use in the transient expression assay were incubated in medium containing 100 mM glucose for 24 h and transferred to medium lacking glucose for 0 to 48 h. Total cellular proteins were extracted and subjected to gel blot analysis using antibodies against SnRK1A, α-amylase, and actin (protein-loading control).
(B) and (D) Total RNAs were extracted from suspension cells (B) and rice embryos (D) as prepared above and subjected to quantitative RT-PCR analyses for the expression of SnRK1A (using primers 1A11 and 1A4) and αAmy3 (using primers 3RT25A and 3RT-R). RNA levels were quantified and normalized to the level of 18S rRNA. The highest mRNA level was assigned a value of 100, and mRNA levels of other samples were calculated relative to this value. Error bars indicate the se for three replicate experiments.
mRNA accumulation profiles of SnRK1A in rice suspension cells and embryos were also determined. Rice suspension cells and embryos were starved of glucose for various lengths of time, and their total RNAs were subjected to quantitative (real-time) RT-PCR analyses. Accumulation of SnRK1A mRNA in two tissues was maintained at similar levels, whereas that of αAmy3 mRNA increased with time, within 48 h after glucose starvation (Figures 1B and 1D).
Together, the studies described above suggest that although the expression of both α-amylase and SnRK1A in rice is coordinately regulated by glucose, α-amylase is regulated at the transcriptional level for mRNA abundance and SnRK1A is regulated at the posttranscriptional level for protein abundance.
SnRK1 protein kinase activity could be determined by phosphorylation of the SAMS peptide (see Supplemental Figure 3A online), a peptide based on the major AMPK phosphorylation site in rat acetyl-CoA carboxylase, which is a relatively specific substrate for AMPK and SNF1 (Hardie et al., 1998; Halford et al., 2003). To determine whether the endogenous SnRK1 protein kinase activity increases under sugar starvation, total soluble proteins were extracted from rice suspension cells and embryos after incubation in +G or –G medium for 24 h, and SnRK1 protein kinase activity was determined using SAMS peptide as the substrate. The SnRK1 protein kinase activity in –G medium was approximately twofold that in +G medium; additionally, in –G medium, the activity was significantly higher in embryos than in suspension cells (see Supplemental Figures 3B and 3C online).
Overexpression of SnRK1A Relieves the Sugar Repression of SRC
Previously, we had shown that αAmy3 SRC is repressed by glucose (Lu et al., 2002; Lee et al., 2003). The role of SnRK1A in the sugar regulation of αAmy3 was further explored in this study. First, to determine whether SnRK1A is capable of relieving the sugar repression of αAmy3 SRC, a rice embryo transient expression assay was performed, using SnRK1A cDNA fused downstream of the maize ubiquitin gene (Ubi) promoter as an effector and αAmy3 SRC fused upstream of a cauliflower mosaic virus (CaMV) 35S minimal promoter (−46 bp upstream of the transcription start site)–Luc construct (35S mp-Luc) as a reporter (see Supplemental Figure 4A online). Dose-response experiments indicated that upon cotransfection of the effector plasmid with the reporter plasmid at a molar ratio of 1:2, SnRK1A relieved the glucose repression of αAmy3 promoter activity (see Supplemental Figure 4B online).
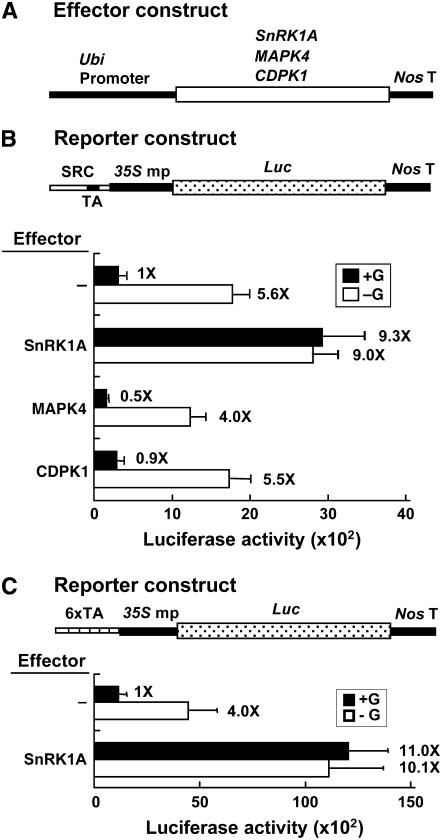
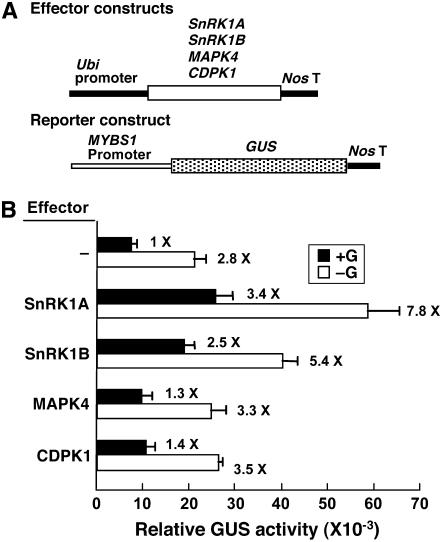
We previously also isolated two rice genes encoding a calcium-dependent protein kinase (CDPK1) and a mitogen-activated protein kinase (MAPK4). Expression of the two protein kinases is induced by sugar starvation (Fu et al., 2002; S.-L. Ho and S.-M. Yu, unpublished results). To determine whether the two protein kinases play any roles in sugar signaling, CDPK1 and MAPK4 cDNAs were fused downstream of the Ubi promoter as effectors (Figure 2A) and the SRC-35S mp-Luc construct was used as a reporter. Transient expression assays in rice embryos showed that only SnRK1A, but not CDPK1 and MAPK4, relieved the glucose repression of SRC (Figure 2B), indicating a specific action of SnRK1A on αAmy3 SRC.
Figure 2.
Overexpression of SnRK1A Relieves the Sugar Repression of SRC.
Rice embryos were cotransfected with effector and reporter plasmids and incubated with 100 mM glucose (+G) or without glucose (−G), and their luciferase activities were assayed. The luciferase activity in rice embryos bombarded with the reporter construct only and in the +G condition was assigned a value of 1×, and other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
(A) The effector contains the Ubi-SnRK1A, Ubi-MAPK4, or Ubi-CDPK1 fusion gene.
(B) Luciferase activity of a reporter containing the SRC-35S mp-Luc fusion gene. SRC contains a duplicated TA box.
(C) Luciferase activity of a reporter containing the 6xTA-35S mp-Luc fusion gene.
To determine the downstream target sequence of SnRK1A in the αAmy3 promoter, six tandem repeats of the 6-bp TA box (TATCCA) (6xTA) fused upstream of the 35S mp-Luc construct were used as a reporter and the rice embryo transient expression assay was performed. SnRK1A overexpression relieved glucose repression of the 6xTA promoter (Figure 2C). The TA box hexamer conferred a higher basal promoter activity than SRC, probably due to the presence of a low level of its binding protein MYBS1, even in the presence of glucose. These results suggest that the TA box could be a downstream target of SnRK1A.
SnRK1A Relieves the Sugar Repression of SRC through the TA Box
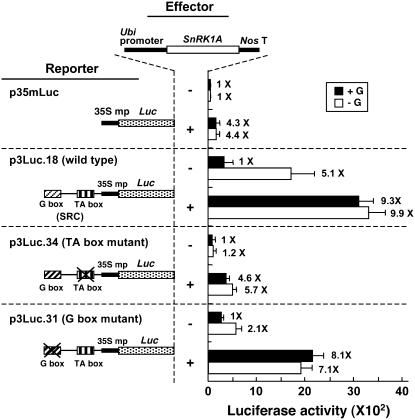
αAmy3 SRC contains a G box and a TA box that are both essential for high activity of SRC under sugar starvation (Lu et al., 1998). To determine whether SnRK1A relieves the sugar repression of SRC through both of these two boxes or only through the TA box, plasmids p3Luc.18, p3Luc.31, and p3Luc.34, containing the wild-type SRC, a mutated G box in SRC, and a mutated TA box in SRC, respectively, fused upstream of the 35S mp-Luc construct (Lu et al., 1998), were used as reporters in rice embryo transient expression assays. Plasmid p35mLuc containing 35S mp-Luc was used as a control reporter, and the Ubi-SnRK1A construct was used as the effector. SnRK1A activated the CaMV35S minimal promoter fourfold (Figure 3), regardless of the presence or absence of glucose, likely due to a general chromatin-remodeling activity of SnRK1A for many promoters, as has been reported for the yeast Snf1 (Kuchin et al., 2000; Lo et al., 2005). In the absence of SnRK1A overexpression, luciferase activities were low for all constructs, except for the wild-type SRC, which was induced by glucose starvation (Figure 3). The TA box appears to be more important than the G box for high activity of SRC and for conferring glucose starvation–induced SRC activity. Overexpression of SnRK1A relieved glucose repression of both wild-type and G box–mutated SRCs, but not of the TA box–mutated SRC. These results confirmed that the TA box is the downstream target of SnRK1A action.
Figure 3.
SnRK1A Relieves the Sugar Repression of SRC through the TA Box.
Rice embryos were cotransfected with plasmids and incubated with 100 mM glucose (+G) or without glucose (−G), and their luciferase activities were assayed. The effector contains the Ubi-SnRK1A fusion gene. The reporters contain Luc expressed under the control of 35S mp only (p35mLuc), wild-type SRC plus 35S mp (p3Luc.18), SRC with a mutated TA box plus 35S mp (p3Luc.34), or SRC with a mutated G box plus 35S mp (p3Luc.31). The luciferase activity in rice embryos bombarded with the reporter construct only and in the +G condition was assigned a value of 1×, and other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
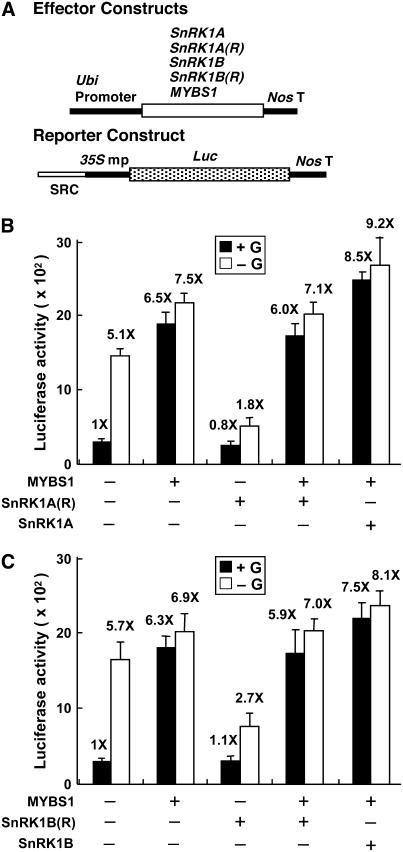
SnRK1 Contains Two Functional Domains That Regulate SRC
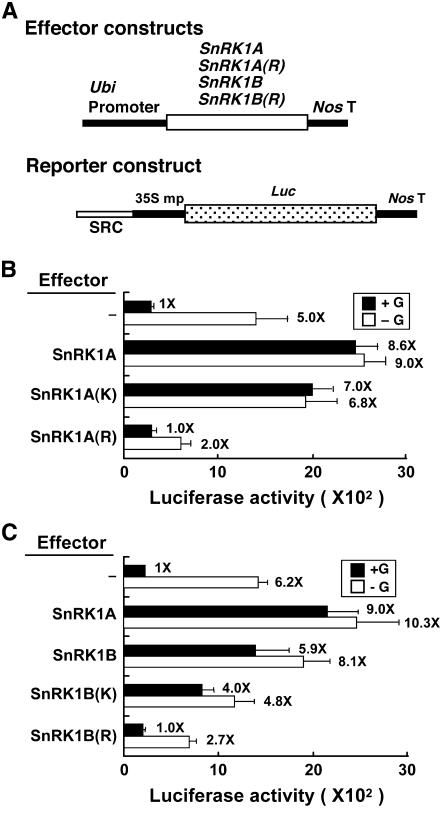
The yeast Snf1 contains two functional domains: an N-terminal protein Ser/Thr kinase domain and a C-terminal regulatory domain (Celenza and Carlson, 1989; Hardie et al., 1998). To study whether SnRK1A contains two functional domains that could regulate SRC, truncated versions of SnRK1A were generated based on information from yeast Snf1. SnRK1A(K), residues 1 to 279, contained the putative protein kinase catalytic domain of SnRK1A; SnRK1A(R), residues 280 to 503, contained the putative regulatory domain responsible for intramolecular interaction with the kinase domain and intermolecular interactions with other components of the SnRK1 complex (see Supplemental Figure 5 online). SnRK1A(K) and SnRK1A(R) were fused downstream of the Ubi promoter and used as effectors (Figure 4A) in rice embryo transient expression assays. The SRC-35S mp-Luc construct was used as a reporter. SnRK1A(K) relieved the glucose repression of SRC activity, similar to full-length SnRK1A (Figure 4B). By contrast, SnRK1A(R) not only failed to relieve the glucose repression of SRC but also repressed SRC activity in the absence of glucose (Figure 4B). SnRK1B(K), residues 1 to 276, and SnRK1B(R), residues 277 to 509, containing the kinase and regulatory domains of SnRK1B, respectively (see Supplemental Figure 5 online), were constructed similarly to SnRK1A(K) and SnRK1A(R). SnRK1B(K) and SnRK1B(R) also relieved the glucose repression of SRC activity and repressed SRC activity in the absence of glucose, respectively (Figure 4C). We were unable to detect the accumulation of full-length and truncated recombinant SnRK1A and SnRK1B, due to the low transfection efficiency of rice embryos by the particle bombardment–mediated DNA delivery system and a lack of antibodies that recognize most of these proteins. Nevertheless, the positive effect of the full-length and kinase domains and the negative effect of the regulatory domains of both SnRK1s could be observed in these studies, suggesting that these proteins were both produced and active.
Figure 4.
SnRK1 Contains Two Functional Domains That Regulate SRC.
Rice embryos were cotransfected with plasmids and incubated with 100 mM glucose (+G) or without glucose (−G), and their luciferase activities were assayed.
(A) The effector contains the Ubi promoter fused upstream of the full-length cDNA or the kinase domain (K) or regulatory domain (R) of SnRK1A and SnRK1B. The reporter contains the SRC-35S mp-Luc fusion gene.
(B) and (C) Luciferase activity in rice embryos. The luciferase activity in rice embryos bombarded with the SRC-35S mp-Luc construct only and in the +G condition was assigned a value of 1×, and other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
SnRK1 Acts Upstream of MYBS1 in Relieving the Sugar Repression of SRC
Previously, we showed that MYBS1 relieves the glucose repression of SRC activity specifically through a direct interaction with the TA box (Lu et al., 2002). To further delineate the relationship of SnRK1A and MYBS1 in the sugar signaling pathway, the rice embryo transient expression assay was performed, using constructs Ubi-MYBS1, Ubi-SnRK1A(R), and Ubi-SnRK1B(R) as effectors and the construct SRC-35S mp-Luc as a reporter (Figure 5A). MYBS1 relieved the glucose repression of SRC activity, while SnRK1A(R) repressed SRC activity in the absence of glucose (Figure 5B). Co-overexpression of MYBS1 and SnRK1A(R) activated SRC activity to a level equivalent to the overexpression of MYBS1 alone, indicating that MYBS1 acts downstream of SnRK1A(R). Co-overexpression of MYBS1 and SnRK1A slightly enhanced SRC activity. Similar results were obtained with co-overexpression of MYBS1 and SnRK1B or SnRK1B(R) (Figure 5C). The level and/or activity of MYBS1 were probably close to saturation in glucose-starved cells; therefore, co-overexpression of SnRK1 and MYBS1 did not confer an additive effect on the activation of αAmy3 SRC activity.
Figure 5.
SnRK1 Acts Upstream of MYBS1 in Relieving the Sugar Repression of SRC.
Rice embryos were cotransfected with plasmids and incubated with 100 mM glucose (+G) or without glucose (−G), and their luciferase activities were assayed. The luciferase activity in rice embryos bombarded with the SRC-35S mp-Luc construct only and in the +G condition was assigned a value of 1×, and other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
(A) The effector contains the Ubi promoter fused upstream of the SnRK1 cDNA, SnRK1(R), or MYBS1 cDNA. The reporter contains the SRC-35S mp-Luc fusion gene.
(B) MYBS1 inhibits the repression of SRC by SnRK1A(R).
(C) MYBS1 inhibits the repression of SRC by SnRK1B(R).
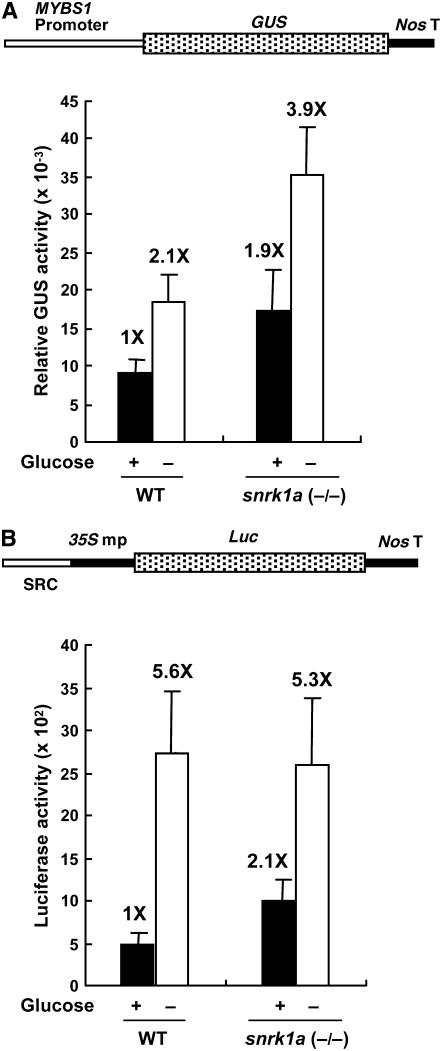
To establish the functional link between MYBS1 and SnRK1A, a 2.5-kb MYBS1 promoter was isolated from the rice genome. This MYBS1 promoter was fused with β-glucuronidase (GUS) as a reporter, constructs Ubi-SnRK1A and Ubi-SnRK1B were used as effectors, and a rice embryo transient expression assay was performed (Figure 6A). In the absence of any effector, MYBS1 promoter activity was repressed 2.8-fold by glucose (Figure 6B). Overexpression of SnRK1A or SnRK1B significantly transactivated the MYBS1 promoter; however, the glucose repression of the MYBS1 promoter remained ∼2.2- to 2.3-fold (Figure 6B). The MYBS1 promoter was also fused with Luc and used as a reporter. However, luciferase activity was too low to be reliable, probably due to the low activity of the MYBS1 promoter and the instability of luciferase. Overexpression of MAPK4 and CDPK1 did not significantly enhance MYBS1 promoter activity (Figure 6B), indicating specific action of SnRK1 on the MYBS1 promoter. These results suggest that SnRK1 acts upstream of MYBS1 in the sugar signaling pathway.
Figure 6.
SnRK1 Transactivates the MYBS1 Promoter.
Rice embryos were cotransfected with plasmids and incubated with 100 mM glucose (+G) or without glucose (−G), and their GUS activities were assayed.
(A) The effector contains the Ubi-SnRK1A, Ubi-SnRK1B, Ubi-MAPK4, or Ubi-CDPK1 fusion gene. The reporter contains the MYBS1-GUS fusion gene.
(B) GUS activity in rice embryos. The GUS activity in rice embryos bombarded with the MYBS1-GUS construct only and in the +G condition was assigned a value of 1×, and other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
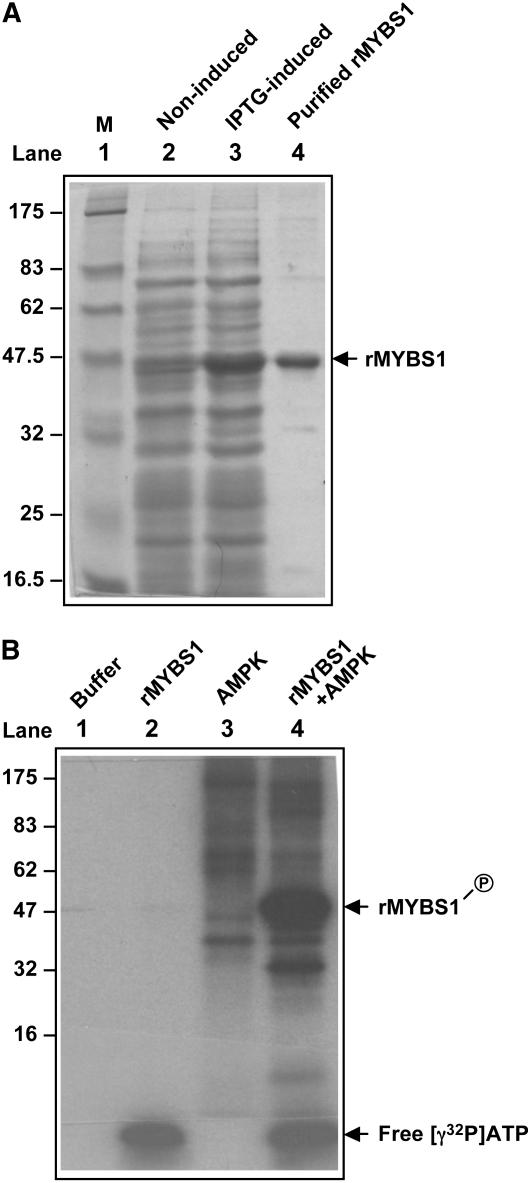
Although the overexpression of SnRK1 did not relieve the glucose repression of the MYBS1 promoter, it completely relieved the glucose repression of αAmy3 SRC. This observation raised a question regarding whether SnRK1 regulates αAmy3 expression mainly through transcriptional regulation of the MYBS1 promoter or also through the posttranslational regulation of MYBS1 activity. AMPK, SNF1, and SnRK1 phosphorylate the Ser or Thr residue in the consensus recognition motifs of their substrates, such as SAMS and AMARA peptides (see Supplemental Figure 3A online) (Hardie et al., 1998; Halford et al., 2003). Although several putative SnRK1 recognition motifs are present in the MYBS1 polypeptide, none of them completely complies with the rule of having conserved hydrophobic and basic residues at all signature positions relative to the predicted phosphorylated Ser or Thr in the consensus recognition motifs. To determine whether MYBS1 could be phosphorylated in vitro by the upstream protein kinase, recombinant MYBS1 (rMYBS1) was expressed in Escherichia coli (Figure 7A) and used as a substrate for phosphorylation by commercially available AMPK, which contains the heterotrimeric protein kinase complex. The autoradiogram of protein gel analysis revealed the incorporation of [γ-32P]ATP into the rMYBS1 (Figure 7B, lane 4), indicating that MYBS1 is a substrate of AMPK in vitro. MYBS1 has a predicted ATP/GTP binding site, which may explain why free [γ-32P]ATP was present in samples containing rMYBS1 after immunoprecipitation (Figure 7B, lanes 2 and 4). This study suggests that MYBS1 might be a phosphorylation target of SnRK1.
Figure 7.
Os MYBS1 Is a Substrate of AMPK in Vitro.
(A) Expression of rMYBS1 in E. coli. Lane 1, protein molecular mass marker (M) in kD; lane 2, total bacterial proteins from noninduced BL21 containing pET-MYBS1; lane 3, total bacterial proteins from isopropylthio-β-galactoside (IPTG)–induced BL21 containing pET-MYBS1; lane 4, rMYBS1 purified from the bacterial proteins shown in lane 3 (arrow).
(B) Phosphorylation of rMYBS1 by AMPK. Lane 1, reaction buffer only; lane 2, reaction buffer containing rMYBS1 only; lane 3, reaction buffer containing AMPK only; lane 4, reaction buffer containing both rMYBS1 and AMPK. Arrows indicate the phosphorylated rMYBS1 and free [γ-32P]ATP.
SnRK1A Is Necessary for MYBS1 and αAmy3 Expression under Sugar Starvation
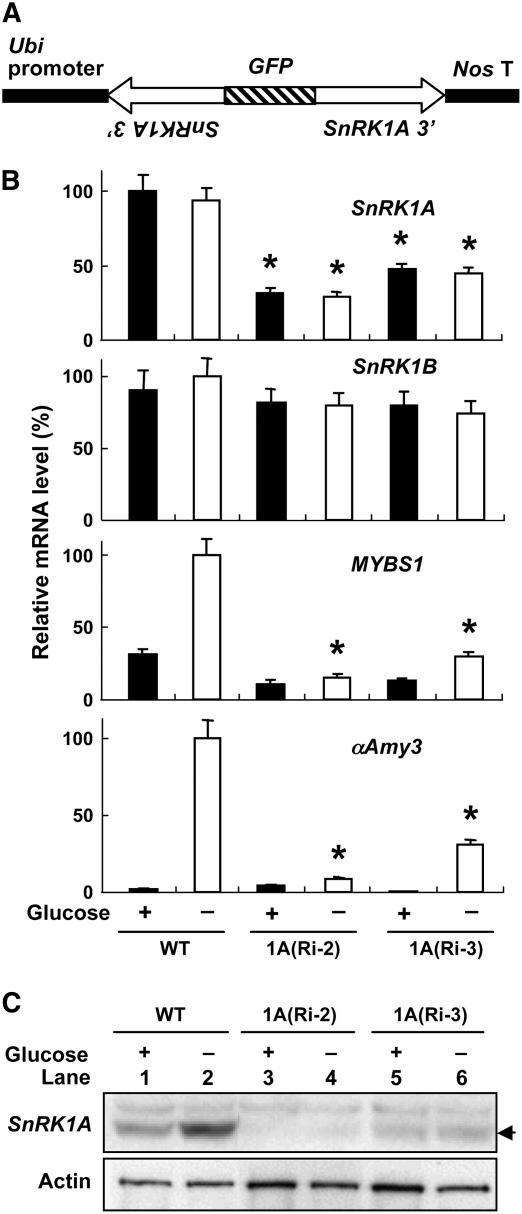
The gain-of-function studies on the role of SnRK1A in sugar signaling (described above) were performed with transient expression assays. To investigate the role of endogenous SnRK1A in sugar signaling, a loss-of-function analysis by a transgenic RNA interference approach was also employed. The 400-bp 3′ untranslated region of SnRK1A cDNA was fused in inverted repeats flanking green fluorescent protein DNA, under the control of the Ubi promoter, and used for rice transformation (Figure 8A). Calli were established from transformed embryos, and quantitative RT-PCR analyses were applied to study the expression patterns of SnRK1A and its downstream genes in response to glucose. The accumulation of SnRK1A mRNA was reduced, while that of SnRK1B mRNA was not affected, in cell lines 1A(Ri-2) and 1A(Ri-3), regardless of the presence or absence of glucose, compared with the wild type (Figure 8B). The accumulation of MYBS1 and αAmy3 mRNA was significantly reduced in two SnRK1A knockdown lines under glucose starvation.
Figure 8.
SnRK1A Is Necessary for MYBS1 and αAmy3 Expression under Sugar Starvation.
(A) SnRK1A-specific RNA interference (RNAi) construct for rice transformation.
(B) Embryonic calli of the wild type and two transformed lines, 1A(Ri-2) and 1A(Ri-3), were incubated with or without 100 mM glucose for 24 h. Total RNA was purified from suspension cells and subjected to quantitative RT-PCR analyses using primers specific for SnRK1A, SnRK1B, MYBS1, αAmy3, and Act1 (internal control), and levels were normalized to 18S rRNA. The highest mRNA level was assigned a value of 100, and the mRNA levels of other samples were calculated relative to this value. Error bars indicate the se for three replicate experiments. Asterisks highlight levels of MYBS1 and αAmy3 mRNAs for comparison.
(C) Suspension cells of lines 1A(Ri-2) and 1A(Ri-3) were cultured in 100 mM glucose-containing medium for 24 h and transferred to glucose-free medium for 24 h. Total cellular proteins were extracted and subjected to gel blot analysis using antibodies against SnRK1A and actin (protein-loading control). The arrow indicates the position of SnRK1A.
Although the accumulation of SnRK1A mRNA in lines 1A(Ri-2) and 1A(Ri-3) was considerably high, the accumulation of SnRK1A protein was barely detected in line 1A(Ri-2) and was significantly reduced in line 1A(Ri-3) (Figure 8C). The reduction of SnRK1A proteins in these two lines correlated with the degree of reduction of MYBS1 and αAmy3 mRNA accumulations under glucose starvation, as shown in Figure 8B. These studies indicate that SnRK1A is required for MYBS1 and αAmy3 expression under glucose starvation.
SnRK1A Is an Upstream Regulator of MYBS1 and αAmy3 in the Sugar Signaling Pathway
Recently, we generated a T-DNA–tagged rice mutant library containing 55,000 transgenic lines and obtained 12,000 T-DNA flanking sequences assigned to the rice genome (Hsing et al., 2007). A snrk1a rice mutant carrying SnRK1A tagged by T-DNA at the ninth exon was identified. Calli were established from embryos, and genomic DNA PCR analysis identified the wild type and snrk1a mutants heterozygous or homozygous for T-DNA inserted alleles in SnRK1A (see Supplemental Figure 4A online). Expression patterns of SnRK1A and its downstream genes in response to glucose were determined by RT-PCR analyses. The SnRK1A 5′ mRNA (the region upstream of the T-DNA inserted site) was detected in heterozygous and homozygous snrk1a mutants, with levels higher than that in the wild type; on the other hand, the SnRK1A 3′ mRNA (the region downstream of the T-DNA inserted site) was detected in the wild type and the heterozygous snrk1a mutant but not in the homozygous snrk1a mutant (see Supplemental Figure 7A online). These results indicated that SnRK1A mRNAs were transcribed but truncated by the insertion of T-DNA at the ninth exon. The loss of the SnRK1A 3′ mRNA, which encodes the regulatory domain of SnRK1A, correlated well with the partial relief of the glucose repression of MYBS1 and αAmy3 (see Supplemental Figure 7A online). The relative mRNA levels of these genes in the wild type and the snrk1a mutants were confirmed by quantitative RT-PCR analyses (see Supplemental Figure 7B online).
Based on the above observation, the SnRK1A 5′ mRNA might encode an active protein kinase. To test this possibility, the truncated SnRK1A mRNA in snrk1a was isolated by RT-PCR (see Supplemental Figure 8A online). Nucleotide sequence analysis indicated that the truncated SnRK1A was fused with the left border and an adjacent CaMV35S enhancer in a reverse orientation in T-DNA. This chimeric gene was transcribed into a mRNA (see Supplemental Figure 8B online). Anti-SnRK1A antibodies, which were generated against a polypeptide containing amino acid residues 347 to 361 of SnRK1A (see Supplemental Figure 1 online), reacted with the C-terminal truncated SnRK1A that was present only in the heterozygous mutant (in low amount) and the homozygous mutant (in high amount) compared with the wild type (see Supplemental Figure 8C online). This result indicated that the chimeric SnRK1A mRNA was indeed translated into a protein containing the SnRK1A kinase domain.
To determine whether the promoter activities of MYBS1 and αAmy3 SRC are affected in the snrk1a mutant, constructs MYBS1-GUS and SRC-35S mp-Luc were transfected into cultured suspension cells of the snrk1a homozygous line and GUS and luciferase activities were determined. Suspension cells were used in this experiment due to a shortage of snrk1a mutant seeds. Both MYBS1 and αAmy3 SRC promoter activities doubled in the presence of glucose (Figures 9A and 9B, respectively), indicating partial relief of glucose repression on these two promoters in this mutant. The extent of repression on SRC decreased from 5.6-fold to 2.5-fold, whereas that on the MYBS1 promoter remained at 2-fold.
Figure 9.
Sugar Repression of MYBS1 and αAmy3 SRC Is Partially Relieved in the Rice snrk1a Mutant.
Constructs MYBS1-GUS (A) and SRC-35S mp-Luc (B) were transfected into cultured suspension cells of a snrk1a homozygous line. Transfected cells were incubated in medium containing or lacking 100 mM glucose for 18 h, and GUS and luciferase activities were determined. The GUS or luciferase activity in the wild-type cells in the presence of glucose was assigned a value of 1X, and the other values were calculated relative to this value. Error bars indicate the se for three replicate experiments.
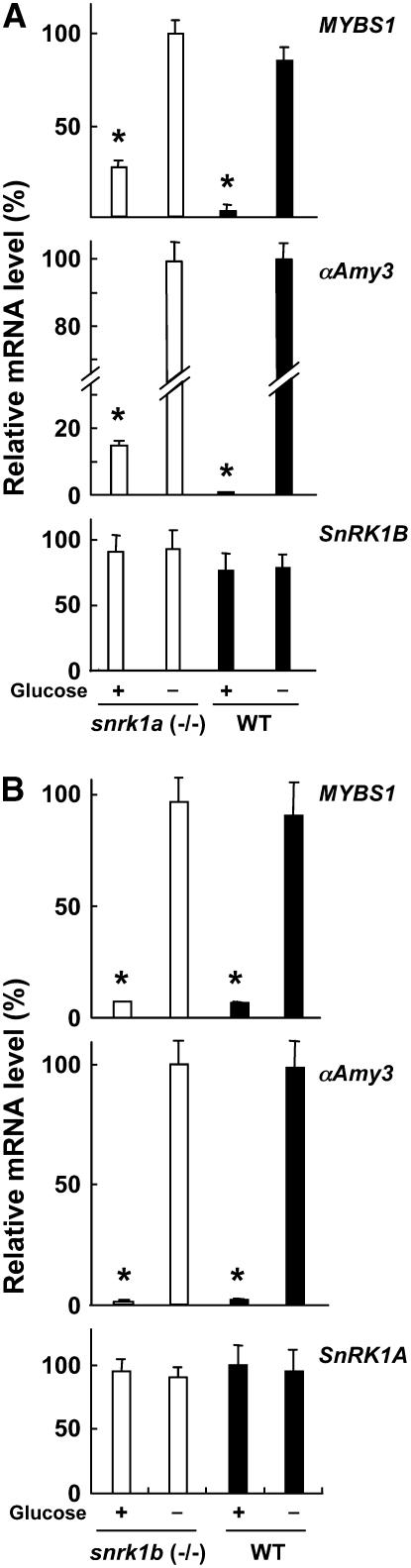
More recently, we also obtained a snrk1b rice mutant from the rice mutant library we have generated (Hsing et al., 2007). SnRK1B was tagged by T-DNA at the fourth intron in the snrk1b mutant (see Supplemental Figure 6B online). Total RNA was purified from embryonic calli of homozygous snrk1a and snrk1b mutants. The accumulation of SnRK1A, SnRK1B, MYBS1, and αAmy3 mRNAs was first determined by RT-PCR analyses (see Supplemental Figure 9 online) and then by quantitative RT-PCR analyses (Figure 10). In the snrk1a mutant, only the SnRK1A 5′ mRNA was detected (see Supplemental Figure 9B online); in the snrk1b mutant, only the 5′ and 3′ truncated SnRK1B mRNAs were detected (see Supplemental Figure 9C online). The level of the 5′ truncated SnRK1B mRNA in the snrk1b mutant was significantly higher than that in the wild type, probably due to the activation of its transcription by the adjacent CaMV35S enhancers in T-DNA. Accumulations of MYBS1 and αAmy3 mRNAs were detected in the snrk1a mutant (Figure 10A) but were barely detected in the snrk1b mutant (Figure 10B) in the presence of glucose, which suggested again that the SnRK1A kinase domain partially relieved the glucose repression of MYBS1 and αAmy3. These studies also suggest that SnRK1A, but not SnRK1B, plays a major role in the sugar signaling pathway regulating MYBS1 and αAmy3 transcription.
Figure 10.
SnRK1A Is an Upstream Regulator of MYBS1 and αAmy3 in the Sugar Signaling Pathway.
Embryonic calli from the wild type and homozygous (−/−) snrk1a (A) and snrk1b (B) mutants were cultured in 100 mM glucose-containing medium for 24 h and transferred to glucose-containing or glucose-free medium for 24 h. Total RNA was purified from cells and subjected to quantitative RT-PCR analysis using primers specific for MYBS1, αAmy3, SnRK1A, and SnRK1B, and levels were normalized to 18S rRNA. The highest mRNA level was assigned a value of 100, and mRNA levels of other samples were calculated relative to this value. Error bars indicate the se for three replicate experiments. Asterisks highlight levels of MYBS1 and αAmy3 mRNAs for comparison.
Abnormal SnRK1A Expression Retards Germination and Seedling Growth
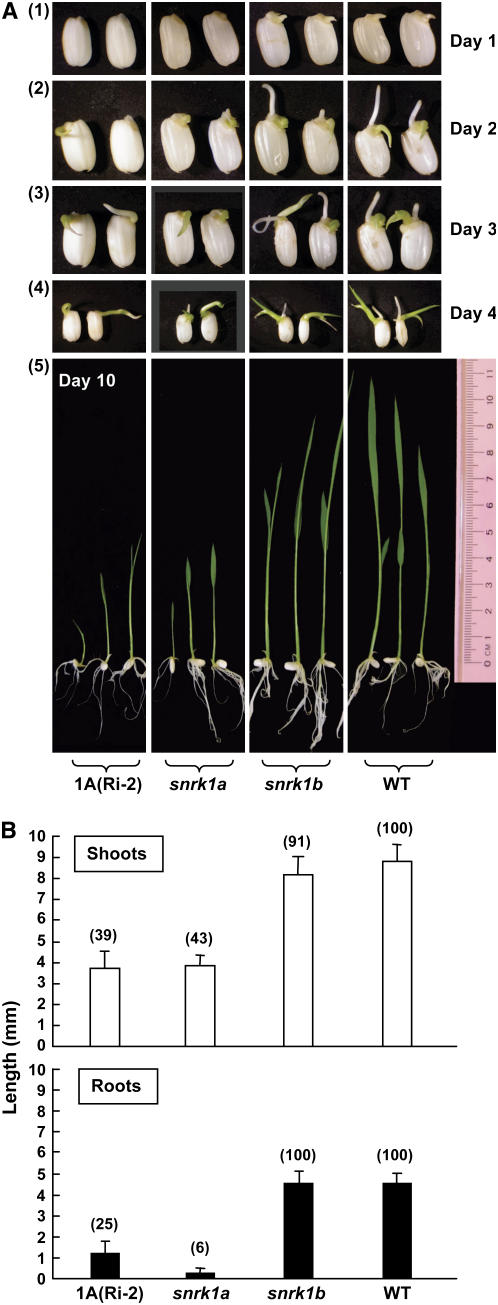
To determine whether SnRK1s control rice growth and development, wild-type, mutant (snrk1a and snrk1b), and SnRK1A RNA interference transgenic seeds were germinated in water for various lengths of time and their phenotypes were compared. Seed germination and seedling growth were retarded in transgenic line 1A(Ri-2) and the snrk1a mutant, but normal seed germination and seedling growth were observed in the snrk1b mutant (Figure 11). At day 1 after seeds were imbibed with water, no growth of shoots and roots was observed for 1A(Ri-2) and the snrk1a mutant, but average shoot and root lengths were 1.0 to 1.2 mm for snrk1b and the wild type. At days 2 to 4, shoot and root lengths were significantly shorter for 1A(Ri-2) and snrk1a compared with snrk1b and the wild type. Shoot and root lengths of wild-type and mutant seedlings were measured quantitatively. In the snrk1a mutant, root growth was more severely retarded than shoot growth. These results suggest that SnRK1A, but not SnRK1B, plays a major role in controlling rice seed germination and seedling growth.
Figure 11.
Abnormal SnRK1A Expression Retards Germination and Seedling Growth.
Rice seeds were incubated in sterile water at 28°C for various lengths of time.
(A) Phenotypes of germinating seeds and seedlings of the wild type, transgenic line 1A(Ri-2), and snrk1a and snrk1b mutant lines at 1 to 4 d (panels 1 to 4) and 10 d (panel 5) after seed imbibition. The magnification of seedling photographs was reduced in panel 4. More photographs of seedlings are shown in Supplemental Figure 10 online.
(B) Lengths of shoots and roots of seedlings measured at day 4. Error bars indicate the se of 10 replicates per line. Numbers in parentheses indicate relative average lengths in percentage.
DISCUSSION
SnRK1A Plays a Key Role in Sugar Signaling Regulating Germination and Seedling Growth
In this study, we investigated the differential functions of two rice SnRK1 proteins and showed that SnRK1A is the major positive regulator for MYBS1 and αAmy3 expression in the sugar signaling pathway in rice. Our studies also demonstrated the important physiological role of SnRK1A in rice, as seed germination and seedling growth were significantly retarded in transgenic rice or mutants expressing abnormal amounts of SnRK1A. SnRK1B was able to regulate αAmy3 SRC activity in the embryo transient expression assays, suggesting that SnRK1B may regulate SRC of downstream gene promoters in the sugar signaling pathway in other tissues or at different developmental stages. Our studies are consistent with the notion that SnRK1A plays a broader role in sugar regulation than SnRK1B, as SnRK1A is uniformly expressed in various growing tissues (including young roots and shoots, flowers, and immature seeds), whereas SnRK1B is mainly expressed in developing seeds (Takano et al., 1998). Histochemical studies have also shown that the SnRK1B promoter directed confined expression of the GUS reporter in the aleurone layer and endosperm cells, which correlates with the appearance of starch granules in these tissues at ∼15 d after flowering; therefore, SnRK1B was proposed to have a special role in carbohydrate metabolism of sink organs in rice (Kanegae et al., 2005).
Future studies on the phenotypes of mutants expressing abnormal amounts of SnRK1A and SnRK1B during entire life cycles may provide more information on the physiological functions of these two proteins in rice growth and development. Furthermore, the study of gene expression profiles in these mutants at different developmental stages (e.g., by microarray analysis) may facilitate the identification of downstream target genes that are regulated by two SnRK1 protein kinases. Consequently, this study has laid a foundation for investigating the mechanisms by which SnRK1A is posttranscriptionally regulated by sugars, interacts with its positive and negative regulators, and regulates downstream gene expression. These studies should lead to a better understanding of how SnRK1 regulates plant growth and development.
Conservation and Divergence in the Regulation and Function of Yeast Snf1 and Plant SnRK1
This study demonstrated the conserved intersubunit and intrasubunit interactions and functions of SnRK1 protein kinases in the sugar signaling pathway in rice. We showed that overexpression of the SnRK1A kinase domain relieved the glucose repression of αAmy3 SRC. A plausible explanation is that the SnRK1A kinase domain itself is capable of activating downstream target factors, similar to kinase domains of the yeast Snf1 and AMPKα1 isoforms, to become constitutively active and could act without the need of an interaction between the Snf1 (α1) regulatory domain and Snf4 (γ) to relieve the Snf1 kinase domain under glucose-repressing conditions (Jiang and Carlson, 1996; Crute et al., 1998). We also found that overexpression of the SnRK1A regulatory domain led to the repression of SRC under glucose starvation. The regulatory domain of the mammalian AMPKα1 subunit also has a dominant negative effect on wild-type AMPKα1 kinase activity, and as the α1 regulatory domain is capable of binding β-γ subunits with an affinity similar to the wild-type α1, it is assumed that the regulatory domain competes with the wild-type α1 for binding to the β-γ subunits (Dyck et al., 1996). This dominant negative effect of the SnRK1 regulatory domain may have resulted from its competition with endogenous SnRK1 for association with other subunits in the SnRK1 protein kinase complex, leading to constitutive autoinhibition of endogenous SnRK1 activity by its own intramolecular interaction under glucose-derepressing conditions.
On the other hand, the regulation and physiological functions of these kinases were also found to have diverged throughout evolution. In yeast, the Snf1 protein kinase activity is dramatically increased upon glucose starvation (Woods et al., 1994; Wilson et al., 1996), but the amount of Snf1 protein is not regulated by glucose (Celenza and Carlson, 1984, 1986). The activity of Snf1 is regulated by interaction with other subunits in the SNF1 protein complex and by phosphorylation by three upstream kinases (Hardie et al., 1998; Hong et al., 2003). In this study, two important findings reveal major differences in sugar regulation between SnRK1 and Snf1. First, the activity of SnRK1 is not significantly increased upon sugar starvation. The increase in the activities of SnRK1 protein kinase appeared to correlate with the increased amounts of SnRK1 protein in rice suspension cells and embryos upon glucose starvation (see Supplemental Figure 3 online). Second, the abundance of SnRK1A is regulated by sugars at the posttranscriptional level. The accumulation of SnRK1A protein was repressed, while that of SnRK1A mRNA was unchanged, by glucose in rice suspension cells and embryos (Figure 1). Differences in amino acid sequences and/or structures between Snf1 and SnRK1A may account for the divergence in their regulation and function.
A Novel Sugar Signaling Pathway in Plants
The sugar signaling pathway connecting the SnRK1 protein kinase with other regulatory components is not well characterized in plants (Polge and Thomas, 2007). In this study, we presented several lines of evidence to show that the SnRK1A protein complex is an upstream positive regulator of MYBS1 and αAmy3 in the sugar signaling pathway in rice. How SnRK1A and MYBS1 are functionally connected is still unclear. SnRK1A in vitro phosphorylates the SAMS peptide that is a relatively specific substrate of the yeast SNF1 and mammalian AMPK protein kinases, indicating that SnRK1A may also regulate target protein activity by phosphorylation. Although MYBS1 was phosphorylated by AMPK in vitro, the phosphorylation of MYBS1 by SnRK1A and an interaction between these two proteins are subjects for further study.
The SnRK1A kinase domain relieved the glucose repression of MYBS1 and αAmy3 expression in a dose-dependent manner, as the degree of relief was higher in the snrk1a homozygous mutant than in the heterozygous mutant. However, the glucose repression of MYBS1 and αAmy3 mRNA accumulation by the overexpressed SnRK1A kinase domain was only partially relieved (Figure 10; see Supplemental Figure 7 online). Similarly, the glucose repression of αAmy3 SRC and MYBS1 promoter activities in the snrk1a homozygous mutant was not completely relieved to the level seen under glucose starvation (Figure 9). Significantly lower activity of the kinase domain than of the complete protein has also been observed in yeast Snf1 (Crute et al., 1998). Consequently, explanations for the partial effect of the SnRK1A kinase domain on the relief of glucose repression must include the fact that, in the presence of glucose, the SnRK1A kinase domain is less active or stable than the wild-type SnRK1A, due to a partial lose of structure or interaction with other components in the SnRK1A complex. Our preliminary study showed that the SnRK1A kinase domain appears to be less stable than the complete protein (K.-W. Lee and S.-M. Yu, unpublished results).
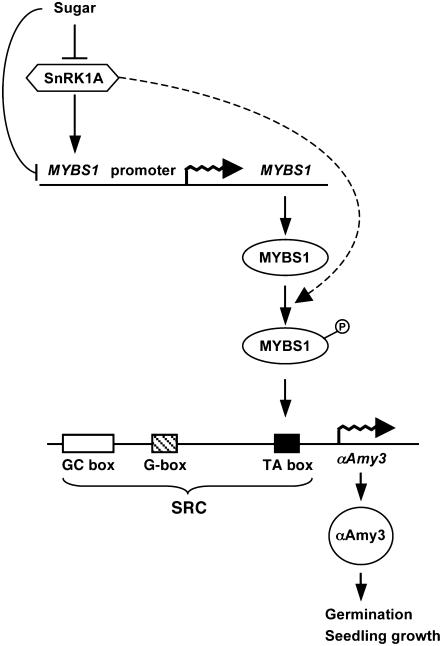
The interaction between MYBS1 and the TA box in the αAmy3 promoter is important for conferring high-level glucose starvation–induced promoter activity. SnRK1A not only acts upstream for glucose starvation–activated expression but also inhibits the glucose repression of MYBS1 and αAmy3. Consequently, our studies have delineated the sugar signaling cascade a few steps upstream of the interaction between MYBS1 and the αAmy3 promoter. A model for the roles of SnRK1A and MYBS1 in sugar signaling in cereals is proposed in Figure 12. Several lines of evidence suggest that sugar regulates MYBS1 and αAmy3 expression through multiple parallel pathways. First, the sugar signal represses MYBS1 promoter activity through a pathway repressing SnRK1A accumulation and activity and another unidentified pathway, as overexpression of SnRK1A could not completely relieve the glucose repression of the MYBS1 promoter. Second, SnRK1A not only activates MYBS1 promoter activity but may also activate MYBS1 protein activity through phosphorylation, as shown by the phosphorylation of MYBS1 by AMPK. Despite the complexity in sugar signaling, SnRK1A plays a key role in this process.
Figure 12.
Proposed Roles of SnRK1 and MYBS1 in Sugar Signaling in Rice.
Details of the model are described in the text. The dashed line indicates the putative pathway.
In summary, the functional link of SnRK1A and MYBS1 as essential components in the sugar signaling pathway regulating SRC in the α-amylase gene promoter provides a new insight into the mechanisms of sugar regulation as well as the physiological relevance of SnRK1A in germination and seedling development in plants.
METHODS
Plant Materials
Rice (Oryza sativa cv Tainung 67) was used in this study. Rice suspension cell culture was established as described (Yu et al., 1991). Cells were subcultured every 7 d by transferring ∼0.5 mL of cells into 25 mL of fresh liquid Murashige and Skoog (MS) medium containing 3% (87 mM) sucrose in a 125-mL flask. Cells were cultured on a reciprocal shaker at 120 rpm and incubated at 26°C in the dark.
Primers
All primers used for the cloning of cDNAs or the construction of plasmids are listed in Table 1.
Table 1.
Primers Used for Plasmid Construction, cDNA Cloning, PCR, RT-PCR, and Real-Time RT-PCR
| Primer | Sequence | Use |
|---|---|---|
| SnRK1A-1F | 5′-ATTGGATCCGAGGGAGCTGGCAGAGATG-3′ | SnRK1A and pUSnf1A(K) |
| SnRK1A-4R | 5′-ATTGGATCCTTAAAGGACTCTCAGCT-3′ | SnRK1A and pUSnf1A(R) |
| SnRK1B-1F | 5′-ATTGGATCCGCATCCATCCTGGTCACTGTGAAA-3′ | SnRK1B and pUSnf1B(K) |
| SnRK1B-4R | 5′-GCGGGATCCGTGTCCTTTATTCATTATTGCAGT-3′ | SnRK1B and pUSnf1B(R) |
| SnRK1A-2R | 5′-ATTGGATCCTTAGTCAGGAGGTGGCACAG-3′ | pUSnf1A(K) |
| SnRK1A-3F | 5′-ATTGGATCCATGGTGCCACCTCCTGACA-3′ | pUSnf1A(R) |
| SnRK1B-2R | 5′-ATTGGATCCCTAGTAACGAGGAAGGCGAATCTG-3′ | pUSnf1B(K) |
| SnRK1B-3F | 5′-ATTGGATCCATGACAGCACAGCAAGCCAAAATG-3′ | pUSnf1B(R) |
| S1-F | 5′-CCGCTCGAGTCTTGTCTTCTTGCTTCCTTTGGTACTTGCC-3′ | pS1GUS |
| S1-R | 5′-CGCGGATCCCGGGTGGTGGATCTCGCACTCTCGC-3′ | pS1GUS |
| 1A1 | 5′-GAATCTTCTCTCGCTCAAGTA-3′ | SnRK1A T-DNA insertion |
| 1A2 | 5′-TCACATTGTGGGTTGATTTCT-3′ | SnRK1A T-DNA insertion |
| 1A4 | 5′-GGGTAACCAATCTGAGT-3′ | SnRK1A 3′ end RT-PCR |
| 1A5 | 5′-CCGCCGCGCGCCGGAGGTCAGTA-3′ | SnRK1A 5′ end RT-PCR |
| 1A6 | 5′-TTACCAGATATCACCTCAGGTGCT-3′ | SnRK1A 5′ end RT-PCR |
| 1A7 | 5′-GCGACAGTTGCCTACTATTTAC-3′ | SnRK1A poly(A) tail and SnRK1A 3′ end RT-PCR |
| 1A11 | 5′-TTCTTAAAGCTCTGCAAGAGC-3′ | SnRK1A 3′ end RT-PCR |
| Q1A5 | 5′-TTATGCCGTTGTCTGCTTC-3′ | SnRK1A real-time RT-PCR |
| Q1A3 | 5′-CCTCTAACGTCTACACACTCCAG-3′ | SnRK1A real-time RT-PCR |
| 35SLB | 5′-GGAATTCAATTCGGCGTTAAT-3′ | SnRK1A KD RT-PCR |
| 1B1 | 5′-TGGCATGTGATTTTAACCTGA-3′ | SnRK1B T-DNA insertion |
| 1B2 | 5′-TATAAGGTCTGGTTAGCCTAC-3′ | SnRK1B T-DNA insertion |
| 1B3 | 5′-CCCCGTGTTGGCTTCAGTG-3′ | SnRK1B 3′ end RT-PCR and real-time RT-PCR |
| 1B4 | 5′-TGATCATACGACTACATTTGTTG-3′ | SnRK1B 3′ end RT-PCR and real-time RT-PCR |
| 1B7 | 5′-TTTTTTAAAGACAAGCTGCGGGAGT-3′ | SnRK1B RT-PCR |
| 1B8 | 5′-GTAACGAGGAAGGCGAATCTG-3′ | SnRK1B RT-PCR |
| S1RTF | 5′-ATGGACGGACATGAGC-3′ | MYBS1 RT-PCR and real-time RT-PCR |
| S1RTR | 5′-GCTTTCACCGGGTGTA-3′ | MYBS1 RT-PCR and real-time RT-PCR |
| 3RT25A | 5′-GTAGGCAGGCTCTCTAGCCTCTAGG-3′ | αAmy3 RT-PCR and real-time RT-PCR |
| 3RT-R | 5′-AACCTGACATTATATATTGCACC-3′ | αAmy3 RT-PCR and real-time RT-PCR |
| Act15 | 5′-CTGATGGACAGGTTATCACC-3′ | Act1 RT-PCR |
| Act13 | 5′-CAGGTAGCAATAGGTATTACAG-3′ | Act1 RT-PCR |
| RB | 5′-AACTCATGGCGATCTCTTACC-3′ | SnRK1A and SnRK1B T-DNA insertion |
| Oligo(dT) anchor | 5′-GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT-3′ | SnRK1A poly(A) tail RT-PCR |
| 18SF | 5′-CCTATCAACTTTCGATGGTAGGATA-3′ | 18S rRNA real-time RT-PCR |
| 18SR | 5′-CGTTAAGGGATTTAGATTGTACTCATT-3′ | 18S rRNA real-time RT-PCR |
Underlined areas indicate restriction sites: CTCGAG, XhoI site; GGATCC, BamHI site.
Plasmids
Yeast strains MCY 1093 (his 4-539, lys 2-801, ura 3-52, suc2+, gal+) and MCY 1846 (lys 2-801, ura 3-52, snf1Δ10) (Celenza and Carlson, 1989) were gifts from Marian Carlson (Columbia University). pAHC 18 contains Luc cDNA fused between the Ubi promoter and the nopaline synthase gene terminator (Bruce et al., 1989). pUGIII contains the rice Ubi promoter–GUS cDNA (Lu et al., 1998). p3Luc.18 contains the SRC (−186 to −82 relative to the transcription start site of αAmy3)-CaMV35S minimal promoter (−46 bp upstream of the transcription site)-Luc chimeric gene (Lu et al., 1998). p3Luc.31 is derived from p3Luc.18 with a mutation in the G box (5′-CGCCTACGTG-3′ → 5′-atgaattcca-3′, with the G box underlined and the mutant sequence in lowercase letters), and p3Luc.34 is derived from p3Luc.18 with a mutation in the TA box (5′-TATCCATATCCAC-3′ → 5′-TATtaggAattcg-3′, with the TA box underlined and the mutant sequences in lowercase letters) (Lu et al., 1998). pUMYBS1 contains the Ubi-MYBS1 construct (Lu et al., 2002).
Isolation of SnRK1 cDNAs
Total RNA was prepared from immature rice seeds harvested at 12 to 15 d after pollination. First-strand cDNAs were synthesized from total RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen). The specific primers used to isolate SnRK1A and SnRK1B cDNAs were designed based on the sequences of OSK1 and OSK24, respectively (Takano et al., 1998). The full-length SnRK1A cDNA (2200 bp) and SnRK1B cDNA (1615 bp) were synthesized by RT-PCR and inserted into the BamHI site in pBluescript KS+, generating pSnRK1A and pSnRK1B.
Plasmid Construction
To make the SnRK1A RNA interference construct for rice transformation, pAHC18 was digested with BamHI to remove the Luc cDNA fragment and blunt-ended. The 400-bp 3′ untranslated region of SnRK1A cDNA was synthesized by PCR, fused in inverted repeats flanking green fluorescent protein DNA, and ligated with truncated pAHC18, generating pUSnRK1A(Ri). pUSnRK1A(Ri) was then digested with HindIII and ligated into binary vector pSMY1H (Ho et al., 2000), generating pAUSnRK1A(Ri).
The kinase and regulatory domains of SnRK1A were predicted based on information from yeast (Jiang and Carlson, 1996). To make constructs for the rice embryo transient expression assay, the kinase domain (amino acids 1 to 279) and the regulatory domain (amino acids 280 to 503) of SnRK1A, and the kinase domain (amino acids 1 to 276) and the regulatory domain (amino acids 277 to 509) of SnRK1B, were synthesized by PCR. The PCR products were inserted into the BamHI site downstream of the Ubi promoter in pAHC18, generating pUSnRK1A(K), pUSnRK1A(R), pUSnRK1B(K), and pUSnRK1B(R). All constructs contained the nopaline synthase gene terminator at 3′ of the effector or reporter gene.
The 2.5-kb MYBS1 promoter region was isolated by PCR using rice genomic DNA as a template. The primer sequences were designed based on the genomic DNA sequence of MYBS1 (BAC clone OJ1005_B10, accession number AP004611). The PCR product containing XhoI and BamHI sites at two ends was translationally fused with the GUS coding region in pUGIII, generating pS1GUS.
Protein Gel Blot Analysis
Total soluble proteins were extracted from cultured rice suspension cells with extraction buffer containing 100 mM K-phosphate buffer, 1 mM EDTA, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 7 mM β-mercaptoethanol, 100 mM NaF, 1 mM NaVO3, 1 mM Na3VO4, 10 mM Na4P2O7, 10 mM N-ethyl-maleimide, and 1× protease inhibitor cocktail (Complete kits; Roche). Total soluble proteins (60 μg) were separated by 10% SDS-PAGE and blotted onto nitrocellulose membranes (Hybond-ECL; Amersham Biosciences). Protein gel blot analysis was performed as described (Yu et al., 1991). Polyclonal rabbit anti-SnRK1A antibodies were diluted 1:5000. Polyclonal rabbit anti-rice α-amylase antibodies (Chen et al., 1994) were diluted 1:10,000. Horseradish peroxidase–conjugated anti-rabbit IgG (Amersham Biosciences) was used as a secondary antibody. Protein signals were detected by chemiluminescence using ECL (Amersham Biosciences).
Anti-SnRK1A Antibodies
SnRK1A-specific antibody was produced against synthetic peptides derived from SnRK1A (5′-SSLAQVTPAETPNSA-3′, residues 347 to 361) (see Supplemental Figure 1 online). Polyclonal anti-SnRK1A antibodies were raised in rabbits and purified by immobilized peptide affinity chromatography (Genesis Biotech).
Rice Embryo Transient Expression Assay
The callus-containing rice embryos were prepared and particle-bombarded as described (Chen et al., 2006). The rice embryos were cobombarded with reporter, effector, and internal control plasmids at a ratio of 2:1:0.25. Because transfection efficiency varies from sample to sample, enzyme activity of an internal control was used to normalize the reporter enzyme activity. When Luc was used as a reporter gene, pUGIII (containing the Ubi-GUS construct) was used as an internal control plasmid. When GUS was used as a reporter gene, pAHC18 (containing the Ubi-Luc construct) was used as an internal control plasmid. Bombarded rice embryos were incubated on MS medium containing 100 mM glucose for 3 h and divided into two halves. Half of the embryos were incubated in MS medium containing 100 mM glucose, and the other half were incubated in MS medium containing in 100 mM mannitol, for 18 h. Total proteins were extracted from embryos with a culture cell lysis reagent buffer [100 mM KH2(PO4), pH 7.8, 1 mM EDTA, 10% glycerol, 1% Triton X-100, and 7 mM β-mercaptoethanol]. GUS and luciferase activity assays were performed as described (Lu et al., 1998). All bombardments were repeated at least four times.
Expression and Purification of Recombinant Proteins
pET-MYBS1 contains MYBS1 cDNA in pET20b (Lu et al., 2002). Escherichia coli strain BL21 (DE3) was transformed with pET-MYBS1 and cultured in Luria-Bertani medium at 37°C until cell density at OD600 reached 0.8. Cells were divided into two flasks each containing or lacking 1 mM IPTG and incubated at 28°C for 8 h. After IPTG induction, the induced and noninduced cells were collected and total proteins were extracted and subjected to SDS-PAGE analysis. Purification of MYBS1 was performed according to the instructions provided by Novagen. Protein concentration was determined with the Bradford reagent (Bio-Rad). The molecular mass of recombinant MYBS1 plus dHA and His tags from pET20b was estimated to be ∼40 kD (365 amino acids).
MYBS1 Phosphorylation Assay
Recombinant MYBS1-dHA (1.5 μg) was mixed with 6 μL of kinase buffer, 1 μL of 0.5 M MgCl2, 0.5 μL of 2 μM ATP, 0.5 μL of [γ-32P]ATP (0.5 μCi, 5000 μCi/mmol), 1 μL of commercial AMPK (Upstate), and 16 μL of water. The reaction was incubated at 30°C for 10 min, and a 30-μL aliquot was incubated with 1 μL of anti-HA antibody, 20 μL of 50% protein G:agarose, and 450 μL of PBS (137 mM NaCl, 2.7 mM KCl, 0.1 mM Na2HPO4, and 2 mM NaH2PO4) with agitation at 4°C overnight. The immunoprecipitated sample was washed with PBST (PBS plus 0.05% Tween 20) by centrifugation (8000g) at 4°C for 2 min. The supernatant was decanted, and the wash was repeated twice. The pellet was resuspended in 20 μL of 5× SDS sample buffer (125 mM Tris, pH 6.8, 4% SDS, 10% glycerol, 0.006% bromphenol blue, and 2% β-mercaptoethanol) and boiled for 5 min. The sample was subjected to SDS-PAGE analysis. The SDS-PAGE product was immobilized on 3M Whatman paper by a vacuum dryer and exposed to x-ray film to detect the protein phosphorylation signal.
Rice Transformation
Plasmid was introduced into Agrobacterium tumefaciens strain EHA101, and rice transformation was performed as described (Chen et al., 2002). Transformed calli were cultured in a liquid MS medium, containing 3% sucrose and 10 μM 2,4-D, for the establishment of suspension cell culture as described (Yu et al., 1991).
Real-Time Quantitative RT-PCR Analysis
First-strand cDNA synthesis was primed with random hexamers (Promega) and catalyzed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) at 37°C for 1.5 h. A 50-fold dilution of the reaction products was then subjected to real-time quantitative RT-PCR analysis using the SYBR Green PCR master mix, an ABI-7000 sequence detection system (Applied Biosystems), and gene-specific primers. Quantitative RT-PCR was analyzed with Microsoft Excel software. After PCR amplifications, all samples were electrophoresed on agarose gels to verify the correct molecular weight of the amplification products.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequences of the Rice SnRK1, Yeast Snf1, and Mammalian AMPK1α Are Conserved.
Supplemental Figure 2. SnRK1A Complements the Yeast snf1 Mutant.
Supplemental Figure 3. The Protein Kinase Activity of SnRK1A Is Repressed by Sugar.
Supplemental Figure 4. SnRK1A Relieves the Sugar Repression of αAmy3 SRC in a Dose-Dependent Manner.
Supplemental Figure 5. Relative Positions of the Putative Functional Domains of SnRK1A and SnRK1B.
Supplemental Figure 6. Genotyping Identified snrk1a and snrk1b Homozygous (−/−) and Heterozygous (+/−) Mutants.
Supplemental Figure 7. Sugar Repression of MYBS1 and αAmy3 Is Partially Inhibited in the snrk1a Mutant Missing the Regulatory Domain of SnRK1A.
Supplemental Figure 8. The Truncated SnRK1A mRNA in the snrk1a Mutant Encodes a Constitutively Active Protein Kinase
Supplemental Figure 9. SnRK1A Is an Upstream Regulator of MYBS1 and αAmy3 in the Sugar Signaling Pathway.
Supplemental Figure 10. Abnormal SnRK1A Expression Retards Germination and Seedling Growth.
Supplementary Material
Acknowledgments
We thank Chyr-Guan Chern and Ming-Jen Fan for technical assistance, Marian Carlson for the yeast strains MCY1093 and MCY1846, and Tuan-Hua David Ho and Harry Iain Wilson for critical review of the manuscript. This work was supported by grants from Academia Sinica and the National Science Council (Grants NSC-91-2311-B-001-093 and NSC-92-2311-B-001-004) of the Republic of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Su-May Yu (sumay@imb.sinica.edu.tw).
Online version contains Web-only data.
References
- Alderson, A., Sabelli, P.A., Dickinson, J.R., Cole, D., Richardson, M., Kreis, M., Shewry, P.R., and Halford, N.G. (1991). Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl. Acad. Sci. USA 88 8602–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao, R.P., Salchert, K., Bako, L., Okresz, L., Szabados, L., Muranaka, T., Machida, Y., Schell, J., and Koncz, C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279 41758–41766. [DOI] [PubMed] [Google Scholar]
- Bruce, W.B., Christensen, A.H., Klein, T., Fromm, M., and Quail, P.H. (1989). Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc. Natl. Acad. Sci. USA 86 9692–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M. (1987). Regulation of sugar utilization in Saccharomyces species. J. Bacteriol. 169 4873–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., and Carlson, M. (1984). Structure and expression of the SNF1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., and Carlson, M. (1986). A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233 1175–1180. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., and Carlson, M. (1989). Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 9 5034–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, M.T., Chao, Y.C., and Yu, S.M. (1994). Novel gene expression system for plant cells based on induction of alpha-amylase promoter by carbohydrate starvation. J. Biol. Chem. 269 17635–17641. [PubMed] [Google Scholar]
- Chan, M.T., and Yu, S.M. (1998. a). The 3′ untranslated region of a rice alpha-amylase gene mediates sugar-dependent abundance of mRNA. Plant J. 15 685–695. [DOI] [PubMed] [Google Scholar]
- Chan, M.T., and Yu, S.M. (1998. b). The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl. Acad. Sci. USA 95 6543–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H., Liu, L.F., Chen, Y.R., Wu, H.K., and Yu, S.M. (1994). Expression of alpha-amylases, carbohydrate metabolism, and autophagy in cultured rice cells are coordinately regulated by sugar nutrient. Plant J. 6 625–636. [DOI] [PubMed] [Google Scholar]
- Chen, P.W., Chiang, C.M., Tseng, T.H., and Yu, S.M. (2006). Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 18 2326–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.-W., Lu, C.-A., Yu, T.-S., Tseng, T.-H., Wang, C.-S., and Yu, S.-M. (2002). Rice alpha-amylase transcriptional enhancers direct multiple mode regulation of promoters in transgenic rice. J. Biol. Chem. 277 13641–13649. [DOI] [PubMed] [Google Scholar]
- Coruzzi, G.M., and Zhou, L. (2001). Carbon and nitrogen sensing and signaling in plants: Emerging ‘matrix effects.’ Curr. Opin. Plant Biol. 4 247–253. [DOI] [PubMed] [Google Scholar]
- Crute, B.E., Seefeld, K., Gamble, J., Kemp, B.E., and Witters, L.A. (1998). Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 273 35347–35354. [DOI] [PubMed] [Google Scholar]
- Dyck, J.R., Gao, G., Widmer, J., Stapleton, D., Fernandez, C.S., Kemp, B.E., and Witters, L.A. (1996). Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic beta and gamma subunits. J. Biol. Chem. 271 17798–17803. [DOI] [PubMed] [Google Scholar]
- Estruch, F., Treitel, M.A., Yang, X., and Carlson, M. (1992). N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S.F., Chou, W.C., Huang, D.D., and Huang, H.J. (2002). Transcriptional regulation of a rice mitogen-activated protein kinase gene, OsMAPK4, in response to environmental stresses. Plant Cell Physiol. 43 958–963. [DOI] [PubMed] [Google Scholar]
- Gancedo, J.M. (1998). Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2001). Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4 387–391. [DOI] [PubMed] [Google Scholar]
- Gibson, S.I. (2000). Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol. 124 1532–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I.A. (1996). Carbohydrate control of gene expression in higher plants. Res. Microbiol. 147 572–580. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Hardie, D.G. (1998). SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 37 735–748. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., Hey, S., Jhurreea, D., Laurie, S., McKibbin, R.S., Paul, M., and Zhang, Y. (2003). Metabolic signalling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 54 467–475. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Paul, M.J. (2003). Carbon metabolite sensing and signalling. Plant Biotechnol. J. 1 381–398. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., Carling, D., and Carlson, M. (1998). The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., and Sakamoto, K. (2006). AMPK: A key sensor of fuel and energy status in skeletal muscle. Physiology (Bethesda) 21 48–60. [DOI] [PubMed] [Google Scholar]
- Hey, S., Mayerhofer, H., Halford, N.G., and Dickinson, J.R. (2007). DNA sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J. Biol. Chem. 282 10472–10479. [DOI] [PubMed] [Google Scholar]
- Ho, S., Chao, Y., Tong, W., and Yu, S. (2001). Sugar coordinately and differentially regulates growth- and stress- related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol. 125 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.L., Tong, W.F., and Yu, S.M. (2000). Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol. 122 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.P., Leiper, F.C., Woods, A., Carling, D., and Carlson, M. (2003). Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100 8839–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.P., Momcilovic, M., and Carlson, M. (2005). Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J. Biol. Chem. 280 21804–21809. [DOI] [PubMed] [Google Scholar]
- Hsing, Y.I., et al. (2007). A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 63 351–364. [DOI] [PubMed] [Google Scholar]
- Hwang, Y.S., Karrer, E.E., Thomas, B.R., Chen, L., and Rodriguez, R.L. (1998). Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 36 331–341. [DOI] [PubMed] [Google Scholar]
- Jiang, R., and Carlson, M. (1996). Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10 3105–3115. [DOI] [PubMed] [Google Scholar]
- Jiang, R., and Carlson, M. (1997). The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17 2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae, H., Miyoshi, K., Hirose, T., Tsuchimoto, S., Mori, M., Nagato, Y., and Takano, M. (2005). Expressions of rice sucrose non-fermenting-1 related protein kinase 1 genes are differently regulated during the caryopsis development. Plant Physiol. Biochem. 43 669–679. [DOI] [PubMed] [Google Scholar]
- Kleinow, T., Bhalerao, R., Breuer, F., Umeda, M., Salchert, K., and Koncz, C. (2000). Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 23 115–122. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Yamamoto, S., Minami, H., Kagaya, Y., and Hattori, T. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 509–540. [DOI] [PubMed] [Google Scholar]
- Kuchin, S., Treich, I., and Carlson, M. (2000). A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos, L., Klein, M., Hofgen, R., and Banfalvi, Z. (1999). Potato StubSNF1 interacts with StubGAL83: A plant protein kinase complex with yeast and mammalian counterparts. Plant J. 17 569–574. [DOI] [PubMed] [Google Scholar]
- Laurie, S., McKibbin, R.S., and Halford, N.G. (2003). Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an alpha-amylase (alpha-Amy2) gene promoter in cultured wheat embryos. J. Exp. Bot. 54 739–747. [DOI] [PubMed] [Google Scholar]
- Lee, Y.-C., Lu, C.-A., Chen, P.-W., Casaretto, J., and Yu, S.-M. (2003). An ABA-responsive bZIP protein, OsBZ8, mediates sugar repression of alpha-amylase gene expression. Physiol. Plant. 119 78–86. [Google Scholar]
- Lo, W.S., Gamache, E.R., Henry, K.W., Yang, D., Pillus, L., and Berger, S.L. (2005). Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas, A., Sos-Hegedus, A., Bimbo, A., and Banfalvi, Z. (2003). Functional diversity of potato SNF1-related kinases tested in Saccharomyces cerevisiae. Gene 321 123–129. [DOI] [PubMed] [Google Scholar]
- Lu, C.A., Ho, T.H., Ho, S.L., and Yu, S.M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C.A., Lim, E.K., and Yu, S.M. (1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273 10120–10131. [DOI] [PubMed] [Google Scholar]
- Man, A.L., Purcell, P.C., Hannappel, U., and Halford, N.G. (1997). Potato SNF1-related protein kinase: Molecular cloning, expression analysis and peptide kinase activity measurements. Plant Mol. Biol. 34 31–43. [DOI] [PubMed] [Google Scholar]
- Muranaka, T., Banno, H., and Machida, Y. (1994). Characterization of tobacco protein kinase NPK5, a homolog of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol. Cell. Biol. 14 2958–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of gibberellin-dependent signalling pathway in barley embryos. Plant Cell 9 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge, C., and Thomas, M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12 20–28. [DOI] [PubMed] [Google Scholar]
- Purcell, P.C., Smith, A.M., and Halford, N.G. (1998). Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J. 14 195–202. [Google Scholar]
- Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57 675–709. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.): S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2 410–418. [DOI] [PubMed] [Google Scholar]
- Shen, W., and Hanley-Bowdoin, L. (2006). Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, J.-J., Jan, S.-P., Lee, H.-T., and Yu, S.-M. (1994). Control of transcription and mRNA turnover as mechanisms of metabolic repression of alpha-amylase gene expression. Plant J. 5 655–664. [Google Scholar]
- Sheu, J.-J., Yu, T.-S., Tong, W.-F., and Yu, S.-M. (1996). Carbohydrate starvation stimulates differential expression of rice alpha-amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271 26998–27004. [DOI] [PubMed] [Google Scholar]
- Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 49–81. [DOI] [PubMed] [Google Scholar]
- Takano, M., Kajiya-Kanegae, H., Funatsuki, H., and Kikuchi, S. (1998). Rice has two distinct classes of protein kinase genes related to SNF1 of Saccharomyces cerevisiae, which are differently regulated in early seed development. Mol. Gen. Genet. 260 388–394. [DOI] [PubMed] [Google Scholar]
- Toyofuku, K., Umemura, T., and Yamaguchi, J. (1998). Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Lett. 428 275–280. [DOI] [PubMed] [Google Scholar]
- Umemura, T., Perata, P., Futsuhara, Y., and Yamaguchi, J. (1998). Sugar sensing and alpha-amylase gene repression in rice embryos. Planta 204 420–428. [DOI] [PubMed] [Google Scholar]
- Wilson, W.A., Hawley, S.A., and Hardie, D.G. (1996). Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6 1426–1434. [DOI] [PubMed] [Google Scholar]
- Woods, A., Munday, M.R., Scott, J., Yang, X., Carlson, M., and Carling, D. (1994). Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269 19509–19515. [PubMed] [Google Scholar]
- Yu, S.M. (1999). Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 121 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.M., Kuo, Y.H., Sheu, G., Sheu, Y.J., and Liu, L.F. (1991). Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J. Biol. Chem. 266 21131–21137. [PubMed] [Google Scholar]
- Yu, S.M., Lee, Y.C., Fang, S.C., Chan, M.T., Hwa, S.F., and Liu, L.F. (1996). Sugars act as signal molecules and osmotica to regulate the expression of alpha-amylase genes and metabolic activities in germinating cereal grains. Plant Mol. Biol. 30 1277–1289. [DOI] [PubMed] [Google Scholar]
- Yu, S.M., Tzou, W.S., Lo, W.S., Kuo, Y.H., Lee, H.T., and Wu, R. (1992). Regulation of alpha-amylase-encoding gene expression in germinating seeds and cultured cells of rice. Gene 122 247–253. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Shewry, P.R., Jones, H., Barcelo, P., Lazzeri, P.A., and Halford, N.G. (2001). Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28 431–441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.