Abstract
Wounding plant tissues initiates large-scale changes in transcription coupled to growth arrest, allowing resource diversion for defense. These processes are mediated in large part by the potent lipid regulator jasmonic acid (JA). Genes selected from a list of wound-inducible transcripts regulated by the jasmonate pathway were overexpressed in Arabidopsis thaliana, and the transgenic plants were then assayed for sensitivity to methyl jasmonate (MeJA). When grown in the presence of MeJA, the roots of plants overexpressing a gene of unknown function were longer than those of wild-type plants. When transcript levels for this gene, which we named JASMONATE-ASSOCIATED1 (JAS1), were reduced by RNA interference, the plants showed increased sensitivity to MeJA and growth was inhibited. These gain- and loss-of-function assays suggest that this gene acts as a repressor of JA-inhibited growth. An alternative transcript from the gene encoding a second protein isoform with a longer C terminus failed to repress jasmonate sensitivity. This identified a conserved C-terminal sequence in JAS1 and related genes, all of which also contain Zim motifs and many of which are jasmonate-regulated. Both forms of JAS1 were found to localize to the nucleus in transient expression assays. Physiological tests of growth responses after wounding were consistent with the fact that JAS1 is a repressor of JA-regulated growth retardation.
INTRODUCTION
Jasmonic acid (JA) is an important effector in plant defense processes and is well known as a regulator of defense gene expression (Devoto and Turner, 2003; Lorenzo and Solano, 2005). However, a series of historically important experiments demonstrated the ability of JA and its methyl ester methyl jasmonate (MeJA) to inhibit plant growth (Yamane et al., 1980; Dathe et al., 1981; Ueda and Kato, 1982), and JA became known to plant biologists as a potential growth regulator. This effect of JA in blocking plant growth was subsequently exploited in many forward genetic screens for plants with a decreased sensitivity to JA. In the first of these screens, the inhibitory effect of MeJA on wild-type Arabidopsis thaliana root growth permitted the isolation of the mutant jasmonic acid–resistant1 (jar1) with decreased sensitivity to MeJA (Staswick et al., 1992). Another early root growth inhibition screen yielded mutants with extreme insensitivity to the bacterial virulence factor coronatine as well as to MeJA (Feys et al., 1994). These coronatine-insensitive (coi) mutants displayed several floral developmental phenotypes, including shorter than wild-type anthers, and provided genetic evidence for the bona fide involvement of JA in growth. Continuing efforts to understand the roles of JA in anther filament elongation in Arabidopsis have led to the identification of genes likely to be involved in filament growth and its regulation (Browse, 2005; Mandaokar et al., 2006). JA signaling has also been found to control female reproductive development in tomato (Solanum lycopersicum), extending the known roles of JA in development (Li et al., 2004). The growth of vegetative tissues is also affected by treatment with jasmonates. Plants treated with MeJA develop shorter petioles than do control plants (Cipollini, 2005), and the short-petiole phenotype is observed in some genetically characterized mutants that accumulate higher than wild-type levels of JA in response to wounding (Bonaventure et al., 2007). Growth inhibition mediated by JA following insult makes sense in that it allows more resource allocation for defense in plants under attack by herbivores (Zavala and Baldwin, 2006).
At the mechanistic level, there are some indications that growth inhibition might occur at the level of the cell cycle. For example, MeJA blocked both epiphyllous bud formation and leaf growth in Bryophyllum diagremontianum, but cotreatment with a cytokinin allowed leaf growth and bud development to proceed, suggesting an inhibitory effect of MeJA on cell division (Saniewski, 1988). More recent studies have shown that treatment with exogenous JA can block both the G1/S and G2/M transitions in the cell cycle (Świątek et al., 2002). Furthermore, the sensitivity of cells to JA was found to be cell cycle–dependent, with cells being less sensitive to JA after S-phase (Świątek et al., 2004). There is also an interesting connection between cell expansion, cell division, and the jasmonate pathway. A null mutation in the COBRA gene strongly affects orientated expansion and cell division in hypocotyls and petioles and causes JA to accumulate to levels far exceeding those in the wild type (Ko et al., 2006). This is where our knowledge of how, at the mechanistic level, MeJA/JA inhibits growth comes to an end. During the defense response and paralleling the effects of attack on the downregulation of growth, JA participates in the modulation of defense gene expression and in the control of tritrophic interactions, whereby herbivore-injured plants can attract predatory insects (Devoto and Turner, 2003; Browse, 2005; Lorenzo and Solano, 2005). JA also stimulates the expression of several JA biosynthesis genes via a positive feedback loop (Laudert and Weiler, 1998; Sasaki et al., 2001). While defense gene expression and JA biosynthesis have received much recent attention, the mechanism of JA-mediated growth inhibition is largely unknown.
Here, we used a MeJA-based growth inhibition screen for transgenic Arabidopsis plants generated after the selection of candidate JA signaling genes from microarray experiments. The objective was to identify genes involved in the growth response to jasmonates and/or in growth responses to conditions that activate the jasmonate pathway (e.g., wounding). This work made use of the allene oxide synthase (aos) jasmonate biosynthesis mutant described by Park et al. (2002). This mutation provides a plant in which JA accumulation is impaired in both resting and wounded leaves. Furthermore, the plants can be maintained from seed to seed as homozygotes for the mutation by spraying developing flowers with MeJA. The strategy used herein was based on four steps whereby (1) microarrays were used to define a list of genes regulated by jasmonates in wounded and unwounded tissues; (2) genes of unknown function from the list were selected and plants overexpressing these genes were constructed; (3) the seeds of these plants were sown on growth medium containing MeJA and plants showing reduced growth inhibition by MeJA were selected; and (4) the candidate genes were characterized further using transcript downregulation by RNA interference (RNAi). Using this approach, one overexpressed cDNA conferred strong resistance to exogenous MeJA. Conversely, transforming plants with RNAi constructs for the gene resulted in hypersensitivity to MeJA. The gene product is likely to act as a key modulator of jasmonate-controlled growth inhibition in response to wounding.
RESULTS
A Microarray-Based Screen for Jasmonate-Regulated Transcripts
Large-scale Arabidopsis microarrays (Hilson et al., 2004) were used to identify candidate genes for JA signaling responses. Transcripts regulated by JA in both wounded and resting leaves were identified by comparison of gene expression in wild-type and aos mutant (Park et al., 2002) plants. Transcript levels were assessed in tissues harvested at 1 h after wounding. We first identified a list of transcripts that were wound-inducible in wild-type plants, choosing a cutoff of twofold change in gene expression and a P value of <0.05 (Student's t test) from three independent replicated experiments. Another list of wound-inducible genes was established for aos plants from three independent replicated experiments. Combining the two lists produced a subset of 814 transcripts that were wound-inducible in wild-type plants but not wound-inducible in aos plants (i.e., these genes were wound-inducible in a jasmonate-dependent manner). In order to produce a shorter and more robust gene list, we performed an additional microarray experiment designed to find transcripts regulated by jasmonates in resting, unwounded leaves. For this, relative transcript levels in the resting leaves of wild-type plants were compared with transcript levels in the resting leaves of aos plants in four biologically independent experiments. This yielded 77 differentially regulated genes that were significantly less expressed (less than a twofold change; P < 0.05) in aos plants. There was an overlap of 35 genes between the two gene lists (814 wound-induced jasmonate-dependent genes and 77 jasmonate-regulated genes in resting leaves).
These 35 genes are listed in Table 1 and represented a starting list from which candidate signaling genes were chosen. The top 15 genes that were the most differentially regulated between unwounded wild-type plants and aos plants (Table 1) were selected, and candidate genes potentially involved in signal transduction were chosen from this short list. This first involved the elimination of genes with known roles in oxylipin biosynthesis (AOS and HYDROPEROXIDE LYASE), a gene annotated as a pseudogene, a gene potentially involved in metabolism (phosphorylase), a gene annotated as an auxin conjugate hydrolase (indole-3-acetic acid-Ala hydrolase), and genes likely to be involved in direct defense (myrosinase binding protein and trypsin inhibitor). This left eight genes, of which seven were selected for the production of transgenic plants. The only gene from the short list that was not selected encoded one of three related expressed proteins. Since they were related, only two of these genes were tested. Plants overexpressing each of these genes were constructed in order to investigate their possible roles in JA-mediated growth responses.
Table 1.
List of Genes Differentially Expressed between aos and Col-0
| Wounding (60 min)
|
||||||
|---|---|---|---|---|---|---|
| Locus | Description | aos:Col-0a | Pb | Col-0:ctla | aos:ctla | Pc |
| At2g24850 | Aminotransferase, putative | 0.126 | 5.1E-05 | 12.58 | 2.60 | 0.0312 |
| At5g42650 | Allene oxide synthase (AOS) | 0.151 | 4.2E-05 | 5.22 | 0.63 | 0.0112 |
| At5g47240 | MutT/nudix family protein | 0.171 | 3.6E-04 | 10.30 | 1.26 | 0.0028 |
| At1g74950 | Expressed protein | 0.178 | 6.3E-03 | 5.72 | 1.11 | 0.0026 |
| At1g52030 | Myrosinase binding protein | 0.185 | 0.0034 | 3.07 | 0.83 | 0.0094 |
| At5g34831 | Pseudogene, hypothetical protein | 0.194 | 3.6E-05 | 10.33 | 1.04 | 0.0229 |
| At4g24350 | Phosphorylase family protein | 0.217 | 2.1E-05 | 4.63 | 0.85 | 0.0006 |
| At2g38750 | Annexin 4 (ANN4) | 0.235 | 0.0008 | 9.79 | 1.05 | 0.0387 |
| At1g72450 | Expressed protein | 0.249 | 0.0257 | 5.58 | 1.58 | 0.0251 |
| At4g15440 | Hydroperoxide lyase (HPL1) | 0.259 | 5.0E-05 | 4.75 | 1.20 | 0.0159 |
| At1g43160 | AP2 domain–containing protein RAP2.6 | 0.280 | 1.2E-05 | 15.19 | 1.16 | 0.0008 |
| At2g38240 | Oxidoreductase, 2OG-Fe-oxygenase | 0.283 | 0.0004 | 35.72 | 1.13 | 0.0052 |
| At2g43530 | Trypsin inhibitor, putative | 0.305 | 0.0019 | 5.57 | 0.85 | 0.0058 |
| At5g13220 | Expressed protein | 0.307 | 0.0012 | 15.60 | 1.02 | 0.0006 |
| At1g51760 | Indole-3-acetic acid-Ala hydrolase (IAR3) | 0.310 | 0.0070 | 12.57 | 1.63 | 0.0017 |
| At3g16470 | Jacalin lectin | 0.335 | 0.0017 | 3.64 | 0.90 | 0.0069 |
| At1g20510 | 4-Coumarate-CoA ligase | 0.342 | 0.0081 | 3.36 | 1.47 | 0.0492 |
| At2g43550 | Trypsin inhibitor, putative | 0.344 | 0.0009 | 2.71 | 0.61 | 0.0074 |
| At1g66760 | MATE efflux family | 0.359 | 0.0344 | 7.65 | 1.56 | 0.0065 |
| At2g39330 | Jacalin lectin | 0.381 | 0.0359 | 3.02 | 0.84 | 0.0044 |
| At5g53750 | Expressed protein | 0.385 | 0.0167 | 11.13 | 1.10 | 0.0002 |
| At4g10390 | Putative protein kinase | 0.394 | 0.0100 | 16.25 | 1.43 | 0.0111 |
| At5g10300 | Hydrolase, α/β-fold family | 0.395 | 0.0012 | 5.32 | 0.83 | 0.0012 |
| At2g46510 | Basic helix-loop-helix (bHLH) family | 0.409 | 0.0411 | 5.02 | 1.53 | 0.0006 |
| At2g39030 | GCN5-related N-acetyltransferase (GNAT) | 0.417 | 0.0142 | 4.97 | 1.08 | 0.0179 |
| At2g22330 | Cytochrome P450, putative | 0.417 | 0.0006 | 4.85 | 1.03 | 0.0040 |
| At4g16760 | Acyl-CoA oxidase (ACX1) | 0.418 | 0.0007 | 2.50 | 1.12 | 0.0334 |
| At1g52890 | No apical meristem (NAM) family | 0.423 | 0.0048 | 5.69 | 0.97 | 0.0015 |
| At4g08170 | Inositol 1,3,4-trisphosphate 5/6-kinase | 0.426 | 0.0056 | 11.33 | 1.08 | 0.0006 |
| At1g32640 | Basic helix-loop-helix protein (RAP-1) | 0.435 | 0.0135 | 3.56 | 1.16 | 0.0358 |
| At5g54170 | Expressed protein | 0.438 | 0.0356 | 11.70 | 1.32 | 0.0003 |
| At4g39980 | Dehydrodeoxyphosphoheptonate aldolase | 0.440 | 0.0015 | 4.98 | 0.99 | 0.0002 |
| At2g20340 | Tyrosine decarboxylase, putative | 0.456 | 0.0087 | 8.12 | 0.88 | 0.0003 |
| At2g32150 | Haloacid dehalogenase-like hydrolase | 0.474 | 0.0016 | 2.76 | 0.90 | 0.0077 |
| At2g29440 | Glutathione S-transferase, putative | 0.478 | 0.0219 | 14.73 | 1.97 | 0.0069 |
Col-0, Columbia-0. The list shows genes that were expressed at a lower level (ratio ≤ 0.5, P < 0.05) in the resting (control [ctl]) leaves of aos than in resting leaves of Col-0 and that were wound-inducible in Col-0 but significantly less or not wound-inducible in aos (P < 0.05). cDNAs for genes listed in boldface were overexpressed in plants for MeJA sensitivity experiments.
Expression ratio.
P values (Student's t test, one-sample hypothesis) were calculated by comparing aos:Col-0 ratios (n = 4) with the expected value of 1, indicating no change in gene expression.
P values (Student's t test, two-sample hypothesis) were calculated by comparing expression ratios from Col-0:ctl (n = 3) with expression ratios from aos:ctl (n = 3) in wounding experiments.
Construction and Testing of Plants Overexpressing Candidate Genes
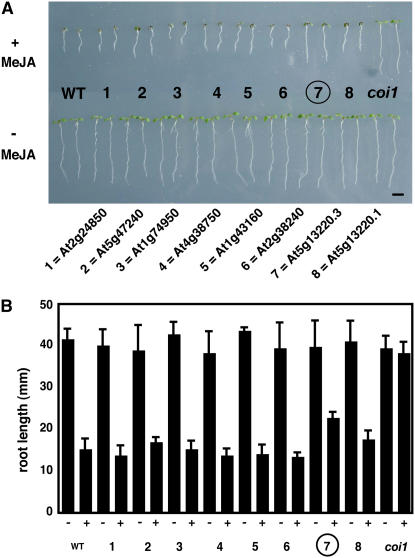
To generate plants overexpressing candidate transcripts, we cloned cDNAs selected from the genes shown in boldface in Table 1 using gene-specific primers (see Methods). Single transcripts were predicted for each of the genes except At5g13220. Multiple transcripts are predicted for this gene (The Arabidopsis Information Resource, http://www.arabidopsis.org) and are predicted to encode two different protein isoforms. Two of the transcripts for this gene were cloned and labeled At5g13220.1 and At5g13220.3, in accordance with the published gene annotation. Each of the eight cDNAs (representing seven genes) were then cloned under the control of the 35S promoter and transformed into Arabidopsis (Weigel and Glazebrook, 2002). T2 seeds (selfed T1 plants) were then screened by growing them on medium containing 25 μM MeJA. MeJA strongly inhibits the growth of wild-type plants but not that of plants insensitive to MeJA, such as the coi1-1 mutant (Feys et al., 1994). One transformant with a cDNA from At5g13220.3 revealed a strong jasmonate-insensitivity phenotype in the assay (Figure 1, group 7). Quantitation of primary root growth confirmed this observation. The roots of the At5g13220.3 transformants grew longer than wild-type roots in the presence of MeJA. By contrast, plants transformed with a second natural cDNA from the At5g13220 gene (At5g13220.1) did not show a strong MeJA-insensitivity phenotype (Figure 1, group 8). This second transcript is predicted to encode a larger protein than the protein from the At5g13220.3 transcript (http://www.Arabidopsis.org). In order to confirm that the expression of At5g13200.1 had been affected, a second transgenic line was tested. Again, this did not recapitulate the effect of the At5g13220.3 transcript (data not shown).
Figure 1.
Screen for MeJA Insensitivity in Plants Transformed with Jasmonate-Regulated cDNAs.
Root growth of transgenic plants in the absence and presence of MeJA (25 μM). Control plants were either wild type, displaying MeJA-inhibited root growth, or coi1-1, displaying insensitivity to MeJA. The intermediate phenotype of the At5g13220.3 overexpressing line is indicated with an encircled 7.
(A) Photographic image of 7-d-old seedlings. Bar = 5 mm.
(B) Primary root lengths of 10-d-old seedlings. −, no MeJA in growth medium; +, MeJA (25 μM) in growth medium (n = 10; error bars indicate sd).
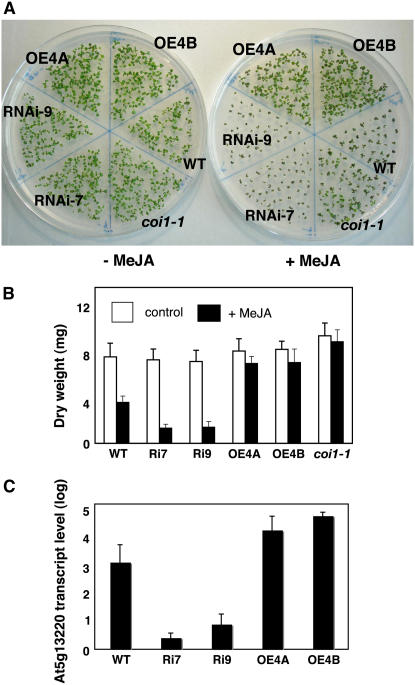
Two independent overexpressing lines for At5g13220.3 (lines OE4A and OE4B) showed a strong MeJA-insensitivity phenotype (Figure 2A). In addition, in order to show that the At5g13220 gene encodes a repressor of MeJA sensitivity, we produced plants in which levels of At5g13220 transcripts were reduced by RNAi. Two independent transgenic plants expressing an RNAi construct for At5g13220.3 (RNAi-7 and RNAi-9) were found to be more sensitive to MeJA than wild-type plants. When grown on MeJA, these plants displayed a greater than wild-type sensitivity to MeJA (Figure 2A). These growth differences among coi1-1, the wild type, and At5g13220.3 transgenic lines were quantitated in plants grown in the presence or absence of MeJA (Figure 2B). The measurements confirmed that overexpression of At5g13220.3 decreased sensitivity to MeJA, whereas RNAi constructs for the gene increased MeJA sensitivity. Quantitative PCR was used to verify that overexpression and RNAi constructs affected transcript levels for At5g13220. In the overexpression lines OE4A and OE4B, transcript levels were ∼8- and 18-fold higher than those in wild-type plants. Conversely, transcript levels in the RNAi-7 and RNAi-9 lines were >200-fold lower than those in wild-type plants (Figure 2C).
Figure 2.
RNAi and Overexpression Constructs for At5g13220 Differentially Affect Sensitivity to MeJA.
(A) Seeds were germinated on agar containing either no MeJA (left plate) or MeJA (25 μM; right plate) for 7 d, at which time they were photographed. Wild-type and a coi1-1 segregating seeds were used as controls for MeJA sensitivity.
(B) Dry weight of aerial tissues for plants grown in the absence or presence of MeJA (25 μM). Plants were grown for 21 d prior to harvest (n = 12; error bars indicate sd). Ri7, RNAi-7; Ri9, RNAi-9; OE4A, overexpression line 4A; OE4B, overexpression line 4B.
(C) Transcript levels relative to EIF4A1 for At5g13220 (all transcript forms) in wild-type and transgenic plants. Quantitative PCR was used to measure relative transcript abundance in 4-week-old soil-grown plants (n = 3; error bars indicate sd). Transcript abundance was not normalized to EIF4A1 transcript abundance.
Specificity and Range of Action of the Repressor of Jasmonate-Inhibited Growth
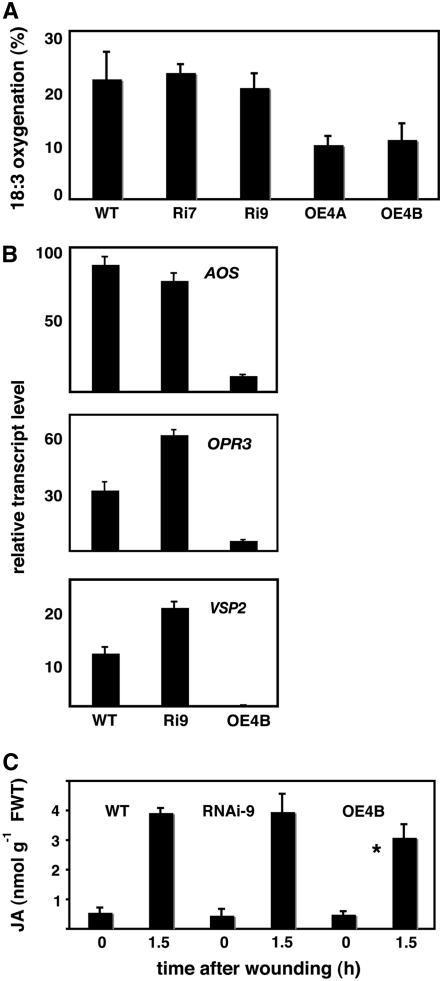
Does the overexpression of At5g13220.3 affect the sensitivity of plants to treatments other than with MeJA? Wild-type, RNAi, and overexpressing plants were grown for 7 d in the absence or presence of NaCl (0.1 M). Salt inhibited primary root growth to a similar extent in all three genotypes. As controls, we used aos seeds. The primary roots of aos displayed slightly reduced growth inhibition relative to the wild type in the presence of NaCl (P = 0.008; see Supplementary Figure 1 online). Similar tests of primary root growth on salicylic acid and on auxin (indole-3-acetic acid) were also conducted. In both cases, overexpression of At5g13220.3 did not affect the plants' sensitivity to these substances (see Supplemental Figure 1 online). Together, the experiments showed that the At5g13220.3 gene product could modulate the plants' growth response to exogenous MeJA and that this was largely specific for MeJA rather than a nonspecific response to other stresses and hormones. However, the results did not show whether endogenous JA-related processes were affected in the absence of exogenous MeJA. The ability of plant tissues to catalyze the first steps in the oxygenation of the jasmonate precursor α-linolenic acid was analyzed in a simple assay (Caldelari and Farmer, 1998) (see Methods). This revealed that the lines OE4A and OE4B overexpressing At5g13220.3 both showed a reduced ability to oxygenate the fatty acid (Figure 3A). Transcript levels in resting leaves for two enzymes of jasmonate biosynthesis, AOS and OXOPHYTODIENOIC ACID REDUCTASE3 (OPR3), as well as for VEGETATIVE STORAGE PROTEIN2 (VSP2) were found to be downregulated in plants overexpressing the repressor. In the cases of OPR3 and VSP2, the levels of the transcripts were slightly higher in RNAi plants (Figure 3B). Quantitative JA measurements revealed no differences in JA levels in the resting leaves of wild-type, overexpressing, and RNAi plants. However, a statistically significant reduction in the level of JA at 90 min after wounding was observed in plants overexpressing At5g13220.3 (Figure 3C).
Figure 3.
Overexpression of At5g13220.3 Affects the Activity of JA Biosynthesis Enzymes, AOS, OPR3, and VSP2 Transcript Levels, and JA Synthesis in Response to Wounding.
(A) Extracts of resting leaves were assayed for their ability to oxygenate the jasmonate precursor α-linolenic acid (18:3) in vitro (n = 4; error bars indicate sd).
(B) Relative transcript levels in resting leaves for two JA biosynthesis genes (AOS and OPR3) and for the VSP2 gene (n = 3; error bars indicate sd).
(C) JA measured in leaves before and after wounding. The wound-induced level of JA at 1.5 h was significantly lower in plants overexpresssing the At5g13320.3 transcript than in wild-type plants (*P = 0.035; n = 3; error bars indicate sd). FWT, fresh weight.
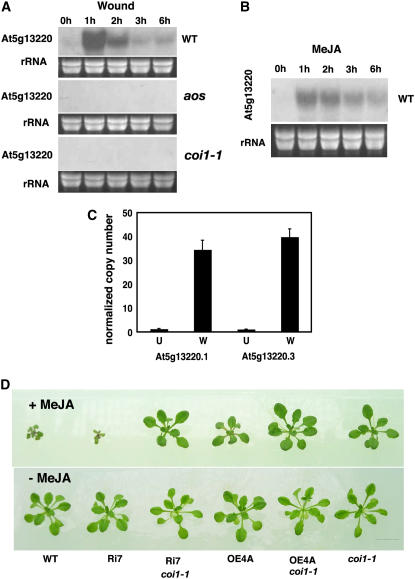
At5g13220 Gene Regulation by the Jasmonate Pathway
From microarray studies, the At5g13220 gene itself was known to be expressed under the control of the JA signal pathway. We examined At5g13220 transcript levels in wild-type plants, in the jasmonate biosynthesis mutant aos, and in the coi1-1 JA perception mutant. The strong wound-inducible expression of the gene was clearly both jasmonate- and COI1-dependent (Figure 4A). When plants were treated with MeJA, transcripts for the At5g13220 gene accumulated transiently, peaking in the experiment at 1 h after treatment and then subsiding to nearly basal levels (Figure 4B). The levels of the At5g13220.1 and At5g13220.3 transcripts normalized to the levels of transcripts for an elongation factor gene were measured in leaves before and 1 h after wounding. Levels of both transcripts increased to similar levels (with ∼30-fold inductions) after wounding (Figure 4C). We then attempted to define the relationship of At5g13220 in JA signaling relative to the COI1 reference gene by generating RNAi-7 coi1-1 and OE4A coi1-1 double mutant plants (see Methods). These plants, along with appropriate controls (wild type, RNAi-7, OE4A, and coi1-1), were grown in parallel in the presence and absence of MeJA. Both the RNAi-7 coi1-1 and the OE4A coi1-1 plants grew like coi1-1 plants in the presence of MeJA (Figure 4D).
Figure 4.
Effects of MeJA and Wounding on At5g13220 Transcript Levels and of MeJA Treatment on At5g13220/coi1-1 Plants.
(A) Mechanical wounding activates At5g13220 expression in wild-type plants but not in the JA biosynthesis mutant aos or the JA perception mutant coi1-1. Plants (5 weeks old) were wounded on the leaf apices (see Methods).
(B) MeJA treatment increases transcript levels. Plants (5 weeks old) were sprayed with a suspension of MeJA (100 μM in water). For all experiments, total RNA (15 μg) preparations were analyzed by RNA gel blotting.
(C) Copy number of transcripts for At5g13220.1 and At5g13220.3 normalized to 100 transcripts for EIF4A1 (n = 6; error bars indicate se). U, unwounded leaves; W, leaves wounded and harvested after 1 h.
(D) Effect of crossing At5g13220.3 overexpressing plants into a background lacking a functional COI1 gene. Plants were grown in the presence (top panel) or absence (bottom panel) of MeJA (25 μM) for 21 d. Bar = 1 cm.
Characteristics and Cellular Localization of the Protein Isoforms
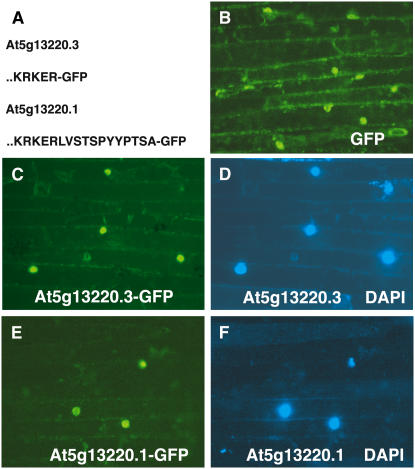
The predicted proteins from the two transcripts were compared (Figure 5A), and this showed that the protein encoded by At5g13220.3 lacked 12 C-terminal amino acids relative to the At5g13220.1 protein. This difference had only a minor effect on the predicted physical properties of the protein (mass and isoelectric point). However, the difference between the two proteins raised the possibility that their intracellular localizations could differ. The cellular localization of the At5g13220.1 and At5g13220.3 proteins was investigated using fusion proteins in which C-terminal green fluorescent protein (GFP) was used as a marker. These constructs were expressed transiently after biolistic delivery to onion (Allium cepa) cells. Both fusion proteins were found to be reproducibly localized to the nucleus of the onion cells in independent experiments (Figures 5B to 5F).
Figure 5.
Cellular Localization of GFP Coupled to At5g13220 Gene Products.
(A) C-terminal sequences encoded by two alternative transcripts of the At5g13220 gene fused to GFP. Only the C-terminal amino acids of the At5g13220 gene products are indicated.
(B) Fluorescence of onion cells expressing GFP constructs.
(C) GFP fluorescence of the At5g13220.3-GFP fusion.
(D) 4′,6-Diamidino-2-phenylindole (DAPI) fluorescence of cells expressing the At5g13220.3-GFP fusion.
(E) GFP fluorescence of the At5g13220.1-GFP fusion.
(F) DAPI fluorescence of cells expressing the At5g13220.1-GFP fusion.
Role of At5g13220 in the Wound Response
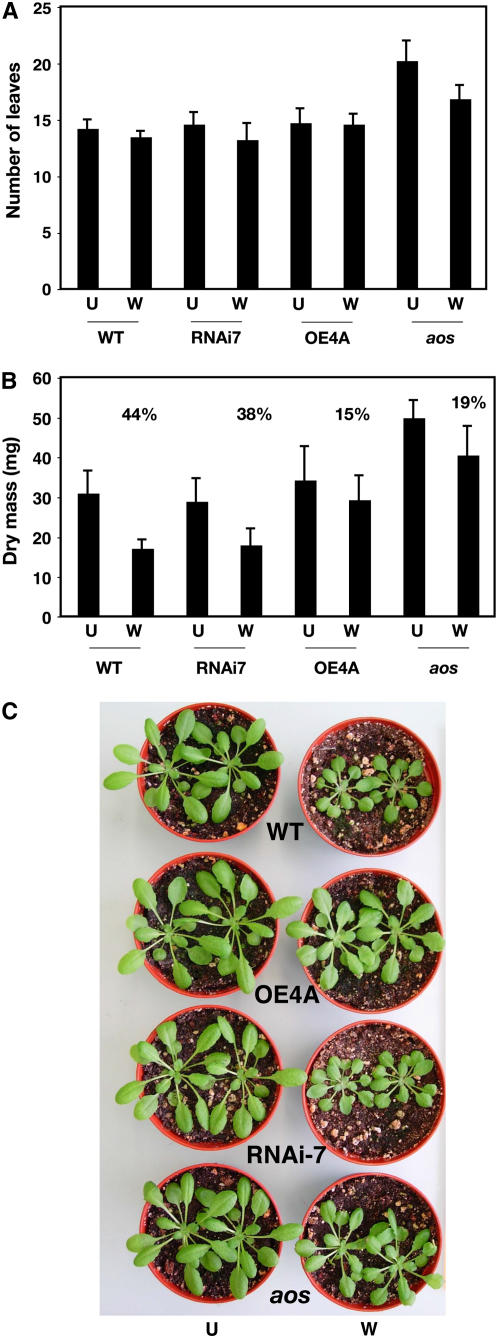
In theory, the modulator of growth inhibition encoded by At5g13200 should affect the plant's growth response to wounding. If this were the case, overexpression of the At5g13220.3 transcript should allow plant growth to continue after wounding, whereas downregulation of the transcript by RNAi should make plants sensitive to wound-induced growth inhibition. In order to test this hypothesis, wild-type plants, aos plants, and transgenic plants with altered levels of the At5g13220.3 transcript were wounded (see Methods) and their growth was assessed. Figure 6 shows the effects of wounding on both the number of leaves and the dry mass of plants. Wounding did not significantly alter the number of leaves produced by each genotype, although nonwounded aos plants reproducibly produced more leaves than other genotypes. Wounding reduced the dry mass of all genotypes, but overexpression of the gene allowed greater postwound growth than seen in wild-type and RNAi plants and revealed a similarly reduced growth inhibition response to the mutant aos (Figure 6B). The difference in postwound growth between wild-type plants and plants overexpressing the At5g13220.3 transcript was visible to the naked eye (Figure 6C).
Figure 6.
Overexpression of At5g13220.3 (JAS1.3) Reduces Wound-Induced Growth Inhibition.
Wild-type, RNAi-7, and OE4A plants as well as the aos mutant were wounded four times on two leaves at 3-d intervals prior to sampling. Plants were 23 d old at the time of the first wound.
(A) Number of leaves over 1 mm long (n = 8; error bars indicate sd). U, unwounded; W, wounded.
(B) Dry mass of plants from the same experiment (n = 8; error bars indicate sd). The reduction in dry mass as a result of wounding for each genotype is given as a percentage.
(C) Visual phenotypes of plants at a similar stage to those used for (A) and (B).
DISCUSSION
Effects of At5g13220 Transcripts on Growth in Response to MeJA Treatment or Wounding
We conducted experiments aimed at finding genes capable of modulating the effects of MeJA on growth in Arabidopsis, and these experiments led us to the At5g13220.3 transcript. When overexpressed in Arabidopsis, this transcript partially repressed the ability of MeJA to inhibit root growth. Conversely, reducing the level of the transcript by RNAi gave the opposing effect: At5g13220 RNAi plants became even more sensitive to exogenous MeJA than wild-type plants. In order to test the specificity of this response for jasmonates, we performed three different experiments. In the first experiment, plants were grown in the presence of NaCl. In Arabidopsis, the inactivation of four genes encoding DELLA proteins permitted increased primary root growth relative to the wild type in the presence of NaCl (Achard et al., 2006). Our results show that modulating the levels of the At5g13220.3 transcript cannot recapitulate the effect of inactivating DELLA protein production. In the other two experiments, we tested the effects of salicylic acid and indole-3-acetic acid on root growth. Salicylic acid can modulate JA responses both at the level of JA biosynthesis enzyme activity (Laudert and Weiler, 1998) and at the level of JA signaling via COI1 (Spoel et al., 2003). Primary root growth in the presence of exogenous salicylic acid was found to be unaffected by modulating the levels of the At5g13220.3 transcript. This transcript, while capable of affecting the growth response to MeJA, does not affect the root growth response to salicylic acid. It is possible that treatments designed to stimulate the activity of the JA pathway might help reveal the effects of the At5g13220.3 transcript on growth in the presence of salicylic acid. Finally, root growth in the presence of indole-3-acetic acid was similar in wild-type plants and plants with altered levels of At5g13220.3 transcripts. This showed that the At5g13220.3 transcript does not appear to encode a major regulator of responses to indole-3-acetic acid.
What, then, is the physiological consequence of altering levels of the At5g13220.3 transcript? To begin to answer this question, wound-induced growth inhibition assays were conducted. In these assays, the numbers of leaves produced in wild-type plants and plants with altered levels of At5g13220 transcript were not strongly affected by wounding, showing that wounding primarily affected growth rather than the differentiation of new leaves. However, the overexpression of the repressor clearly reduced the effect of wounding on growth arrest that is seen in the wild type. The overexpressing plants responded to multiple wounds in a manner similar to the aos mutant—that is, their growth continued after wounding and their dry mass was reduced by only 15% as a consequence of wounding. This can be compared with the 44% reduction in dry mass for wounded wild-type plants. This reproducible experiment confirmed that the transcript can modulate the wound inhibition of growth, and this will now allow a deeper mechanistic study of this phenomenon. For example, a search for interaction partners for JAS1 might also provide clues to how the protein modulates growth inhibition in response to wounding. It should also be possible to identify additional transcripts that are differentially regulated in wounded wild-type plants and wounded overexpressor plants. The failure of RNAi directed against At5g13220 to strongly alter wound-induced growth arrest may be due to the possibility that other related genes intervene in wound-induced growth arrest. In conclusion, our results suggest that At5g13220.3 is a mediator that can strongly affect growth responses after wounding. It will be worthwhile to investigate the possibility of using transcripts such as At5g13220.3 to reduce plant growth inhibition after insect or pathogen damage. If this strategy works, such an application could, theoretically, have agronomic potential in reducing yield loss as a consequence of attack.
The effects of overexpression of At5g13220.3 were not exclusively restricted to altering postwound growth. Wound-induced JA synthesis was reduced, albeit weakly. Also, fresh leaf extracts from overexpressing plants showed a reduced capacity to oxygenate linolenic acid, the principal source of JA. These latter results suggested that the expression of JA biosynthesis genes might have been affected by overexpressing At5g13220.3. This was indeed observed for two genes (AOS and OPR3). The expression of VSP2 was also reduced in leaves of overexpressor plants, suggesting that, in resting tissues, the effects of overexpressing At5g13220.3 extend beyond affecting JA synthesis. However, no strong visual phenotype of overexpressing the gene was seen in resting plants. Furthermore, these results provide the first molecular markers indicating the consequences of At5g13220.3 overexpression. No effects of the gene, whether overexpressed or downregulated by RNAi, led to strong effects on plant defense responses. Transgenic plants infected with the fungus Botrytis cineria showed no alteration in disease symptoms relative to wild-type plants (data not shown). The gene, therefore, does not seem to be directly involved in defense against this pathogen. In summary, the primary effects of overexpressing At5g13220.3 are on growth responses to wounding, but there is clear evidence for some impact on other JA-related processes, in particular JA biosynthesis and VSP2 gene expression. These data raise the possibility that a certain threshold level of JA, reachable in the wild-type plants but not in At5g13220.3 overexpressing plants, must be produced to facilitate growth inhibition after wounding.
Functional Identification of a Jas Motif
Using the MyHits workbench (Pagni et al., 2004) and in particular the Jacop, M-coffee, profile-search, Dotlet, and Jalview tools, the complete proteome of Arabidopsis (The Arabidopsis Information Resource release 7) was searched for sequences similar to that of At5g13220. The most related proteins found were characterized by basic pI values and by relatively weak overall homology except in two conserved motifs. The first is the Zim motif, which is detected by Pfam model PF06200 (Finn et al., 2006) and was recently renamed (Vanholme et al., 2007). The second short motif was identified through our experiments since it was truncated in the short protein isoform encoded by At5g13220.3. This motif was called Jas for jasmonate-associated, with At5g13220.1 being JAS1.1. Its sequences for genes related to JAS1.1 are listed in Table 2 and are characterized by a central S-L-x(5)-K-R-x(2)-R conserved pattern. In many of the gene products, this is flanked by a conserved Pro (P) on the N-terminal side and a conserved Pro-Tyr (PY) dipeptide on the C-terminal side (the first eight genes listed in Table 2). An interesting feature of JAS1-related proteins is that they do not form a well-defined family. They can be described as part of a larger superfamily of Zim-Jas proteins. In every protein listed in Table 2, a Zim motif is followed on the C-terminal side by the Jas motif (with the exception of two isoforms of At1g70700 in which Jas is missing). These 22 proteins from 14 genes can be grouped into 7 cohorts representing individual genes or small groups of closely related genes.
Table 2.
Predicted Arabidopsis Proteins Related to JAS1 and Containing a Zim Motif followed by a Jas Motif, and Sequence of This Jas Motif
| Protein | Sequence of the Jas Motif | Comments |
|---|---|---|
| At5g13220.1 | PIARRKSLQRFLEKRKERLVSTSPY | JAS1 isoforms 2 and 3 are the same protein lacking part of the Jas motif |
| At5g13220.2 | PIARRKSLQRFLEKRKER* | |
| At5g13220.3 | PIARRKSLQRFLEKRKER* | |
| At1g70700.1 | Isoforms 1 and 2 are distinct proteins lacking the Jas motif | |
| At1g70700.2 | ||
| At1g70700.3 | PQARKASLARFLEKRKERLMSAMPY | |
| At3g17860.1 | PLARKASLARFLEKRKERVTSVSPY | |
| At3g17860.2 | PLARKASLARFLEKRKERVTSVSPY | |
| At3g17860.3 | PLARKASLARFLEKRKERVTSVSPY | |
| At1g48500.1 | PQTRKASLARFLEKRKERVINVSPY | |
| At3g43440.1 | PIARRRSLQRFFEKRRHRFVHTKPY | At3g43440.1 is a recently duplicated gene with two Jas motifs |
| At3g43440.1 | PIARRRSLQRFLEKRRDRSTKPDGS | |
| At5g20900.1 | PIARRHSLQRFLEKRRDRLVNKNPY | |
| At1g19180.1 | PIARRASLHRFLEKRKDRVTSKAPY | |
| At1g19180.2 | PIARRASLHRFLEKRKDRVTSKAPY | |
| At1g74950.1 | PIARRASLHRFLEKRKDRITSKAPY | |
| At1g30135.1 | KASMKKSLQSFLQKRKIRIQATSPY | |
| At2g34600.1 | KASMKRSLHSFLQKRSLRIQATSPY | |
| At1g17380.1 | RIARRASLHRFFAKRKDRAVARAPY | |
| At1g72450.1 | RIARRASLHRFFAKRKDRAVARAPY | |
| At4g14713.1 | QANRKVSLQRYREKRKDRKFSKAKK | PEAPOD proteins PPD1 and PPD2 |
| At4g14713.2 | QANRKVSLQRYREKRKDRCIYILTL | |
| At4g14720.1 | QANRKVSLQRYLEKRKDRRFSKTKK |
Fully conserved residues are shown in boldface and underlined, and partially conserved residues are shown in boldface. Asterisks indicate C-terminal amino acids. The different protein cohorts were defined to be made of sequences that are globally homologous. The proteins can be aligned along their entire lengths within a group.
All of the proteins listed in Table 2 are predicted to be relatively small (131 to 353 amino acids), and all are basic. Only At2g34600.1 encodes a protein with a predicted pI < 8 (pI = 7.59), while all other genes predict proteins with pI values ranging from 8.2 to 10.7 (mean for the whole group listed in Table 2 is pI = 9.7). There is considerable sequence diversity among the genes listed in Table 2. Most of this occurs at the N termini of the proteins as well as between the Zim and Jas motifs. This suggests that the part of the superfamily listed in Table 2 might have been under selection for sequence diversity. Other genes with Zim and Jas-like motifs are present in the Arabidopsis genome. The N-terminal moiety of the CCT motif in GATA-type transcription factors (e.g., At4g24470) is reminiscent of the Jas motif. However, proteins from these genes have extremely weak homology with those listed in Table 2 and tend to be acidic; all have pI values of <7.2. These members of the Zim-Jas superfamily are not listed in Table 2. Apart from JAS1.3 (encoded in At5g13220.3), none of the other JAS1-like genes has a known function, with the exception of At4g14713 and At4g14720, which encode the PEAPOD proteins PPD1 and PPD2, respectively. In vegetative tissues, PEAPOD proteins regulate lamina size, leaf curvature, and trichome branching (White, 2006). While abnormal leaf curvature has been observed in a genetically characterized mutant displaying overactive JA biosynthesis (Bonaventure et al., 2007), there is no evidence that PPD proteins are regulated by jasmonates. However, the fact that two types of protein from Table 2 (PPD and JAS1) affect growth responses might indicate that other members have related functions affecting growth.
Expression of Genes Related to JAS1
Information on the expression of 12 of the 14 genes listed in Table 2 was available in Genevestigator (Zimmermann et al., 2004) and is displayed in Supplemental Figure 2 online. Wounding, bacterial (Pseudomonas syringae pv tomato), and fungal (B. cineria) pathogenesis, as well as exposure to MeJA, all increased the levels of several transcripts. Other treatments leading to enhanced levels of the transcripts included exposure to silver nitrate (an elicitor of reactive oxygen species [ROS]) or the ROS ozone. ROS are known to stimulate jasmonate signaling (Orozco-Cardenas et al., 2001). Salt stress also increased levels of transcripts for several of the genes, and this might be related to salt-induced ROS production (Zhu, 2001). Among the treatments leading to downregulation of the levels of several of the transcripts was treatment with syringolin, a probable virulence factor from P. syringae pv syringae (Michel et al., 2006). This is of interest since it implies that syringolin might interfere with jasmonate signaling through some JAS1-like genes. Agrobacterium tumefaciens infection also reduced the levels of some of the transcripts, and one of the features of the superfamily of genes is that it responds to diverse biotic stresses. Interestingly, cycloheximide treatment strongly enhanced the levels of several Zim-Jas transcripts, including those of JAS1. One interpretation of this is that, in the presence of cycloheximide, these genes are negatively regulated by proteins that are constantly turned over. When the synthesis of these putative regulators is blocked by cycloheximide, the transcription of genes occurs even in the absence of further protein synthesis. The upregulation of At5g13220 by wounding may signify that its proteins are being replaced to maintain correct cellular levels.
Possible Roles of the Repressor in Jasmonate Signaling
In order to be active as a signal, JA is modified by conjugation to hydrophobic partner molecules such as the amino acid Ile (Staswick and Tiryaki, 2004). Signaling is effected through the action of the COI1 protein, which is part of a jasmonate-regulated ubiquitin E3 ligase that is thought to target regulatory proteins for destruction by proteolysis (Devoto et al., 2002; Xu et al., 2002). How would JAS1.3 function in this context? Our results showed that the protein exerts its effects primarily on postwound vegetative growth rather than by affecting a broad range of jasmonate-sensitive processes such as fertility. It is possible that the protein acts as a downstream nuclear mediator repressing signaling through COI1. Increasing the level of the At5g13220.3 protein would reduce output from COI1, whereas decreasing its level would allow increased signaling through COI1. This would explain our results with the overexpression lines, in which plants have reduced sensitivity to MeJA. In this model, COI1 would be necessary for the action of overexpressed At5g13220.3. This was indeed observed both at the level of gel blotting for At5g13220 transcripts accumulating in response to wounding and in coi1-1 At5g13220.3 overexpressing plants. The C terminus of At5g13220 transcripts is rich in basic amino acids that sometimes serve as cleavage sites for plant proteinases (Ryan et al., 2002). The At5g13220-derived proteins might be substrates for proteolysis, and one possibility is that the two isoforms may be differentially proteolyzed by small proteinases or modified by larger preproteolytic complexes such as ubiquitin ligases, including that containing COI1. Indeed, COI1 is predicted to target transcriptional regulators for proteolysis (Devoto and Turner, 2003). JAS1 could be such a target. We found evidence that both JAS1.1 and JAS1.3 are targeted to the nucleus, and the proteins are likely to be functional in this compartment. Since the expression of JAS1.3 affects transcript levels for at least three genes, it is possible that the protein somehow modulates transcription.
One interesting finding that emerges from this study is the fact that, of two transcripts from the same gene (At5g13220), only one altered plant growth after wounding. The product from the At5g13220.3 gene is predicted to shorten the protein by 12 amino acids relative to the protein predicted from At5g13220.1. However, both transcript forms accumulate in response to wounding, suggesting that both protein forms may be produced in the cell. While the short protein form may operate to limit the extent of growth inhibition after wounding, we do not know why the longer At5g13220.1 transcript is produced. One possibility is that a balance between the long and short versions of the protein is required for the correct regulation of growth responses to wounding. However, overexpressing At5g13220.1, which should have affected this balance, did not give a phenotype. It should be noted that three Zim-Jas motif–encoding genes appeared in our microarray-derived short list of candidates of interest (Table 1): At5g13220, At1g72450, and At1g74950. This latter gene was also tested in our experiments but did not significantly alter root growth sensitivity to MeJA (Figure 1). This result is consistent with our results for At5g13220, in which only the truncated form At5g13220.3, lacking a complete Jas motif, showed strong biological activity. This raises the question of why so many JAS1-like genes are differentially regulated in response to wounding. Growth regulation in response to insult is extremely complex and presumably must be coordinated between cell types. It is possible that some of these proteins have cell type–specific roles in growth or are involved in other aspects of the jasmonate response, such as the control of defense gene expression and/or JA biosynthesis and modification.
Conclusion
These results reveal that an isoform of a protein of previously unknown function can modify growth in response to wounding and that this process requires jasmonate signaling. The gene identifies a related group of 14 genes from a larger superfamily of Zim-Jas–like genes. Together, the 14 related genes are currently predicted to produce 22 protein isoforms, some of which lack Jas motifs. It should be emphasized that the JAS1.3 protein identified in this study is unlikely to be the only regulator of the growth inhibition response to wounding. It is possible that other proteins listed in Table 2 may be negative or positive regulators of jasmonate-signaled growth. Indeed, correctly controlling growth in response to a wound is a complex process, and many other genes must participate in this process. Some might be downregulated by wounding and would not have been identified by the approach we undertook. Among transcripts that are upregulated by wounding are the other jasmonate-response genes identified in this study (Table 1). They can also be investigated for their possible roles in the postwound growth response and in other aspects of jasmonate signaling.
METHODS
Plant Materials and Chemicals
Arabidopsis thaliana accessions Col-0, the jasmonate synthesis mutant aos (Park et al., 2002), and the coi1-1 jasmonate perception mutant (Feys et al., 1994) were grown on soil at 20°C and 70% RH, with 10 h of light at 120 μmol·m−2·s−1. Fully expanded rosette leaves of plants were crushed across the apical lamina with a forceps, which effectively wounded 30% of the leaf area. Plants were harvested at various times after wounding and immediately frozen in liquid nitrogen. Chemicals were from Sigma-Aldrich.
RNA Extraction and Microarrays
Plants were grown under short-day conditions (10 h of light) for 6 weeks prior to wounding the apical 30% of leaves (between 10 and 15 leaves per plant), which were then incubated in the light for another 1 h prior to RNA extraction. RNA extraction, cDNA synthesis/labeling, and array hybridization were performed as described by Reymond et al. (2000). A microarray containing 25,000 gene-specific tags was used (Hilson et al., 2004). Three biologically independent experiments compared transcript levels in wounded and resting wild-type leaves. At P = 0.05, the false discovery rate (FDR) (Storey and Tibshirani, 2003) was calculated to be 15.5%. In similar experiments (three biologically independent replicates), transcript profiles were compared in resting and wounded aos leaves. For these experiments, the FDR at P = 0.05 was 23.0%. To compare resting levels of transcripts in wild-type and aos leaves, four independent experiments were performed. This list comprised 77 genes (FDR of 83.7% at P = 0.05). This FDR value is high because the overall transcription pattern of the two genotypes is very similar. Details of array scanning, normalization, and data analysis are listed in the public access database ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), in which full experimental data are accessible. The experiment description and data have been deposited in ArrayExpress with accession number E-ATMX-9.
Generation and Selection of Transgenic Plants
Gene-specific primers were used to amplify cDNAs for the genes of interest (boldface in Table 1): At2g24850, 5′-GAAGAACTCAAACGCAAGACAGA-3′ and 5′-CCTGATAATTAAGGAATTACGCAC-3′; At5g47240, 5′-CCTTCTCCTTCTTCTTTTTACCAC-3′ and 5′-AGTCTACTGAGAGTACCGGTTCGA-3′; At1g74950, 5′-CTCCAATTTCTCAATCGAAACG-3′ and 5′-TGGATTATATTAATGGCCATCAA-3′; At2g38750, 5′-AACCAAGAGCCGGAAATCAA-3′ and 5′-ATCACTCATTGTCACAATCGCA-3′; At1g43160, 5′-CAAGAATCATAAACGAGCCAG-3′ and 5′-CCCATGTATAGTCATAACATTCG-3′; At2g38240, 5′-TCCACTACATTTCTCTCCTTGAGT-3′ and 5′-AAACCTCAAGAGAGAGCAGCAA-3′; At5g13220.1 and At5g13220.3, 5′-CCATTTATTCATCTCAAAACCCA-3′ and 5′-TCACGGGATAAGAGAGAGCTTCT-3′. The cDNAs were amplified from reverse transcription products with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The sequences were then verified by DNA sequencing. For overexpression, the resulting cDNAs were cloned into the vector pMDC32 (Curtis and Grossniklaus, 2003), which was then transferred into Agrobacterium tumefaciens. The gene-specific tag CATMA5a11430 was cloned into the Agricola vector (Hilson et al., 2004) for RNAi of At5g13220. Plants were transformed by floral dipping (Weigel and Glazebrook, 2002). Seeds from these plants were sown on agar containing 40 μg/mL hygromycin (pMDC72 vector) or glufosinate (Agricola vector) for selection.
Growth Inhibition Assays
For root growth assays, the T2 seeds of transformants were sterilized with 5% sodium hypochlorite and plated on half-strength Murashige and Skoog medium containing MeJA (25 μM) and hygromycin (40 μg/mL) and grown for 4 d at 22°C. Control plants (wild type and coi1-1) were plated on the same medium (with 25 μM MeJA) without hygromycin. At 4 d, hygromycin-resistant seedlings of transformants and wild-type and coi1-1 seedlings were transferred onto fresh medium containing MeJA (25 μM) for a further 3 d. In parallel, for the control experiment (bottom row in Figure 1A), the transformants were plated on half-strength Murashige and Skoog medium containing hygromycin (40 μg/mL) and grown for 4 d. Control plants (wild type and coi1-1) were plated on the same medium without hygromycin. Homozygous coi1-1 plants were selected by their visual phenotype: no anthocyanin accumulation near the region of the apical meristem. At 4 d, hygromycin-resistant seedlings of transformants and wild-type and coi1-1 seedlings were transferred onto fresh medium for a further 3 d. Quantitative root length measurements were obtained in a similar way except that plants were grown for 7 d after transfer from selective medium onto medium either lacking or containing MeJA. For further characterization of the MeJA sensitivity of At5g13220 transgenic plants, seeds (T3) were plated directly onto medium containing MeJA (25 μM) or medium lacking MeJA for control plates. Plants were then grown for 7 d in continuous light (Figure 2A). Longer term plant growth in the presence of MeJA (Figure 2B) was assessed in a similar way. The T3 seeds of transgenic lines as well as wild-type and coi1-1 seeds were first grown in Petri dishes with (25 μM) or without MeJA for 7 d and then transferred to fresh medium with (25 μM) or without MeJA for a further 14 d under continuous light. Primary root growth inhibition in the presence of NaCl was assayed according to Achard et al. (2006). Seeds were germinated on medium lacking or containing 0.1 M NaCl and grown vertically for 7 d. Similarly, root growth was assessed in the presence or absence of exogenous salicylic acid (50 μM) or indole-3-acetic acid (0.1 and 1 μM). Primary root length was measured on scanned images with ImageJ software (Abramoff et al., 2004).
To assess the position of At5g13220.3 in signaling, coi1-1 homozygous plants were fertilized with pollen from the RNAi-7 line or from the OE4A overexpression line. F1 plants were grown to obtain selfed seeds (F2). The seeds of wild-type, RNAi-7, OE4A, and coi1-1 plants were germinated on 25 μM MeJA and grown for 7 d. coi1-1 plants were selected for their insensitivity to MeJA. By contrast, the F2 seeds of RNAi-7 coi1-1 and OE4A coi1-1 were selected for the transgenes by plating on 25 μg/L glufosinate (RNAi-7) or 40 μg/L hygromycin (OE4A) plus 25 μM MeJA for 7 d. RNAi-7 coi1-1 and OE4A coi1-1 were selected for their double resistance to antibiotic and MeJA. After 7 d, the seedlings of all genotypes were transferred into 4-liter growth boxes with fresh medium containing MeJA (25 μM) and grown for another 14 d. This experiment was repeated four times. Control plants, grown in the absence of MeJA, were selected as follows. Transgenes were selected on glufosinate (RNAi-7) or hygromycin (OE4A). After one of the experiments, the presence of homozygous coi1-1 alleles was verified in five RNAi-7 coi1-1 and five OE4A coi1-1 plants (with five homozygous coi1-1 plants and five wild-type plants as controls) with cleaved-amplified polymorphic sequence markers from Xie et al. (1998). For wound-induced growth inhibition assays with soil-grown plants (Figure 6), 23-d-old plants were wounded three times at 3-d intervals. For each wounding, ∼30% of the apical part of two leaves was crushed with forceps. Different leaves were wounded each time, and plants were harvested at 3 d after the last wound (i.e., at the age of 33 d). Control plants were touched but not wounded.
Real-Time PCR Quantitation and RNA Gel Blotting
To measure the relative levels of transcript for the gene At5g13220 in various transgenic plants (Figure 2), total RNA (15 μg; purified according to Reymond et al., 2000) was copied to cDNA with the SuperScript II first strand synthesis system using oligo(dT)21 according to the manufacturer's instructions. The FullVelocity SYBR Green method (Stratagene) was used. Primers were designed with a Tm of 60°C. The primers used were 5′-ATCCCGATTTCTCCGGTCCA-3′ and 5′-ACTTTCTCCTTGCGATGGGAAGA-3′. The reference transcript was the eukaryotic translation initiation factor 4A1a (EIF4A1 or TIF4A1) amplified from the primers 5′-CCAGAAGGCACACAGTTTGATGCA-3′ and 5′-AGACTGAGCCTGTTGAATCACATC-3′. Real-time PCR was performed with an Mx3000P spectrofluorimentric thermal cycler (Stratagene). Transcript levels were measured relative to EIF4A1 without normalization. A similar approach was taken to measure the relative levels of transcripts from At5g13220.1 and At5g13220.3 in unwounded leaves and leaves at 1 h after wounding (Figure 5). One microgram of RNA was used to produce cDNA. The transcripts for At5g13220.1 and At5g13220.3 were quantified relative to transcripts from EIF4A1. The following primers were used: At5g13220.1, 5′-GAAGCGCAAGGAGAGATTAG-3′; At5g13220.3, 5′-AAGGAGAGGTAATGATTCTTCAACAAT-3′; At5g13220, the same reverse primer was used for both transcripts, 5′-AGTAGGTAACGTAATCTCC-3′; Elf4A.1, 5′-CCAGAAGGCACACAGTTTGATGCA-3′ and 5′-GATGTGATTCAACAGGCTCAGTCT-3′. Values for transcript levels were from six 10-fold dilutions of RNA (0.01 fg to 1 pg) and At5g13220 transcripts normalized to 100 EIF4A1 transcripts. The experiment was repeated with independent biological replicates and gave similar results.
To measure transcripts encoded by JA marker genes, TaqMan probe and primer assays were obtained from Applied Biosystems (custom assays): AOS (At5g42650, assay At02314438_s1); OPR3 (At2g06050, assay At02232505_m1); and VSP2 (At5g24770, assay At02304127_g1). The two reference genes were Elongation Factor1α (EF-1α) (At1g07930, assay At02350528_s1) and Actin7 (At5g09810, assay At02335714_g1). A cDNA template for PCR was generated with a single RT reaction: 1 μg of RNA was mixed with RNase-free water, 2.5 μL of random hexamers (0.1 μg/μL; Invitrogen), and 1 μL of deoxynucleotide triphosphate mix (10 μM) to a final volume of 12 μL, then incubated at 65°C for 5 min. To this, 4 μL of 5× first-strand buffer, 2 μL of 0.1 M DTT, 1 μL of RNasin Plus RNase inhibitor, and 40 units/μL (Promega) and 1 μL (200 units) of SuperScript II (Invitrogen) were added; the mixture was incubated at 42°C for 60 min. PCR was performed in a 10-μL reaction volume. PCR plates (384 wells) were set up with a Freedom Evo liquid-handling robot (Tecan) and then run on a 7900HT sequence detection system (Applied Biosystems). Each 10-μL reaction contained 1× TaqMan Universal PCR Master Mix (Applied Biosystems) and 2 μL of template cDNA (the reverse transcription reaction was previously diluted 1:20). The reaction was initiated by activation of the polymerase at 95°C for 10 min, followed by 45 two-step amplification cycles consisting of 15 s of denaturation at 95°C and a 60-s extension at 60°C. Threshold cycle values for triplicate reactions of the normalization and test genes were obtained using SDS2.2 (Applied Biosystems). Analysis of the reference gene expression stability was done using the geNorm script (Vandesompele et al., 2002). The two reference genes EIF4A1 and β Actin7 were found to be stable across treatments, with an average gene stability measure (M) of 0.39 (the lower the M value, the more stable a reference gene is; the accepted threshold is 1.5), with a coefficient of variance of 13.5%. Normalized relative gene expression values were calculated using the qBase package (version 1.2.3; http//:medgen.ugent.be/qBase). RNA gel blot analysis of At5g13220 was as described (Reymond et al., 2000). Gels lanes were loaded with 15 μg of total RNA. For probe preparation, the gene-specific tag sequence of At5g13220 (www.catma.org) was PCR-labeled with digoxigenin (Roche Biochemicals) according to the manufacturer's instructions using primers 5′-CTCGTTTCGGGAACTGTTCCTATG-3′ and 5′-ATTCTGGCCAAAGAGCTTTGGTCT-3′. Probe detection was performed using the DIG luminescent detection kit (Roche) according to the manufacturer's protocol.
Protein Localization Assays
For protein localization assays, the pMDC84 vector (Curtis and Grossniklaus, 2003) was used for C-terminal translational fusions to At5g13220 isoforms .1 and .3. The At5g13220.1-GFP fusion was constructed with At5g13220.1 amplified with the following primers: 5′-attB1-CAAAGAAGATGTCGAAAG-3′ and 5′-attB2-GGCCGATGTCGGAT-3′. For the At5g13220.3-GFP fusion, the primers 5′-attB1-CAAAGAAGATGTCGAAAG-3′ and 5′-attB2-CCTCTCCTTGCGCTT-3′ were used. Plasmids were precipitated onto gold beads and transformed into onion (Allium cepa) epidermis as described (Mueller et al., 1997). Nuclei were localized by staining with DAPI dilactate. Microscopy was performed with a Leica Microsystems DM5000B microscope equipped with a Leica DFC420 camera used to visualize DAPI and a Leica DFC340F camera for GFP fluorescence. The experiment was repeated with a second batch of onions and gave the same results.
Oxylipin Assays
To test the ability of leaf extracts to oxygenate α-linolenic acid (18:3) and to generate jasmonate precursors, expressed leaf juice (2 μL) from the leaves of 5-week-old plants was incubated with the fatty acid for 5 min (Caldelari and Farmer, 1998). JA in leaves was measured by gas chromatography with an oxygen-18 JA internal standard as described by Mueller et al. (2006). The time point of 90 min was chosen since this is when the peak JA level is attained in response to a single wound inflicted with forceps over the apical 30% of the leaf surface (Reymond et al., 2000).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Primary Root Growth Measured after Vertical Growth on Medium Containing NaCl, Salicylic Acid, or Indole-3-Acetic Acid.
Supplemental Figure 2. Expression Characteristics of Zim-Jas Genes.
Supplementary Material
Acknowledgments
We thank M. Bueno (DNA Array Facility, Lausanne) for excellent technical support; V. Rodriguez, C. Hardtke, H. Weber, G. Bonaventure, and S. Goepfert (University of Lausanne) for helpful comments; C. Darimont, C. Davoine, and C. Nussbaumer (University of Lausanne) for technical support; and B. Kunstner (University of Lausanne) for help maintaining plants. This work was supported by the Swiss National Science Foundation (Project 3100A0-101711) and by a University of Lausanne genomics grant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Edward E. Farmer (edward.farmer@unil.ch).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Achard, P., Cheng, H., De Grawe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. [DOI] [PubMed] [Google Scholar]
- Bonaventure, G., Gfeller, A., Proebsting, W.M., Hoerstensteiner, S., Chételat, A., Martinoia, E., and Farmer, E.E. (2007). A gain of function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J. 49 889–898. [DOI] [PubMed] [Google Scholar]
- Browse, J. (2005). Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 72 431–456. [DOI] [PubMed] [Google Scholar]
- Caldelari, D., and Farmer, E.E. (1998). Rapid assays for the coupled cell free generation of oxylipins. Phytochemistry 47 599–604. [Google Scholar]
- Cipollini, D. (2005). Interactive effects of lateral shading and jasmonic acid on morphology, physiology, seed production, and defense traits in Arabidopsis thaliana. Int. J. Plant Sci. 166 955–959. [Google Scholar]
- Curtis, M., and Grossniklaus, U. (2003). A Gateway TM cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe, W., Roensch, H., Preiss, A., Schade, W., Sembdner, G., and Schreiber, K. (1981). Endogenous plant hormones of the broad bean, Vicia faba L. (−)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 153 530–535. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32 457–466. [DOI] [PubMed] [Google Scholar]
- Devoto, A., and Turner, J.G. (2003). Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann. Bot. (Lond.) 92 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Beneditti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D., et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Res. 34 D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.-H., Kim, J.H., Jayanty, S.S., Howe, G.A., and Han, K.-H. (2006). Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J. Exp. Bot. 57 2923–2936. [DOI] [PubMed] [Google Scholar]
- Laudert, D., and Weiler, E.W. (1998). Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signaling. Plant J. 15 675–684. [DOI] [PubMed] [Google Scholar]
- Li, L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pickersky, E., and Howe, G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8 532–540. [DOI] [PubMed] [Google Scholar]
- Mandaokar, A., Thines, B., Shin, B., Lange, B.M., Choi, G., Koo, Y.J., Yoo, Y.J., Choi, Y.D., and Browse, J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46 984–1008. [DOI] [PubMed] [Google Scholar]
- Michel, K., Abderhalden, O., Bruggmann, R., and Dudler, R. (2006). Transcriptional changes in powdery mildew infected wheat and Arabidopsis leaves undergoing syringolin-triggered hypersensitive cell death at infection sites. Plant Mol. Biol. 62 561–578. [DOI] [PubMed] [Google Scholar]
- Mueller, L.A., Hinz, U., Uzé, M., Sautter, C., and Zryd, J.-P. (1997). Biochemical complementation of the betalain biosynthetic pathway in Portulaca grandiflora by a fungal 3,4-dihydroxyphenylalanine dioxygenase. Planta 203 260–263. [Google Scholar]
- Mueller, M.J., Mene-Saffrane, L., Grun, C., Karg, K., and Farmer, E.E. (2006). Oxylipin analysis methods. Plant J. 45 472–489. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas, M.L., Narvaez-Vasquez, J., and Ryan, C.A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13 179–191. [PMC free article] [PubMed] [Google Scholar]
- Pagni, M., Ioannidis, V., Cerutti, L., Zahn-Zabal, M., Jonbgeneel, C.V., and Falquet, L. (2004). MyHits: A new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32 W332–W335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-H., Halitschke, R., Kim, B.H., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31 1–12. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A., Pearce, G.A., Scheer, J., and Moura, D.S. (2002). Polypeptide hormones. Plant Cell 14 (suppl.): S251–S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saniewski, M. (1988). The effect of methyl jasmonate and benzyladenine of epiphyllous bud formation in Bryophyllum diagremontianum. Bull. Acad. Pol. Sci. Biol. 36 39–44. [Google Scholar]
- Sasaki, Y., et al. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8 153–161. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Świątek, A., Azmi, A., Stals, H., Inze, D., and Van Onckelen, H. (2004). Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett. 572 118–122. [DOI] [PubMed] [Google Scholar]
- Świątek, A., Lenjou, M., Van Bockstaele, D., Inze, D., and Van Onckelen, H. (2002). Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 128 201–211. [PMC free article] [PubMed] [Google Scholar]
- Ueda, J., and Kato, J. (1982). Inhibition of cytokinin-induced plant growth by jasmonic acid and its methyl ester. Physiol. Plant. 54 473–497. [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12 239–244. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- White, D.R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 13238–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, H., Sugawara, J., Susuki, Y., Shimamura, E., and Takahashi, N. (1980). Syntheses of jasmonic acid related compounds and their structure-activity relationship on the growth of rice seedlings. Agric. Biol. Chem. 44 2857–2864. [Google Scholar]
- Zavala, J.A., and Baldwin, I.T. (2006). Jasmonic acid signalling and herbivore resistance traits constrain regrowth after herbivore attack in Nicotiana attenuata. Plant Cell Environ. 29 1751–1760. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2001). Plant salt tolerance. Trends Plant Sci. 6 66–71. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- Thines et al. (2007) and Chini et al. (2007) have reported finding JAZ proteins that repress jasmonate responses and are degraded in a COI1-dependent manner in jasmonate signaling. Their list includes JAS1, which, in their nomenclature, is JAZ10.
- Chini, A., Fonseca, S., Fernández, G., Adie, G., Chico, J.M., Lorenzo, O., García-Casado, G., López-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (July 18, 2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature http://dx.doi.org/10.1038/nature06006. [DOI] [PubMed]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (July 18, 2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature http://dx.doi.org/10.1038/nature05960. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.