Abstract
The synergid cells within the female gametophyte are essential for reproduction in angiosperms. MYB98 encodes an R2R3-MYB protein required for pollen tube guidance and filiform apparatus formation by the synergid cells. To test the predicted function of MYB98 as a transcriptional regulator, we determined its subcellular localization and examined its DNA binding properties. We show that MYB98 binds to a specific DNA sequence (TAAC) and that a MYB98–green fluorescent protein fusion protein localizes to the nucleus, consistent with a role in transcriptional regulation. To identify genes regulated by MYB98, we tested previously identified synergid-expressed genes for reduced expression in myb98 female gametophytes and identified 16 such genes. We dissected the promoter of one of the downstream genes, DD11, and show that it contains a MYB98 binding site required for synergid expression, suggesting that DD11 is regulated directly by MYB98. To gain insight into the functions of the downstream genes, we chose five genes and determined the subcellular localization of the encoded proteins. We show that these five proteins are secreted into the filiform apparatus, suggesting that they play a role in either the formation or the function of this unique structure. Together, these data suggest that MYB98 functions as a transcriptional regulator in the synergid cells and activates the expression of genes required for pollen tube guidance and filiform apparatus formation.
INTRODUCTION
The angiosperm female gametophyte contains two synergid cells that lie adjacent to the egg cell and are essential for reproduction. During the angiosperm fertilization process, a pollen tube grows into one of the synergid cells, ceases growth, ruptures, and releases its two sperm cells into this cell. The synergid cell that interacts with the pollen tube typically undergoes cell death before or upon pollen tube penetration. The two sperm cells then migrate to and fuse with the egg cell and central cell, and ultimately, the fertilized female gametophyte gives rise to the embryo and endosperm of the seed (reviewed in Lord and Russell, 2002; Weterings and Russell, 2004).
During the final stages of pollen tube growth, a pollen tube grows along the placental surface, along the surface of the ovule's funiculus, into the ovule's micropyle, and finally into the female gametophyte. To determine whether the female gametophyte plays a role in pollen tube guidance, several groups have analyzed pollen tube growth in Arabidopsis thaliana mutants defective in embryo sac development. These studies showed that the female gametophyte is required for pollen tube guidance at two stages: guidance from the placenta to the funiculus (funicular guidance phase) and guidance from the funiculus to the micropyle (micropylar guidance phase) (reviewed in Higashiyama, 2002; Johnson and Preuss, 2002; Higashiyama et al., 2003).
To determine which cells within the female gametophyte produce the pollen tube attractant(s), Higashiyama and colleagues (2001) analyzed pollen tube attraction by Torenia ovules in which specific cells of the female gametophyte were removed by laser ablation. Pollen tubes were attracted by ovules lacking the central cell and egg cell but not by ovules lacking the synergid cells, indicating that the synergid cells are the source of a pollen tube attractant. Recently, the maize (Zea mays) synergid cell–expressed gene Zm EA1 was shown to be required for pollen tube guidance (Marton et al., 2005), but its precise role in this process still needs to be determined (McCormick and Yang, 2005).
At the micropylar pole, the synergid cell wall is extensively thickened and elaborated, forming a structure referred to as the filiform apparatus (see Supplemental Figure 1 online). In Arabidopsis and many other species, the filiform apparatus has numerous finger-like projections into the synergid cytoplasm. Several functions for the filiform apparatus have been proposed, including pollen tube reception, import of metabolites, and export of the pollen tube attractant(s). However, a specific function of the filiform apparatus in pollen tube reception or transport has not been demonstrated (reviewed in Willemse and van Went, 1984; Huang and Russell, 1992; Higashiyama, 2002).
Genetic screens have identified several mutants that appear to be defective in synergid cell function. In sirene (Rotman et al., 2003) and feronia (Huck et al., 2003) mutants, wild-type pollen tubes enter mutant female gametophytes but fail to cease growth and release their contents. Although the corresponding genes have not been identified, they likely encode molecules involved in synergid cell–pollen tube interaction. gametophytic factor2 (gfa2) female gametophytes fail to undergo synergid cell death following pollination. GFA2 encodes a J domain–containing protein required for mitochondrial function, suggesting that functional mitochondria are required for synergid cell death (Christensen et al., 2002). Additional mutants affected in synergid function appear to be present in a recently identified collection of female gametophyte mutants (Pagnussat et al., 2005).
Genes functioning in the synergid cells have also been identified through expression-based screens. The Arabidopsis synergid-expressed genes include MYB98 (Kasahara et al., 2005) and the 18 genes listed in Table 1. Additional synergid-expressed genes have been identified in maize (Cordts et al., 2001; Le et al., 2005; Marton et al., 2005; Yang et al., 2006).
Table 1.
Synergid-Expressed Genes Discussed in This Article
| Genea | AGIb | Expression in the Female Gametophyte | Predicted Functionc |
|---|---|---|---|
| DD2 | At5g43510 | SCs, (EC), (CC) | Defensin-like protein (CRP) |
| DD3 | At3g56610 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD4 | At5g42955 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD8 | At5g52975 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD11 | At1g52970 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD12 | At2g21655 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD17 | At5g34885 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD18 | At1g45190 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD27 | At3g05460 | SCs, CC | Sporozoite surface protein–related |
| DD31 | At1g47470 | SCs | Hypothetical protein (CRP) |
| DD32 | At3g17080 | SCs, (EC), (CC) | Self-incompatibility protein–related (CRP) |
| DD34 | At4g07515 | SCs, (EC), (CC) | Expressed protein (CRP) |
| DD35 | At5g12380 | SCs | Putative annexin |
| DD39/QRT3 | At4g20050 | SCs, (EC) | Polygalacturonase |
| DD42 | At2g20660 | SCs, (EC), (CC) | Rapid alkalinization factor family protein14 |
| DD46 | At1g22015 | SCs, CC | Galactosyltransferase family protein |
| DD56 | At4g30590 | SCs, (EC), (CC) | Plastocyanin-like domain–containing protein |
| DD67 | At5g11940 | SCs, (EC), (CC) | Subtilase family protein |
CC, central cell; CRP, Cys-rich protein; EC, egg cell; SC, synergid cell. Parentheses indicate weak expression.
Genes were identified by Steffen et al. (2007).
Arabidopsis Genome Initiative number.
Based on The Arabidopsis Information Resource annotations (www.arabidopsis.org).
MYB98 encodes a MYB family protein and is predicted to function as a transcriptional regulator. Within the female gametophyte, MYB98 is expressed predominantly in the synergid cells. myb98 female gametophytes are defective in pollen tube guidance: wild-type pollen tubes grow from the placenta to the funiculus but then fail to grow into the micropyle, indicating that the myb98 mutation affects the micropylar guidance phase of pollen tube guidance. In addition, the filiform apparatus of myb98 mutants lacks the finger-like projections observed in wild-type synergid cells. However, with the exception of the filiform apparatus defect, myb98 synergid cells appear normal, suggesting that MYB98 regulates the expression of a specific set of genes that function in pollen tube guidance and filiform apparatus formation (Kasahara et al., 2005). Here, we provide evidence for this model. We show that MYB98 is localized to the nuclei of the synergid cells, binds DNA at a specific sequence, and is required for the expression of a battery of synergid-expressed genes. We also show that at least one of the downstream genes is likely a direct target of MYB98. Finally, we show that many of the downstream genes encode proteins that are secreted into the filiform apparatus. Together, these data suggest very strongly that MYB98 activates a branch of the synergid gene regulatory network that is required for pollen tube guidance and filiform apparatus formation.
RESULTS
MYB98 Binds DNA and Is Localized to the Nucleus
MYB98 is predicted to encode a MYB family transcription factor (Stracke et al., 2001). If this prediction is correct, MYB98 should bind DNA and be localized to the nucleus. To determine the subcellular localization of MYB98, we analyzed transgenic plants containing a construct (ProMYB98:GFP-MYB98) comprising the MYB98 promoter, a green fluorescent protein (GFP) coding sequence, and the entire MYB98 coding sequence. This construct was sufficient to rescue the myb98-1 mutant phenotype, suggesting that localization of this fusion protein mimics that of endogenous MYB98. Rescue was established by generating plants homozygous for both the myb98-1 mutation and the ProMYB98:GFP-MYB98 construct (see Methods); these plants had full seed set.
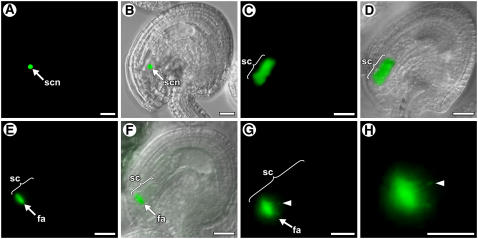
As shown in Figures 1A and 1B, in a mature, unfertilized, wild-type female gametophyte (stage FG7) (Christensen et al., 1997), GFP-MYB98 was localized to the nuclei of the two synergid cells.
Figure 1.
Localization of GFP Fusion Proteins in Wild-Type Ovules.
(A), (C), (E), (G), and (H) show fluorescence images of the GFP signals. (B), (D), and (F) show fluorescence bright-field overlay images. fa, filiform apparatus; sc, synergid cell; scn, synergid cell nucleus. Bars = 20 μm in (A) to (F) and 10 μm in (G) and (H).
(A) and (B) Localization of GFP-MYB98 in a mature female gametophyte (stage FG7). The GFP signal is localized to the nuclei of the synergid cells. Only one synergid nucleus is visible; the other is out of the focal plane.
(C) and (D) Localization of DD34-GFP in a mature female gametophyte (stage FG7). The GFP signal is present throughout the synergid cell.
(E) and (F) Localization of DD4-GFP in a mature female gametophyte (stage FG7). The GFP signal is localized to the filiform apparatus.
(G) and (H) Localization of DD11-GFP in a mature female gametophyte (stage FG7). These are high-magnification micrographs showing that the GFP signal is localized to the filiform apparatus. Arrowheads show a finger-like projection.
We used electrophoretic mobility shift assays (EMSAs) to determine whether MYB98 binds DNA. We previously showed that MYB98's Myb domain (R2 and R3 Myb repeats) is highly similar to that of mammalian c-Myb (Kasahara et al., 2005), suggesting that MYB98 binds DNA to a sequence similar to that of c-Myb. Several studies have shown that c-Myb binds to the consensus sequence YAACNG (Y = thymine or cytosine, N = any nucleotide) and that the AAC residues are critical for binding specificity (Biedenkapp et al., 1988; Weston, 1992; Tanikawa et al., 1993; Oda et al., 1998).
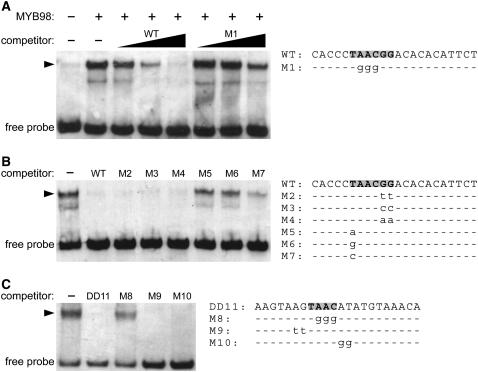
To determine whether MYB98 binds to the c-Myb consensus sequence, we generated the MYB98 Myb domain in Escherichia coli and analyzed interaction with a labeled oligonucleotide containing the sequence TAACGG using EMSAs. As shown in Figure 2A, addition of the MYB98 Myb domain caused a shift in the mobility of the labeled oligonucleotide, indicating that MYB98 binds to DNA. To determine whether this binding is specific for the sequence TAACGG, we added unlabeled oligonucleotides to the binding reaction. As shown in Figure 2A, the wild-type oligonucleotide competed fully, whereas an oligonucleotide containing mutations in the core AAC residues (M1) competed much less effectively. These data indicate that MYB98 binds to DNA in a sequence-specific manner.
Figure 2.
MYB98 Binds to the Sequence YAAC in Vitro.
All panels are from EMSAs using the MYB98 Myb domain and oligonucleotides containing wild-type and mutant versions of the MYB98 binding site. Arrowheads indicate the mobility shifts that occur upon the addition of the MYB98 protein. The minor bands below the major bands are present in a constant ratio to the major bands and may represent the binding of partially degraded MYB98 protein to the oligonucleotides. The sequences of the oligonucleotides used in these reactions are shown at right.
(A) MYB98 binds to the c-Myb consensus binding site. Lane 1 contains the labeled wild-type oligonucleotide alone. Lanes 2 to 8 contain the labeled wild-type oligonucleotide plus MYB98 protein. Lanes 3 to 5 contain unlabeled wild-type oligonucleotide in 1×, 10×, and 100× molar excess, respectively. Lanes 6 to 8 contain unlabeled mutated (M1) oligonucleotide in 1×, 10×, and 100× molar excess, respectively. The unlabeled wild-type oligonucleotide competes partially at 1× to 10× and fully at 100× molar excess. The unlabeled M1 oligonucleotide competes weakly at 100× molar excess.
(B) MYB98 binds to the sequence YAAC. All lanes contain labeled wild-type oligonucleotide plus MYB98 protein. Lanes 2 to 8 contain unlabeled oligonucleotides at 25× molar excess. Oligonucleotides M2, M3, and M4, which all contain mutations at positions 5 and 6, compete fully with the wild-type oligonucleotide. Oligonucleotides M5 and M6, in which the nucleotide at position 1 has been changed to an adenine or a guanine, fail to compete with the wild-type oligonucleotide. Oligonucleotide M7, in which the nucleotide at position 1 has been changed to a cytosine, competes at a reduced level.
(C) MYB98 binds to TAAC−144. All lanes contain labeled wild-type oligonucleotide (oligonucleotide DD11) plus the MYB98 protein. Lanes 2 to 5 contain unlabeled oligonucleotides at 25× molar excess. The wild-type oligonucleotide (DD11) competes fully. Oligonucleotide M8, in which the nucleotides in the core AAC residues have been mutated, fails to compete. Oligonucleotides M9 and M10, in which nucleotides outside the core AAC residues have been mutated, compete fully.
To further define the MYB98 DNA binding sequence, we performed a series of competition assays in which binding of MYB98 to a labeled oligonucleotide containing the sequence TAACGG was competed with a 25-fold excess of unlabeled oligonucleotides containing mutations at various positions. As shown in Figure 2B, oligonucleotides containing mutations at positions 5 and 6 (M2, M3, and M4) competed as effectively as the wild-type oligonucleotide, an oligonucleotide containing a cytosine at position 1 (M7) competed but did so less effectively than the wild-type oligonucleotide, and oligonucleotides containing an adenine or a guanine at position 1 (M5 and M6) did not compete with the wild-type oligonucleotide. Together, these data suggest that MYB98 binds to the sequence YAAC with a preference for TAAC.
In summary, MYB98 is localized to the nucleus of the synergid cells and binds DNA at a specific sequence. These data suggest very strongly that MYB98 functions as a transcriptional regulator in the synergid cells.
MYB98 Is Not Autoregulated
Autoregulation is a common feature of transcription factor gene regulation (Freeman, 2000; de Folter and Angenent, 2006). Upstream of the MYB98 coding region are several putative MYB98 binding sites, suggesting that MYB98 may regulate its own transcription. To test for this, we compared the expression of ProMYB98:GFP in MYB98/MYB98 and MYB98/myb98-1 pistils. In plants hemizygous for the ProMYB98:GFP construct, the percentage of female gametophytes expressing GFP was similar in MYB98/MYB98 (48.4%; n = 279) and MYB98/myb98-1 (44.9%; n = 285) pistils. Additionally, within MYB98/myb98-1 pistils, the intensity of GFP fluorescence was similar among gametophytes. These data suggest that MYB98 does not regulate its own expression.
Identification of 16 Genes Downstream of MYB98
The results presented above suggest that MYB98 regulates the expression of genes within the synergid cell. To test this further, we determined whether any of the synergid-expressed genes listed in Table 1 are downregulated in myb98 female gametophytes. We used real-time RT-PCR to assay the expression of these 18 genes in MYB98/MYB98 and myb98-1/myb98-1 pistils. As shown in Table 2, all except DD35 showed reduced expression in myb98-1/myb98-1 pistils relative to MYB98/MYB98 pistils, suggesting that the other 17 genes require MYB98 for expression.
Table 2.
Real-Time RT-PCR Analysis of 18 Synergid-Expressed Genes
| Gene Name | CTMYB98/MYB98 Pistils | CTmyb98-1/myb98-1 Pistils | Fold Change |
|---|---|---|---|
| DD2 | 19.2 | 28.8 | 776 |
| DD3 | 21.6 | 25.3 | 13 |
| DD4 | 18.7 | 32.4 | 13,308 |
| DD8 | 23.8 | 27.7 | 15 |
| DD11 | 24.2 | 32.1 | 239 |
| DD12 | 20.9 | 26.3 | 42 |
| DD17 | 25.7 | 39.0 | 10,086 |
| DD18 | 22.2 | >40 | 228,207 |
| DD27 | 23.4 | 26.1 | 6.5 |
| DD31 | 25.4 | 34.1 | 416 |
| DD32 | 29.5 | >40 | 1,448 |
| DD34 | 30.3 | 38.1 | 223 |
| DD35 | 24.9 | 25.8 | 1.9a |
| DD39/QRT3 | 24.7 | 27.9 | 9.2 |
| DD42 | 25.9 | 29.7 | 14 |
| DD46 | 25.9 | 29.8 | 15 |
| DD56 | 25.1 | 33.1 | 256 |
| DD67 | 26.5 | 33.3 | 111 |
CT, threshold cycle.
Not significantly different from 1.0.
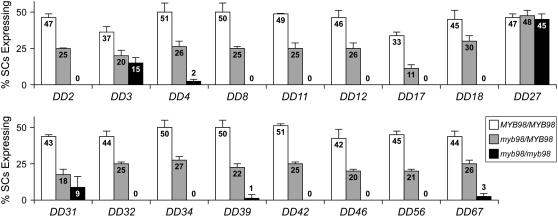
To validate these expression patterns, we analyzed the expression of the corresponding promoter:GFP constructs for these 17 genes in MYB98 and myb98-1 female gametophytes. Using crosses, we generated plants hemizygous for the promoter:GFP construct and homozygous for the myb98-1 mutation. We then scored the number of female gametophytes expressing GFP in the synergid cells in these plants as well as in the parental line. As shown in Figure 3, the number of female gametophytes expressing ProDD27:GFP in the synergid cells was approximately equal in MYB98/MYB98 and myb98-1/myb98-1 pistils. By contrast, the other 16 genes showed reduced expression of the corresponding promoter:GFP construct in myb98-1/myb98-1 pistils (Figure 3). Together, these data indicate that MYB98 is required for the expression of these 16 genes during synergid cell development.
Figure 3.
Expression of Synergid-Expressed Genes in MYB98 and myb98-1 Female Gametophytes.
Percentage of synergid cells (SCs) expressing the corresponding promoter:GFP construct in MYB98/MYB98 (white bars), myb98-1/MYB98 (gray bars), and myb98-1/myb98-1 (black bars) pistils. All plants scored were hemizygous for the promoter:GFP construct. Error bars indicate sd.
Genes Downstream of MYB98 Encode Proteins That Localize to the Filiform Apparatus
The predicted functions of the 16 genes downstream of MYB98 are listed in Table 1. More than half (9 of 16 genes) encode proteins of unknown function. All except one (DD34) encode proteins with predicted N-terminal signal peptides. Most (11 of 16 genes) of the genes downstream of MYB98 encode small, Cys-rich proteins (Table 1), 6 of which belong to two families: DD11, DD18, and DD31 are within the CRP3700 subgroup; and DD4, DD17, and DD34 are within the CRP3740 subgroup (Silverstein et al., 2007).
To gain insight into the functions of the genes downstream of MYB98, we obtained and analyzed T-DNA mutants from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). Of the 16 genes, T-DNA lines were available for 6 genes (DD4, DD18, DD31, DD39/QRT3, DD56, and DD67). In all cases, the T-DNAs segregated normally (e.g., 1:2:1 within the progeny of self-pollinated heterozygous plants) and the heterozygous and homozygous mutant plants exhibited full seed set. These data suggest that the female gametophyte is not affected by these six mutations. The absence of mutant phenotypes for these T-DNA lines is likely due to functional redundancy, as all are members of large gene families.
As a second approach, we selected six genes (DD2, DD4, DD11, DD12, DD32, and DD34) and determined the subcellular localization of the encoded proteins. DD34 was chosen because it lacks a putative signal peptide (discussed above). The other five have putative signal peptides. We analyzed transgenic lines expressing C-terminal GFP fusion proteins in the synergid cells. Each transgene construct contained the upstream regulatory sequences and the complete coding region fused with a GFP coding sequence. We analyzed the localization patterns in mature, unfertilized female gametophytes (stage FG7). As shown in Figures 1C and 1D, DD34-GFP, which lacks an N-terminal signal peptide, is distributed throughout the cytoplasm. By contrast, as shown in Figures 1E to 1H and Supplemental Figures 2A to 2F online, with the other five fusion proteins, which all contain N-terminal signal peptides, strong GFP signals were associated with the filiform apparatus and GFP was not detected in the cytoplasm. These data suggest that DD34 is a cytoplasmic protein, whereas the DD2, DD4, DD11, DD12, and DD32 proteins are secreted into the filiform apparatus.
The results presented above suggest that within the synergid cells, secretion into the filiform apparatus is the default secretory pathway. To test this, we analyzed transgenic plants containing a protein fusion construct (ProMYB98:SP-GFP) comprising the MYB98 promoter, the coding sequence of a signal peptide (from DD11), and a GFP coding sequence. As shown in Supplemental Figures 2G and 2H online, this fusion protein was localized to the filiform apparatus. Thus, an N-terminal signal peptide appears to be sufficient to localize a protein to the filiform apparatus in the synergid cells.
The DD11 Promoter Contains a MYB98 Binding Site Necessary for Synergid Expression
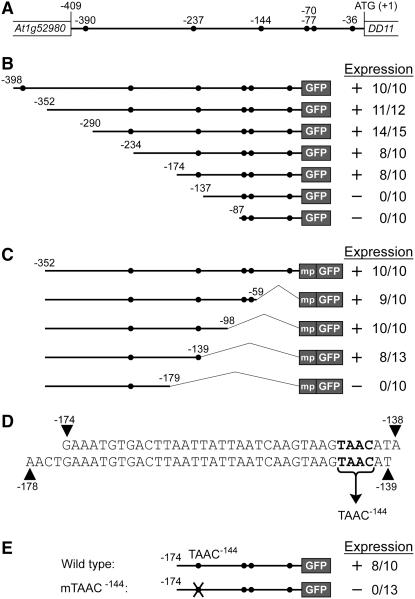
To identify in vivo MYB98 DNA binding sites, as well as direct targets of MYB98, we chose one gene and dissected its promoter. We chose DD11 because the location of the adjacent upstream gene (Figure 4A) suggests that the regulatory sequences necessary for synergid expression are located within a small (409 bp) region. Consistent with this, we previously showed that a promoter fragment containing 398 bp upstream of the translational start site is sufficient to direct expression to the synergid cells (Steffen et al., 2007). In addition, as shown in Figure 4A, the DD11 promoter contains six YAAC sites, which are potential MYB98 binding sites (Figure 2); therefore, this gene could be a direct target of MYB98.
Figure 4.
Identification of Sequences Necessary for Synergid Expression within the DD11 Promoter.
(A) Schematic of the genomic region encompassing the DD11 promoter including the upstream gene (At1g52980). Closed circles represent putative MYB98 binding sites (YAAC). All contain the sequence TAAC, and they are designated TAAC−390, TAAC−237, TAAC−144, TAAC−77, TAAC−70, and TAAC−36.
(B) and (C) Expression of the 5′ (B) and 3′ (C) promoter deletion constructs. Schematics of the constructs are shown at left. The 5′ promoter deletion constructs (B) contain 5′ end points of between −398 and −87 bp relative to the translational start, a 3′ end point of +21 bp relative to the translational start, and are fused with a GFP coding region. The 3′ promoter deletion constructs (C) contain a 5′ end point of −352 bp relative to the translational start, 3′ end points of between −1 and −179 bp relative to the translational start, and are fused with a 35S minimal promoter (mp) and a GFP coding region. Expression levels of the corresponding constructs in transgenic plants are shown at right.
(D) Sequences of the regions identified as necessary for synergid expression in the 5′ (top) and 3′ (bottom) deletions of the DD11 promoter. These regions contain a single TAAC site (TAAC−144; boldface).
(E) Expression of the mTAAC−144 construct. A schematic of this construct is shown at left. mTAAC−144 contains a mutation in the core AAC residues (TAAC mutated to TGGG) of the MYB98 binding site and is otherwise identical to the −174 5′ promoter deletion construct (B). Expression levels of the corresponding constructs in transgenic plants are shown at right.
In (B), (C), and (E), expression levels are indicated as follows: + indicates that GFP was detected in the synergid cells in some transgenic plants, − indicates that GFP was not detected in the synergid cells in any transgenic plants, the numerator of the ratio indicates the number of transgenic lines expressing GFP, and the denominator of the ratio indicates the total number of transgenic lines analyzed.
To determine whether any of these YAAC sites, or any other sequences, are necessary for DD11 expression, we generated a series of 5′ (Figure 4B) and 3′ (Figure 4C) deletions of the DD11 promoter, fused these with a GFP coding region, introduced these promoter:GFP constructs into wild-type Arabidopsis plants, and analyzed ≥10 transgenic lines per construct. With the 5′ deletion series, a promoter fragment containing 174 bp upstream of the translational start site was sufficient to drive the expression of GFP in the synergid cells, but removal of an additional 37 bp (to position −137) abolished expression (Figure 4B). With the 3′ deletion series, a promoter fragment containing −352 to −139 bp relative to the translational start site was sufficient to drive the expression of GFP in the synergid cells, but removal of an additional 40 bp from the 3′ end (to position −179) abolished expression (Figure 4C).
The regions identified in the 5′ and 3′ deletions are compared in Figure 4D. These regions overlap by 36 bp, at positions −174 to −139, suggesting that this 36-bp region contains sequences necessary for synergid expression. As shown in Figure 4D, this region contains a single putative MYB98 binding site (TAAC) at position −144. We refer to this site as TAAC−144.
To determine whether MYB98 can bind to TAAC−144, we performed EMSAs with the MYB98 Myb domain and an oligonucleotide containing a region of the DD11 promoter encompassing this site. As shown in Figure 2C, MYB98 bound to this oligonucleotide, and this binding was competed with the wild-type competitor and competitors with mutations outside the core AAC residues (oligonucleotides M9 and M10) but not with competitors bearing mutations within the core AAC residues (oligonucleotide M8). These data indicate that MYB98 binds to TAAC−144 in vitro.
To determine whether TAAC−144 is necessary for DD11 expression, we generated a construct (mTAAC−144) in which this site is mutated in the context of a promoter fragment containing 174 bp upstream of the translational start site and introduced this construct into wild-type plants. As shown in Figure 4E, the wild-type promoter fragment was sufficient to drive the expression of GFP in the synergid cells. By contrast, the same construct containing a mutation in TAAC−144 was not expressed in the synergid cells. Together, these data indicate that the DD11 promoter contains a MYB98 binding site necessary for synergid expression.
DISCUSSION
The synergid cells are required for pollen tube guidance and fertilization and have several structural specializations, including a filiform apparatus. We are dissecting the gene regulatory network of the Arabidopsis synergid cell to understand how this important cell type acquires its unique features and functions. Here, we show that MYB98 functions as a transcription factor, that 16 genes lie downstream of MYB98 in the synergid gene regulatory network, and that at least 1 of these genes, DD11, is a direct target of MYB98. We further show that many of the genes downstream of MYB98 encode proteins that are secreted into the filiform apparatus, suggesting that they play a role in the formation or function of this structure. Together, these data establish that MYB98 activates a branch of the synergid gene regulatory network required for pollen tube guidance and filiform apparatus formation.
MYB98 Encodes a Synergid Cell Transcription Factor
MYB98 is a member of the R2R3-MYB gene family, which comprises 125 genes in Arabidopsis (Stracke et al., 2001) and 80 to 84 genes in rice (Oryza sativa) (Jia et al., 2004; Jiang et al., 2004). R2R3-MYB proteins are thought to function as transcription factors because they contain a structurally conserved DNA binding domain referred to as the Myb domain. Consistent with this, many R2R3-MYB proteins have been shown to function in transcriptional regulation (Stracke et al., 2001; Dias et al., 2003). Here, we show that MYB98 is localized to the nuclei of the synergid cells (Figures 1A and 1B) and is required for the expression of a battery of genes in the synergid cells (Figure 3). We further show that MYB98 binds to a specific DNA sequence (Figure 2) and that a MYB98 binding site is necessary for synergid expression (Figure 4E). Together, these results strongly suggest that MYB98 functions as a transcriptional regulator within the synergid cells.
MYB98 Positively Regulates a Battery of Genes within the Synergid Cells
We previously identified a battery of synergid-expressed genes (Steffen et al., 2007), including MYB98 (Kasahara et al., 2005). However, the relationships among these genes within the synergid gene regulatory network were not established. Here, we show that 16 of these genes are downregulated in myb98 synergid cells (Table 2, Figure 3), establishing that they are positively regulated by MYB98. These data also show that MYB98 regulates a large number of genes, at least 16 and presumably more.
Five of the genes downstream of MYB98 (DD3, DD4, DD31, DD39/QRT3, and DD67) are expressed in the absence of MYB98, albeit at much reduced levels (Figure 3; see Supplemental Figure 3 online). The dramatically reduced expression of these genes in myb98 female gametophytes indicates that MYB98 is a central regulator of these genes. The low but detectable expression of these genes in myb98 female gametophytes indicates that other synergid factors can compensate, in part, for the absence of MYB98. Thus, these genes appear to be activated in the synergid cells by multiple factors.
MYB98 Binds to the Sequence TAAC and Directly Regulates DD11
The genes downstream of MYB98 could be regulated either directly or indirectly by MYB98. As a first step toward understanding MYB98 regulation of the downstream genes, we identified MYB98 binding sites in vitro and in vivo. In vitro, MYB98 binds to the sequence YAAC (Y = thymine or cytosine), with a higher affinity for TAAC (Figure 2B). The sequence YAAC is contained within the binding sites of several other R2R3-MYB proteins, including WEREWOLF (Koshino-Kimura et al., 2005; Ryu et al., 2005), MYB2 (Abe et al., 1997), GAMyb (Gubler et al., 1995), MYBGA (Chen et al., 2006), C1 (Roth et al., 1991), P (Grotewold et al., 1994), and MYB.Ph3 (Solano et al., 1997).
To identify in vivo MYB98 binding sites, we dissected the promoter of the DD11 gene. This analysis identified a 36-bp region that contains a MYB98 binding site at position −144 (TAAC−144). Mutations in the core AAC residues of TAAC−144 abolish both MYB98 binding (Figure 2C) and synergid expression (Figure 4E). These data suggest very strongly that MYB98 directly regulates the DD11 gene, although we cannot eliminate the possibility that MYB98 activates the expression of another MYB protein that binds to TAAC−144 in the DD11 promoter.
The MYB98 binding site (TAAC) is unlikely to confer synergid expression on its own. Analysis using Patmatch (http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl) indicates that this sequence is present one or more times in the promoter of essentially every gene within the Arabidopsis genome. Therefore, it is likely that additional cis elements are required to confer synergid-specific expression. Consistent with this, the DD11 promoter contains six TAAC sites (Figure 4A), and only TAAC−144 was shown to be necessary for synergid expression. None of the other TAAC sites appear to be functional: TAAC−77, TAAC−70, and TAAC−36 do not compensate for the loss of TAAC−144 in construct mTAAC−144 (Figure 4E); TAAC−237 does not compensate for the loss of TAAC−144 in the −179 3′ promoter deletion construct (Figure 4C); and deletion of TAAC−390 does not affect expression (Figure 4B). These observations suggest that only TAAC−144 is functional. TAAC−144 may contain adjacent sequences required for optimal MYB98 binding or may be located at an optimal distance from other cis elements. Several other R2R3-MYB proteins, including WEREWOLF and GLABRA1 (Schiefelbein, 2003) and C1 (Grotewold, 2005), interact in a combinatorial fashion with cofactor proteins.
Currently, we do not know whether the other 15 genes are regulated directly or indirectly by MYB98. All of these genes contain multiple TAAC sites in their promoters. However, as discussed above, the same is true for essentially every gene in the Arabidopsis genome, and our analysis of the DD11 promoter has shown that not all TAAC sites are functional; thus, the presence of the TAAC sites in the downstream genes is not indicative of direct regulation. Promoter analysis of the other downstream genes will be required to determine which, if any, are direct targets.
Functions of the Genes Downstream of MYB98
myb98 synergid cells have defects in pollen tube guidance and filiform apparatus formation (Kasahara et al., 2005), suggesting that some or all of the downstream genes play a role in these processes. Consistent with this, five of the MYB98-regulated genes we identified encode proteins that localize to the filiform apparatus. Furthermore, 10 additional genes encode proteins containing putative N-terminal signal peptides, and an N-terminal signal peptide is sufficient to localize GFP to the filiform apparatus, suggesting that some of these untested proteins may also be localized to the filiform apparatus.
Many of the MYB98-regulated genes could feasibly encode a pollen tube attractant. Several of these genes are predicted to encode proteins involved in cell signaling. DD32 encodes a protein similar to the S proteins from Papaver. These proteins are expressed in the stigma, interact with incompatible pollen, and induce a Ca2+-dependent signaling pathway that results in cell death of the incompatible pollen tubes (Thomas et al., 2003; Thomas and Franklin-Tong, 2004). DD2 is a member of the defensin-like family (Thomma et al., 2002). This family encodes proteins similar to defensins, which are small (typically 45 to 50 amino acids), Cys-rich, basic, secreted peptides with conserved secondary structure that function as antimicrobial peptides (Thomma et al., 2002) and signaling molecules (Li et al., 2001; Dresselhaus, 2006). DD42 is predicted to encode a rapid alkalinization factor, and a related protein, RALF, is a secreted polypeptide that may function in signaling (Pearce et al., 2001; Haruta and Constabel, 2003). DD67 encodes a subtilase, and members of this family are involved in the generation of extracellular peptide signals (Berger and Altmann, 2000). In addition, many of the genes of unknown function may encode signaling molecules. Most encode small, Cys-rich proteins with putative N-terminal signal peptides, suggesting that they may be secreted and may play a role in cell signaling (reviewed in Dresselhaus, 2006).
Several of the MYB98-regulated genes may play a role in filiform apparatus formation. Two of these genes are predicted to encode cell wall–modifying proteins. DD39/QRT3 encodes a polygalacturonase involved in the degradation of pectin in plant cell walls (Rhee et al., 2003). DD46 is predicted to encode a galactosyltransferase, and related proteins in Arabidopsis modify cell wall polysaccharides (Edwards et al., 1999; Li et al., 2004). Although the localization of these proteins has not been determined, they may be secreted into the filiform apparatus, because they also have putative N-terminal signal peptides (discussed above). These proteins may function to form or stabilize the specialized cell wall of the filiform apparatus.
The filiform apparatus dramatically increases the surface area of the plasma membrane at the micropylar pole of the synergid cells (see Supplemental Figure 1 online) and is associated with an elaborated endoplasmic reticulum, Golgi stacks, and numerous vesicles. Based on these structural features, it has been proposed that the filiform apparatus facilitates the transport of substances into and out of the synergid cells (Willemse and van Went, 1984; Huang and Russell, 1992). In support of this proposal, micropylar exudates likely from the synergid cells have been observed in Gasteria verrucosa (Franssen-Verheijen and Willemse, 1993). Here, we provide additional evidence. We show that five proteins are secreted into the filiform apparatus (Figures 1E to 1H; see Supplemental Figure 2 online). In addition, it is likely that some of the other 10 signal peptide–containing proteins are also secreted into the filiform apparatus (discussed above). These data indicate that the filiform apparatus functions in transport and that it is the site at which numerous proteins are secreted from the synergid cells.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were surface-sterilized with chlorine gas and germinated on plates containing 0.5× Murashige and Skoog salts (Sigma-Aldrich M-9274), 0.05% MES, 1.0% sucrose, and 0.8% Phytagar (Life Technologies). Ten-day-old seedlings were transferred to Scott's Redi-Earth or Sunshine Mix No. 2 supplemented with MiracleGro and grown under 24 h of illumination. The following T-DNA lines were used: DD4 (SALK_073152), DD18 (SALK_117305), DD31 (SALK_114435), DD39/QRT3 (SAIL_912_G05), DD56 (SALK_023946), and DD67 (SALK_067858).
Plant Transformation
T-DNA constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis plants (accession Columbia-0) were transformed using a modified floral dip procedure (Clough and Bent, 1998). Transformed progeny were selected by germinating surface-sterilized T1 seeds on growth medium containing antibiotics (30 μg/mL kanamycin sulfate or 5 μg/mL BASTA) supplemented with 15 μg/mL cefotaxime. Resistant seedlings were transplanted to soil at 10 d after germination. Transgene identity was verified by PCR using a gene-specific primer and a T-DNA–specific primer.
Expression and Purification of MYB98
A 372-bp region spanning the R2R3 DNA binding domain of MYB98 was amplified from a MYB98 cDNA clone (Kasahara et al., 2005) using primers 98CF (5′-TTACGCCAAGCTCATATGTCATCATGGAAAGAAAC-3′) (introduces a NdeI site) and 98CR (5′-GGAGCTCGAATTCGGATCCTAATACTTAGATCTAC-3′) (introduces a BamHI site). The resulting fragment was ligated into pET28a(+) (Novagen) at the NdeI and BamHI sites to generate clone pET28a.98. The resulting construct was sequenced and introduced into Escherichia coli strain BL21 (DE3) pLysS (Stratagene).
The transformed bacteria were grown to a density of 1.5 at OD600 at 37°C in 50 mL of Luria-Bertani medium, isopropyl-1-thio-β-galactopyranoside was added to a final concentration of 100 nM, and the bacterial culture was incubated at room temperature for 1 h. The culture was then centrifuged to pellet the bacteria, the supernatant was removed, and the bacterial pellet was resuspended in 2 mL of HIS-Bind buffer (20 mM Tris-Cl, 0.5 M NaCl, and 5 mM imidazole, pH 8.0) plus 25 μL of E. coli protease inhibitors (Sigma-Aldrich).
To isolate the recombinant protein, the cells were disrupted using a probe sonicator and centrifuged at 20,000g for 15 min. One hundred microliters of nickel-nitrilotriacetic acid agarose slurry (GE/Amersham) was then added to the supernatant. This mixture was incubated with gentle rocking for 20 min at 4°C. The mixture was then centrifuged, the supernatant was removed, and the pellet was washed three times, once with 1 mL of HIS-Bind buffer followed by two washes with 1 mL of HIS-Wash buffer (20 mM Tris-Cl, 0.5 M NaCl, and 60 mM imidazole, pH 8.0). The pellet was then resuspended in 200 μL of HIS-Elute buffer (20 mM Tris-Cl, 0.5 M NaCl, and 1 M imidazole, pH 8.0) for 10 min at room temperature. This solution was then centrifuged, and the supernatant was dialyzed in storage buffer (20% glycerol, 0.5 mM DTT, 6 mM MgCl2, 50 mM KCl, 0.1 mg/mL BSA, 10 mM Tris-Cl, and 1 mM EDTA, pH 8.0) using a Centriprep centrifugal filter unit with an Ultracel YM-3 membrane (Millipore).
EMSAs
Oligonucleotides for the gel mobility shift assays were obtained from Operon (www.operon.com) and made double-stranded by annealing sense and complementary oligonucleotides in TEN buffer (10 mM Tris-Cl, 1 mM EDTA, and 0.1 M NaCl, pH 8.0) by heating to 95°C for 5 min and slowly cooling to room temperature (∼30 min). Double-stranded oligonucleotides were labeled with digoxigenin using the Roche digoxigenin gel shift kit, second generation (Roche), following the manufacturer's protocol. The binding reactions were performed in binding buffer (10% glycerol, 0.5 mM DTT, 6 mM MgCl2, 50 mM KCl, 0.1 mg/mL BSA, 10 mM Tris-Cl, and 1 mM EDTA, pH 8.0) for 15 min at room temperature. For all reactions, 77 fM of digoxigenin-labeled oligonucleotides, the appropriate concentration of unlabeled oligonucleotides, and 20 ng of protein were mixed in a 10-μL binding reaction. The binding reactions were then incubated on ice for 5 min and were resolved on 1-mm-thick 8% native polyacrylamide gels in 0.5× TBE buffer (44.5 mM Tris base, 44.5 mM boric acid, and 1 mM EDTA, pH 8.0) at 250 V for 20 min using the mini Protean 3 system (Bio-Rad). The oligonucleotides were transferred to Hybond-N+ nylon transfer membranes (GE/Amersham) by electroblotting at 400 mA for 30 min in 0.5× TBE. The oligonucleotides were cross-linked to the nylon membranes using a UV Stratalinker 1800 (Stratagene) at the standard setting (120,000 μJ/cm2). Digoxigenin-labeled oligonucleotides were detected according to the manufacturer's protocol (Roche).
Analysis of GFP Expression Patterns
Plants containing GFP constructs were analyzed using a Zeiss Axioplan compound microscope. GFP was excited using a UV lamp and detected with the 38 HE EGFP filter set; images were captured using an Axiocam MRm REV2 camera with the AxioVision software package version 4.5 (Zeiss). Analysis was performed at 24 h after emasculation of stage 12c flowers (Christensen et al., 1997). For analysis of GFP fluorescence in the synergid cells, we followed the dissection procedure outlined by Steffen et al. (2007).
Protein Fusion Constructs
To generate the MYB98 protein fusion construct, we amplified the promoter region (−686 bp from the start of translation) using primers p98EF1 and p98ER1. This PCR product was cloned into pEGAD (Cutler et al., 2000) using the SacI and AgeI sites (replacing the 35S promoter), resulting in construct ProMYB98:GFP. Next, the MYB98 coding region was amplified from a cDNA clone (Kasahara et al., 2005) using the primers 98Fegad and 98Regad. This fragment was cloned into ProMYB98:GFP using EcoRI and BamHI, resulting in construct ProMYB98:GFP-MYB98. Sequences of primers are listed in Supplemental Table 1 online.
To determine whether the GFP-MYB98 protein fusion complements the myb98-1 phenotype, we introduced the ProMYB98:GFP-MYB98 construct into myb98-1/myb98-1 plants by Agrobacterium-mediated transformation. In the T1 generation, we observed rescue of the pollen tube guidance defect in at least 50% of ovules in 17 independent transformants. In addition, in the T2 generation, we identified plants homozygous for both the myb98-1 mutation and the ProMYB98:GFP-MYB98 construct; these plants had full seed set.
To generate the protein fusion constructs for DD2, DD4, DD11, DD12, and DD32, we amplified the promoter regions and coding regions using the following primers: DD2F and DD2R for DD2, DD4F and DD4R for DD4, DD11F and DD11R for DD11, DD12F and DD12R for DD12, and DD32F and DD32R for DD32. Sequences of primers are listed in Supplemental Table 1 online. In all cases, the PCR primers added unique restriction enzyme sites that were used to ligate the PCR products into the pBI101.gfp vector. These constructs were introduced into wild-type Arabidopsis plants by Agrobacterium-mediated transformation, and at least 10 transgenic lines were analyzed per construct.
To generate the protein fusion construct for the DD11 signal peptide (ProMYB98:SP-GFP), we amplified 700 bp of the MYB98 regulatory region using the following primers: 5941-98F and 5941-98R3. This PCR product was cloned into pFGC5941 (Kerschen et al., 2004) using the EcoRI and NcoI sites (replacing the 35S promoter), resulting in the construct pFGC98. We amplified the GFP coding sequence from pBI101.gfp using the following primers: GFPF1 and GFPR1. This PCR product was cloned into pGFC98 using the BamHI and XbaI sites, resulting in the construct pFGC98-GFP. We synthesized complementary oligonucleotides encompassing the putative N-terminal signal peptide from DD11 (DD11-SP1, 5′-CGCGCCATGGAGAAAGCAATTCTCATAACGTTTCTCATAGCCACCACATCGATGGTTTATCAAACTG-3′; DD11-SP2, 5′-GATCCAGTTTGATAAACCATCGATGTGGTGGCTATGAGAAACGTTATGAGAATTGCTTTCTCCATGG-3′). These oligonucleotides were annealed creating compatible overhangs, which were used to clone the DD11 signal peptide into pGFC98-GFP using the AscI and BamHI sites, resulting in the construct ProMYB98:SP-GFP. All constructs were sequence-verified. The putative DD11 signal peptide consists of the following amino acid sequence: 5′-MEKAILITFLIATTSMVYQT-3′ (PSORT, http://psort.nibb.ac.jp/). This construct was introduced into wild-type Arabidopsis plants by Agrobacterium-mediated transformation, and 17 transgenic lines were analyzed.
Analysis of Promoter:GFP Expression in MYB98/MYB98, MYB98/myb98, and myb98/myb98 Pistils
We first identified single-locus lines for each reporter construct. For each promoter:GFP construct, T2 seeds from multiple T1 lines were plated on growth medium containing kanamycin sulfate (30 μg/mL), the resistant:sensitive ratio was scored, and lines with a resistant:sensitive ratio of 3:1 were transplanted to soil. Eight plants from each line were scored for the percentage of synergid cells expressing GFP to confirm that the reporter construct was present at only a single locus and to identify homozygous plants.
Plants homozygous for the promoter:GFP transgenes were used in a cross with myb98-1/myb98-1 pollen. The resulting F1 plants were hemizygous at the promoter:GFP locus and heterozygous at the MYB98 locus. F1 plants were used as pollen donors in a second cross with myb98-1/myb98-1 females. The resulting F1 plants were genotyped by PCR to identify plants hemizygous for the promoter:GFP locus and homozygous for the myb98-1 mutation using primers described by Kasahara et al. (2005).
When scoring the percentage of female gametophytes with GFP fluorescence in the synergid cells, at least three pistils were analyzed from at least three individuals per generation.
Promoter Deletion Constructs
Promoter fragments were generated by PCR using the primers listed in Supplemental Table 2 online. The primers introduced unique restriction enzyme sites. The PCR products were digested and ligated with pBI101.GFP (for 5′ deletions) or 35SmpGFP (for 3′ deletions) (Yadegari et al., 2000). All constructs were sequence-verified. The 5′ deletion constructs were fused with a GFP coding region. The 3′ deletion constructs were fused with the minimal promoter:GFP construct (mpGFP), which contained a GFP coding region fused at its 5′ end with a minimal promoter derived from the cauliflower mosaic virus 35S promoter (Benfey and Chua, 1990).
Real-Time RT-PCR
Pistils were harvested from ms1/ms1 and myb98-1/myb98-1 plants and placed immediately into liquid nitrogen. Pistils were harvested from the oldest stage 12 flower (Smyth et al., 1990) on each plant as well as the next two older flowers.
RNA was extracted from the pistil tissue using the Qiagen RNeasy kit following the manufacturer's protocol (www.qiagen.com). DNA contamination was removed from samples using the Ambion TURBO DNA-free DNase kit following the manufacturer's protocol. After DNase treatment, RNA samples were repurified using the Qiagen RNeasy kit following the manufacturer's protocol. Aliquots of RNA (1 μg) were reverse-transcribed using the RETROscript kit (Ambion) following the manufacturer's protocol.
For real-time RT-PCR, PCR was performed using the Roche FastStart DNA Master SYBR Green I master mix (www.roche.com) in a volume of 10 μL on a Roche LightCycler system. The PCR mixture consisted of 0.5 μL of cDNA, 0.5 μM primers, and 1× master mix. For each real-time RT-PCR run, ACTIN2 (At3g18780) was used as an internal control to normalize for any pipetting error of the cDNA template. The PCR program consisted of 95°C for 5 min followed by 45 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 10 s. To determine the specificity of the PCR, the amplified products were subjected to melt curve analysis using the machine's standard method as well as run on a gel. The reported threshold cycle values are averages of two independent trials (biological replicates). For DD18 and DD32, expression was not detected in myb98-1 ovules. In these cases, a threshold cycle value of 40 was used to calculate fold change. All real-time RT-PCR primers were the same as reported by Steffen et al. (2007).
Identification of TAAC Sites within the Arabidopsis Genome
We used Patmatch 1.1 (http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl) to search for the sequence TAAC within the Locus Upstream Sequences −500bp (DNA) database. This analysis identified 28,180 genes containing this sequence in their promoters.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Depiction of the Synergid Cells and the Filiform Apparatus.
Supplemental Figure 2. Localization of GFP Fusion Proteins in Wild-Type Ovules.
Supplemental Figure 3. Intensity of the GFP Signal from ProDD3:GFP in MYB98 and myb98-1 Synergid Cells.
Supplemental Table 1. Primers Used to Generate the Protein Fusion Constructs.
Supplemental Table 2. Primers Used to Generate the 5′ and 3′ Deletions of the DD11 Promoter.
Supplementary Material
Acknowledgments
We thank Ramin Yadegari, Jaimie Van Norman, and members of the Drews laboratory for critical review of the manuscript. We thank Ramin Yadegari for providing the pBI101.GFP and 35SmpGFP vectors. We thank the ABRC for providing the T-DNA insertion alleles and pFGC5941. This work was supported in part by a National Science Foundation grant (Grant IOB-0542953) to G.N.D. and by a National Institutes of Health Developmental Biology Training Grant (Grant 5T32 HD-07491) appointment to J.A.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gary N. Drews (drews@bioscience.utah.edu).
Online version contains Web-only data.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250 959–966. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Biedenkapp, H., Borgmeyer, U., Sippel, A.E., and Klempnauer, K.H. (1988). Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335 835–837. [DOI] [PubMed] [Google Scholar]
- Chen, P.W., Chiang, C.M., Tseng, T.H., and Yu, S.M. (2006). Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 18 2326–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cordts, S., Bantin, J., Wittich, P.E., Kranz, E., Lorz, H., and Dresselhaus, T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25 103–114. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter, S., and Angenent, G.C. (2006). Trans meets cis in MADS science. Trends Plant Sci. 11 224–231. [DOI] [PubMed] [Google Scholar]
- Dias, A.P., Braun, E.L., McMullen, M.D., and Grotewold, E. (2003). Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 131 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus, T. (2006). Cell-cell communication during double fertilization. Curr. Opin. Plant Biol. 9 41–47. [DOI] [PubMed] [Google Scholar]
- Edwards, M.E., Dickson, C.A., Chengappa, S., Sidebottom, C., Gidley, M.J., and Reid, J.S. (1999). Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19 691–697. [DOI] [PubMed] [Google Scholar]
- Franssen-Verheijen, M.A.W., and Willemse, M.T.M. (1993). Micropylar exudate in Gasteria (Aloaceae) and its possible function in pollen tube growth. Am. J. Bot. 80 253–262. [Google Scholar]
- Freeman, M. (2000). Feedback control of intercellular signalling in development. Nature 408 313–319. [DOI] [PubMed] [Google Scholar]
- Grotewold, E. (2005). Plant metabolic diversity: A regulatory perspective. Trends Plant Sci. 10 57–62. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta, M., and Constabel, C.P. (2003). Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol. 131 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T. (2002). The synergid cell: Attractor and acceptor of the pollen tube for double fertilization. J. Plant Res. 115 149–160. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). Pollen-tube guidance: Beacons from the female gametophyte. Curr. Opin. Plant Biol. 6 36–41. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293 1480–1483. [DOI] [PubMed] [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140 233–292. [Google Scholar]
- Huck, N., Moore, J.M., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130 2149–2159. [DOI] [PubMed] [Google Scholar]
- Jia, L., Clegg, M.T., and Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 5 R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M.A., and Preuss, D. (2002). Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Kasahara, R.D., Portereiko, M.F., Sandaklie-Nikolova, L., Rabiger, D.S., and Drews, G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschen, A., Napoli, C.A., Jorgensen, R.A., and Muller, A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566 223–228. [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura, Y., Wada, T., Tachibana, T., Tsugeki, R., Ishiguro, S., and Okada, K. (2005). Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 46 817–826. [DOI] [PubMed] [Google Scholar]
- Le, Q., Gutierrez-Marcos, J.F., Costa, L.M., Meyer, S., Dickinson, H.G., Lorz, H., Kranz, E., and Scholten, S. (2005). Construction and screening of subtracted cDNA libraries from limited populations of plant cells: A comparative analysis of gene expression between maize egg cells and central cells. Plant J. 44 167–178. [DOI] [PubMed] [Google Scholar]
- Li, P., Chan, H.C., He, B., So, S.C., Chung, Y.W., Shang, Q., Zhang, Y.D., and Zhang, Y.L. (2001). An antimicrobial peptide gene found in the male reproductive system of rats. Science 291 1783–1785. [DOI] [PubMed] [Google Scholar]
- Li, X., Cordero, I., Caplan, J., Molhoj, M., and Reiter, W.D. (2004). Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol. 134 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, E.M., and Russell, S.D. (2002). The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18 81–105. [DOI] [PubMed] [Google Scholar]
- Marton, M.L., Cordts, S., Broadhvest, J., and Dresselhaus, T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307 573–576. [DOI] [PubMed] [Google Scholar]
- McCormick, S., and Yang, H. (2005). Is there more than one way to attract a pollen tube? Trends Plant Sci. 10 260–263. [DOI] [PubMed] [Google Scholar]
- Oda, M., Furukawa, K., Ogata, K., Sarai, A., and Nakamura, H. (1998). Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 276 571–590. [DOI] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Pearce, G., Moura, D.S., Stratmann, J., and Ryan, C.A., Jr. (2001). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.Y., Osborne, E., Poindexter, P.D., and Somerville, C.R. (2003). Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B.A., Goff, S.A., Klein, T.M., and Fromm, M.E. (1991). C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell 3 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F., and Faure, J.E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13 432–436. [DOI] [PubMed] [Google Scholar]
- Ryu, K.H., Kang, Y.H., Park, Y.H., Hwang, I., Schiefelbein, J., and Lee, M.M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132 4765–4775. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6 74–78. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A., Moskal, W.A., Jr., Wu, H.C., Underwood, B.A., Graham, M.A., Town, C.D., and Vandenbosch, K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51 262–280. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Fuertes, A., Sanchez-Pulido, L., Valencia, A., and Paz-Ares, J. (1997). A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 272 2889–2895. [DOI] [PubMed] [Google Scholar]
- Steffen, J.G., Kang, I.-H., Macfarlane, J., and Drews, G.N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51 281–292. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Tanikawa, J., Yasukawa, T., Enari, M., Ogata, K., Nishimura, Y., Ishii, S., and Sarai, A. (1993). Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA 90 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S., Osman, K., de Graaf, B.H., Shevchenko, G., Wheeler, M., Franklin, C., and Franklin-Tong, N. (2003). Investigating mechanisms involved in the self-incompatibility response in Papaver rhoeas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., and Franklin-Tong, V.E. (2004). Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 429 305–309. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Cammue, B.P., and Thevissen, K. (2002). Plant defensins. Planta 216 193–202. [DOI] [PubMed] [Google Scholar]
- Weston, K. (1992). Extension of the DNA binding consensus of the chicken c-Myb and v-Myb proteins. Nucleic Acids Res. 20 3043–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings, K., and Russell, S.D. (2004). Experimental analysis of the fertilization process. Plant Cell 16 (suppl.): S107–S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 159–196.
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Kaur, N., Kiriakopolos, S., and McCormick, S. (2006). EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224 1004–1014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.