Abstract
The rice (Oryza sativa) genome contains 18 copies of genes of the ARGONAUTE (AGO) family. Although AGO members play important roles in RNA-mediated silencing during plant development, a family member that is specifically involved in sexual reproduction has not been identified in plants. We identified the rice AGO gene MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) from the analysis of seed-sterile mutants. In the mel1 mutant, chromosome condensation was arrested at early meiotic stages and irregularly sized, multinucleated, and vacuolated pollen mother cells (PMCs) frequently appeared in developing anthers. In addition, histone H3 lysine-9 dimethylation of pericentromeres was rarely reduced and modification of the nucleolar-organizing region was altered in mel1 mutant PMCs. The mutation also affected female germ cell development. These results indicate that the germ cell–specific rice MEL1 gene regulates the cell division of premeiotic germ cells, the proper modification of meiotic chromosomes, and the faithful progression of meiosis, probably via small RNA–mediated gene silencing, but not the initiation and establishment of germ cells themselves.
INTRODUCTION
In plants, sexual reproduction is the most important step in increasing the genetic diversity of offspring. In contrast with animals, germ cell differentiation of most flowering plants is initiated after the completion of flower organ development, a terminal phase of ontogeny. In Arabidopsis thaliana, the MADS box transcription factor AGAMOUS (AG) triggers reproductive organ development (Mizukami and Ma, 1997; Honma and Goto, 2001). AG also functions during the maturation of stamens and carpels, as well as in their early development, and activates the SPOROCYTELESS (SPL) gene (also known as NOZZLE) (Ito et al., 2004), encoding a key regulator of sporogenesis (Schiefthaler et al., 1999; Yang et al., 1999). In plant reproductive organogenesis, sporogenesis is characterized by the differentiation of the hypodermis into archesporial cells, which serve as primordial germ cells. In anthers, archesporial cells are thought to divide periclinally to give rise to parietal cells toward the exterior and to sporocytes or sporogenous cells (SCs) toward the interior (Davis, 1966; Scott et al., 2004). Parietal cells finally differentiate into inner anther walls, including the tapetum, and SCs undergo several rounds of mitotic division and differentiate to acquire a meiotic cell fate. By contrast, in both Arabidopsis and rice (Oryza sativa), the ovule develops a single SC at the hypodermis of the ovule primordium. Parietal cell fate is determined by the receptor kinase EXTRA SPOROGENOUS CELLS/EXCESS MICROSPOROCYTES1 in Arabidopsis (Canales et al., 2002; Sorensen et al., 2002; Zhao et al., 2002) and MULTIPLE SPOROCYTE1 (MSP1) in rice (Nonomura et al., 2003). However, the plant signaling network that regulates these early developmental processes remains poorly understood.
The discovery of RNA-mediated gene silencing pathways or RNA interference (RNAi) highlighted the fundamental role of small RNAs in eukaryotic gene regulation and antiviral defense (reviewed in Vaucheret, 2006). Recent studies in several metazoans have revealed the importance of RNAi machinery in germline maintenance and function. The Drosophila gene PIWI encodes a protein of the ARGONAUTE (AGO) family that was identified to be required specifically for spermatogenesis and oogenesis (Lin and Spradling, 1997; Cox et al., 1998; Saito et al., 2006). Another member of the PIWI subfamily, AUBERGINE, is also required for Drosophila oocyte maturation (Kennerdell et al., 2002). The Caenorhabditis elegans genome contains 27 AGO family genes, at least five of which are required for fertility (Yigit et al., 2006). In mice, three PIWI subfamily members, MIWI, MILI, and MIWI2, play roles in spermatogenesis (Carmell et al., 2007). Recent evidence has indicated that Drosophila PIWI functions in nuclear RNA silencing by associating with repeat-associated small interfering RNAs derived from repetitive targets (Saito et al., 2006) and that MIWI and MILI associate with PIWI-interacting RNAs, which are highly abundant in testis (Aravin et al., 2006; Grivna et al., 2006). This phenomenon of multiple AGO proteins joining a sexual reproduction pathway suggests that their functions are spatially and temporally subdivided during the reproduction of multicellular eukaryotes.
The Arabidopsis genome contains 10 members of the AGO family (Morel et al., 2002), 4 of which have been characterized with respect to biological function: AGO1 and PNH/ZLL/AGO10 are involved in shoot meristem maintenance (Moussian et al., 1998; Lynn et al., 1999; Vaucheret 2006), AGO4 is involved in RNA-directed DNA methylation and silencing of a small class of transposons (Zilberman et al., 2003), and ZIP/AGO7 is involved in the juvenile–adult transition in vegetative development (Hunter et al., 2003; Peragine et al., 2004). Plant reproduction also requires RNAi machinery, in which AGO1 functions in effecting the full expression of LFY, AP1, and AG, encoding transcription factors to determine meristem identity, flowering transition, and/or flower organ identity (Kidner and Martienssen, 2005). In addition, AGO1 plays a central role in the posttranscriptional gene silencing of CLF, encoding a Polycomb group protein that maintains the repression of both KNOX genes and the homeotic genes AG and AP3 in vegetative leaves (Goodrich et al., 1997; Katz et al., 2004), and in pollen development (Kidner and Martienssen, 2005). However, in plants, no AGO proteins that act specifically in germ cell development have been identified to date.
The cultivated rice (O. sativa subsp japonica cv Nipponbare) genome includes a set of 18 annotated AGO genes. However, the only rice AGO homolog whose function has been characterized is Os PNH/ZLL (Nishimura et al., 2002). This study demonstrates that the AGO family member MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) functions specifically in the germ cell lineage in rice.
RESULTS
MEL1 Encodes an ARGONAUTE Family Protein
To better understand the genetic network that supports sexual reproduction in flowering plants, seed-sterile mutant lines of rice were isolated from a pool of mutants induced by somatic culture and regeneration (Hirochika et al., 1996; Yamazaki et al., 2001). Of 600 lines that segregate sterile plants, all of which develop normally in the vegetative phase (see Supplemental Figure 1A online), 80 lines exhibited the mutant phenotypes in meiosis and/or reproductive organogenesis (data not shown). Anthers of sterile mutants in the line NC0005 displayed abnormal meiosis, in which cell growth arrested mostly in early prophase I (see below). The sterile phenotype segregated as a single recessive mutation in the offspring of heterozygous plants (fertile:sterile = 225:70, χ2 = 0.254 for a ratio of 3:1, 0.50 < P < 0.70). When homozygous sterile plants were pollinated with wild-type pollen, only 0.79% of mutant flowers set mature seeds, suggesting that the mutation also affected megagametogenesis. This MEL1 gene was isolated by means of the Tos17 tag, an endogenous Ty1/copia-like retrotransposon involved in the rice genome (Hirochika et al., 1996).
Genomic DNA was extracted from 54 fertile and 19 sterile NC0005 plants, digested with XbaI restriction enzyme, and used for DNA gel blot analysis with a Tos17 partial sequence as a probe. Of the >10 bands of transposed Tos17, an ∼8-kb band was found to be completely linked to the mel1 sterile phenotype (see Supplemental Figure 1B online). Isolation and sequencing of the flanking sequence revealed that Tos17 was inserted into the gene encoding an AGO family protein (Figure 1), which could be discriminated by containing PAZ (for Piwi, Argonaute, and Zwille) and PIWI domains according to the InterPro protein family database (http://www.ebi.ac.uk/interpro/index.html). This gene was given the locus identifier Os03g0800200 by the Rice Annotation Project Database (http://rapdb.lab.nig.ac.jp/index.html). Next, the pKN16 binary plasmid, which contained an 18-kb XhoI/BamHI DNA fragment with the entire coding region of the candidate gene and its upstream sequence, was used for complementation analysis; of 30 mel1/mel1 plants carrying pKN16, 22 plants underwent recovery of the pollen and seed sterility phenotypes (average seed fertility = 20.3 ± 19.7%), while all nine mel1/mel1 plants carrying the empty vector generated completely sterile spikelets only (see Supplemental Figure 2 online). The pKN16 plasmid contained two other putative genes upstream of MEL1. Os03g0800400 on the sense strand was truncated in pKN16 and therefore is thought to be unrelated to the mel1 phenotype. However, pKN16 contained the entire length of Os03g0800300 on the antisense strand, but there was no difference in the 3.0-kb genomic sequence covering this putative gene between mel1/mel1 and +/+ siblings when three plants were used in each experiment (data not shown). Thus, we concluded that the Tos17 insertion into the rice AGO gene caused the mel1 sterile phenotype.
Figure 1.
Rice MEL1 Had a Typical AGO Structure.
(A) Schematic diagram of the structure of the rice MEL1 protein, which was composed of 1058 amino acids. Arrowheads indicate the site of Tos17 insertion in the respective allelic lines.
(B) Alignment of the PIWI domains of Os MEL1 (accession number AB297928), Os PNH (ABO81950), At PNH (AAD40098), At AGO1 (AAC18440), At AGO4 (NP_565633), Ce CSR1 (NP_001040938), and Dm PIWI (NP_476875). Amino acids conserved in more than three proteins are highlighted with black boxes. The D, D, and H residues, which are key metal-coordinating residues involved in RNase H activity (Yigit et al., 2006), are marked with asterisks beneath the sequences.
The Tos17-flanking sequence database (http://tos.nias.affrc.go.jp/) revealed that the MEL1 gene was tagged by >20 insertions of Tos17. We confirmed that two allelic lines, NF5040 and ND6051, segregated seed-sterile plants with pollen mother cells (PMCs) that arrested at early meiosis I, the same as the mel1 phenotype (data not shown). In all three allelic lines, Tos17 was inserted upstream of or within the region encoding the PIWI domain (Figure 1A), generating in-frame stop codons (data not shown). The PIWI domain plays an essential role in gene silencing triggered by small RNAs such as short interfering RNAs and microRNAs (miRNAs) (Carmell et al., 2002). Insertion of Tos17 into such an important domain suggests that all allelic lines used in this study might segregate null mutants. The NC0005 allele of the MEL1 gene was designated mel1-1 and was used for all of the experiments described below.
The 3,450-bp full-length MEL1 cDNA was isolated by cDNA library screening and 5′ rapid amplification of cDNA ends (RACE) technology from young rice panicles. The MEL1 transcript was detected in young panicles, but not in vegetative organs, by semiquantitative RT-PCR (Figure 2A). In mutant anthers, the amount of MEL1 transcripts was ∼70% less than that in wild-type anthers (Figure 2B). In addition, the RT-PCR products in the mutant were 72 bp smaller than those in the wild type. Sequencing showed that the size reduction was due to aberrant elimination of the eighth exon, which carried the Tos17 insertion in the mel1-1 allele (data not shown). These results suggest that the mel1 mutant expresses only aberrant mRNAs that are longer or shorter than those in the wild type owing to Tos17 insertion or altered splicing.
Figure 2.
Rice MEL1 Is Expressed Specifically in Reproductive Organs.
(A) RT-PCR analysis of MEL1 (top) and Actin (bottom) as a positive control in vegetative and reproductive organs. P1 to P5 indicate stages of panicle development; a P1 panicle is 5 mm long or less, P2 is ∼10 mm, P3 is ∼30 mm, P4 is ∼60 mm, and P5 is ∼100 mm.
(B) RT-PCR (15 cycles) products for four loci, MEL1, PAIR1, PAIR2, and Actin, were compared for wild-type (+/+) and mel1-1 mutant (−/−) anthers, respectively, at meiosis I for a mixture of 0.5- to 0.8-mm anthers. RT-PCR without reverse transcriptase (−) was performed as a negative control. The signal intensity ratio of the mel1-1 mutant signal to the wild-type signal, in which the ratio for the Actin locus was set as 1.0, is shown at right for each locus.
MEL1 Is Distinct from Other Plant AGOs
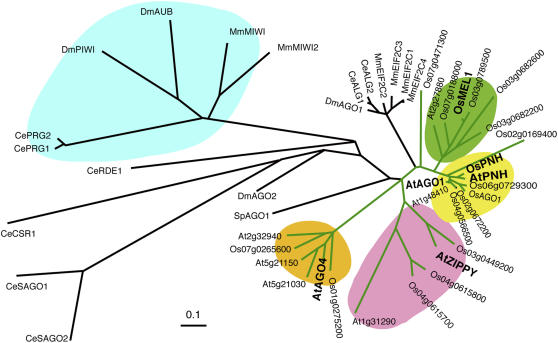
The rice genome (cv Nipponbare) contains 18 AGO family members, including MEL1. The MEL1 cDNA encoded a putative protein of 117 kD and 1058 amino acids. In addition to the PAZ and PIWI domains, the N terminus of MEL1 had Gly-rich (54.6% of amino acids 4 to 36) and Ala-rich (39.6% of amino acids 131 to 178) stretches (Figure 1A). The Gly-rich region is also found in the N terminus of Arabidopsis AGO1 (Bohmert et al., 1998). The PSORT program (Nakai and Kanehisa, 1992) predicted that the MEL1 protein would localize in the cytoplasm. Phylogenetic analysis using the PAZ and PIWI domains for rice, Arabidopsis, C. elegans, Drosophila melanogaster, and mouse revealed that all plant AGOs were in the same branch that included several animal and yeast AGO members but were phylogenetically distinguished from the PIWI subfamily, which contains germline-specific insect and animal AGOs (Figure 3). The plant AGO group was further divided into four subgroups: AGO1, ZIPPY, AGO4, and MEL1 (Figure 3). In the PAZ and PIWI domains, MEL1 had 66.6 and 68.3% identity to rice and Arabidopsis AGO1. The Asp, Asp, and His residues, which are key metal-coordinating residues involved in RNase H activity (Yigit et al., 2006), were conserved in the PIWI domain of MEL1 (Figure 1B).
Figure 3.
Phylogenetic Tree Constructed Using the Peptide Sequences of PAZ and PIWI Domains.
For plants, all AGO family members in Oryza sativa (Os) and Arabidopsis thaliana (At) were included (connected with green lines). In addition, functionally known AGOs from Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Mus musculus (Mm), and Schizosaccharomyces pombe (Sp) were included (connected with black lines). The plant locus identifiers correspond to the Arabidopsis Genome Initiative codes for Arabidopsis and the Rice Annotation Project codes for rice. Accession numbers are as follows: NP_510322 (Ce ALG-1), NP_493837 (Ce ALG-2), CAA98113 (Ce PRG-1), AAB37734 (Ce PRG-2), AAF06159 (Ce RDE-1), NP_523734 (Dm AGO1), NP_476875 (Dm PIWI), AAF49619 (Dm AGO2), CAA64320 (Dm AUB), NP_067286 (Mm MIWI), AY135692 (Mm MIWI2), XP_001257025 (Mm EIF2C1), NP_694818 (Mm EIF2C2), NP_700451 (Mm EIF2C3), NP_694817 (Mm EIF2C4), AAD40098 (At PNH), ABO81950 (Os PNH), ABO81951 (Os AGO1), CAA19275 (Sp AGO1), NP_001040938 (Ce CSR-1), NP_504610 (Ce SAGO-1), and NP_490758 (Ce SAGO-2). The PIWI subfamily, which functions specifically in germline development in insects and animals, is highlighted in blue. The plant subfamilies Os MEL1 (AB297928), At AGO1 (AAC18440), At ZIPPY (AAQ92355) and At AGO4 (NP_565633) are highlighted in green, yellow, red, and orange, respectively.
MEL1 mRNA Is Expressed Specifically in Germ Cells
In situ expression of MEL1 mRNAs was investigated in young wild-type panicles. Faint MEL1 mRNA signals first appeared in the hypodermis of developing stamen primordia (Figure 4A). Interestingly, hypodermal MEL1 signals appeared as stepping stones along transverse stamen axes (arrowheads in Figures 4B and 4C). In subsequent stages, MEL1 expression was clearly detected in four microsporangia (Figures 4D and 4E). The signals became more intense in male SCs when anther walls became three-layered, but they were not detected in the anther walls themselves (Figures 4F and 4G). In the female organ, the MEL1 signals appeared in the hypodermal layer of the ovule primordium after the carpel began to develop (Figure 4C). Subsequently, a single megaspore mother cell (MMC) became clearly marked by the MEL1 signal when inner integuments began to develop (Figures 4F and 4H). The signals in both male and female organs disappeared after the SCs entered meiosis and gametogenesis (Figures 4I to 4K). The signals did not appear in the shoot apex (Figure 4L) or in young flowers probed with a sense strand RNA as a negative control (Figure 4M). These results show that rice MEL1 mRNA is expressed specifically in male and female archesporial cells and SCs before meiosis but not in the nursery cells supporting SCs.
Figure 4.
MEL1 mRNA Is Expressed Specifically in Archesporial Cells and SCs in Male and Female Reproductive Organs in Rice.
(A) Faint signals (arrow) appeared in the hypodermis of developing stamen primordia (st).
(B) Intense stepping stone signals appeared along stamen axes (arrowheads). By contrast, no signal was observed in the nucellar primordium (np). ca, carpel.
(C) A faint signal appeared in the hypodermis of the nucellus (nu; arrow) when the carpel began to develop and stamens differentiated into four-lobed organs.
(D) When the carpel had enclosed the nucellus, the programmed position of archesporial cells clearly corresponded to MEL1 signals. an, anther.
(E) A magnified view of the area shown by the dashed square in (D).
(F) to (H) When anthers were three-layered, the MEL1 signal clearly marked SCs, but not anther walls, epidermis (ep), endothecium (en), or secondary parietal cells (spc) ([F], magnified in [G]). In the nucellus with developing inner integument (ii), a single SC was clearly stained ([F], magnified in [H], in which the nucellus and the inner integument are outlined).
(I) The MEL1 signal was absent until the onset of meiosis. ta, tapetum.
(J) When the female SC entered meiosis, no MEL1 signal was observed in either male or female reproductive organs. The completion of enclosure of the nucellus by the inner integument indicates that the female meiocytes had entered meiosis (Itoh et al., 2005). oi, outer integument; pi, pistil.
(K) No MEL1 signal was detected in microspores (ms).
(L) No MEL1 signal was detected in shoot apical meristem (sm) or plastchron (pl).
(M) A negative control in which a flower section at the same stage as that shown in (C) was probed with the sense strand of MEL1 mRNA.
Bars = 100 μm.
Loss-of-Function Mutation Disrupts the Development of Archesporial Derivatives
Histological analysis of the anthers was performed using transmission electron microscopy (TEM). In three-layered anthers in which SCs had undergone premeiotic mitosis, the shape and size of the SCs were identical in wild-type and mutant plants, whereas the mutant SCs were slightly more vacuolated (Figures 5A and 5B). At this stage, indirect immunofluorescence of whole-mounted anthers showed that the mutant SCs formed normal phragmoplasts (Figure 6A), cytoskeletal structures held by two arrays of microtubule bundles that determine the position of the cytokinetic plane (Verma, 2001). The mass of cytosol, nuclei, and nucleoli of the mutant SCs became prominently larger than in other somatic cells, as in wild-type SCs (Figures 5A and 5B), suggesting that the mel1 mutation did not affect the initiation, establishment, and early mitotic division of germ cells. When anther walls transitioned from three- to four-layered, at which time the tapetal cells had differentiated, densely stained mitochondria became conspicuous in the cytoplasm of mutant SCs, unlike in the wild-type SCs (Figures 5A and 5B). In wild-type SCs, most mitochondria were composed of indistinct cristae and less-stained matrix (Figure 5C) (Mamun et al., 2005). On the other hand, the inner wall cells of wild-type and mutant anthers had mitochondria with a very similar appearance, with inflated cristae and densely stained matrix (Figures 5D and 5F). The densely stained mitochondria in mutant SCs closely resembled those in the surrounding somatic cells (Figure 5E). In the leptotene stage, at which the wild-type SCs had matured into PMCs to undergo meiosis and the tapetum had completely differentiated (Figure 5G), abnormally fused PMCs frequently appeared, and vacuoles in the PMCs were ruptured in the mutant (Figure 5H). In addition to the SC aberration, inner wall cells also became vacuolated and often developed extra cell layers (arrowheads in Figure 5H). The mutant PMCs still carried compact and densely stained mitochondria (Figure 5J), while the wild-type PMCs carried faintly stained mitochondria (Figure 5I). These observations suggest that the mel1 mutation affects both archesporial derivatives (i.e., the SCs and inner wall cells of anthers).
Figure 5.
Histological Analysis of Wild-Type (Top Row) and mel1-1 (Bottom Row) Sporogenesis in Anthers by TEM Observation.
(A) to (F) When anther walls transitioned from three- to four-layered, densely stained mitochondria became conspicuous in the cytoplasm of mutant SCs ([B] and [E]), whereas those in the wild type were faintly stained ([A] and [C]). The densely stained mitochondria in mutant SCs (E) closely resembled the appearance of those in nursery somatic cells of the wild type (D) and the mutant (F). en, endothecium; no, nucleolus; nu, nucleus; spc, secondary parietal cells. Bars = 2 μm.
(G) and (H) In the leptotene stage, at which the wild-type SC had matured into PMCs to undergo meiosis and the tapetum (ta) had completely differentiated (G), abnormally fused PMCs frequently appeared in mel1-1 anthers (the position of an incomplete cell wall is indicated by arrows, and an aberrantly terminated wall is magnified in the inset in [H]), and vacuoles in PMCs were ruptured (H). In addition, the inner anther wall, including the tapetum, middle layer (ml), and endothecium, was vacuolated and extra-layered (arrowheads) (H). ep, epidermis. Bars = 10 μm.
(I) and (J) In the leptotene stage, mitochondria were faintly stained in wild-type PMCs (open arrowheads in [I]) but densely stained in mel1-1 PMCs (closed arrowheads in [J]). A faintly stained structure that was heterogeneous to electron-dense nucleolar materials frequently appeared in mel1-1 PMCs (asterisk in [J]). Both wild-type and mutant PMCs formed axial elements on meiotic chromosomes (arrows in [I] and [J]). ne, nuclear envelope; v, vacuole. Bars = 1 μm.
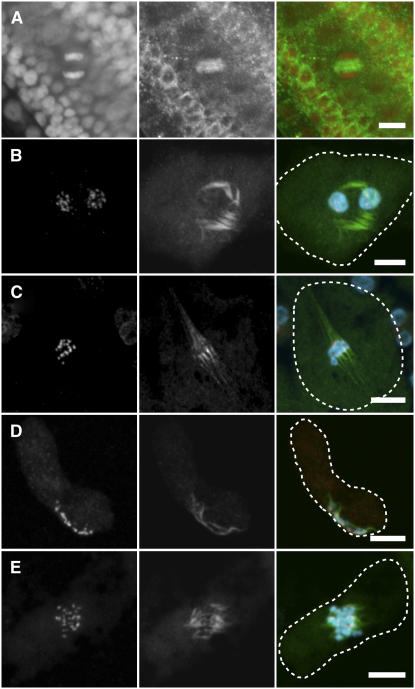
Figure 6.
Indirect Immunofluorescence of α-Tubulin for Wild Type and mel1-1 PMCs.
(A) In three-layered mel1-1 mutant anthers, the sporogenous cells underwent premeiotic mitosis and formed normal phragmoplasts at the midzone of telophase cells; counterstaining of chromatin with propidium iodide (left) and α-tubulin (middle) is pseudocolored with red and green, respectively, and merged (right).
(B) to (E) The centromeres (Os CenH3; left) and α-tubulin (middle) are pseudocolored in red and green, respectively, and merged (right) with counterstained chromatin (blue). PMCs are outlined with dashed lines.
(B) In the mel1-1 PMCs arrested at premeiosis, aberrant microtubule bundles elongated around the nuclei but seemed to lose polarity.
(C) At meiotic metaphase I in wild-type PMCs, bipolar and well-elongated spindle microtubule bundles captured the centromeric regions of the homologous chromosome pairs.
(D) and (E) The mel1-1 PMCs that escaped from meiotic arrest formed bipolar but much shorter spindles than wild-type PMCs.
Bars = 10 μm.
Subsequent stages of anther and ovule development were optically sectioned using confocal laser scanning microscopy (CLSM). When wild-type anthers underwent meiosis I, aberrant PMCs with respect to shape and size were observed in the mutant anthers (see Supplemental Figure 3 online). These PMCs frequently formed aberrant microtubule bundles around nuclei arrested at premeiosis or meiotic prophase I (Figure 6B). After the wild-type anther passed through meiosis and microgametogenesis, most of the mutant PMCs still arrested at meiosis and failed to produce tetrad spores, resulting in complete male sterility. Even though the PMCs escaped meiotic arrest, bipolar but much shorter spindles than found in the wild type were established at metaphase I (Figures 6C to 6E). Female gametogenesis was also affected in the mel1-1 mutant. In wild-type ovules, a single embryo sac was clearly observed. By contrast, in mel1-1 mutant ovules, female gametogenesis was arrested at various stages, the premeiosis, meiosis, and tetrad stages (see Supplemental Figure 3 online). In some mutant ovules, the embryo sac was completely eliminated. These observations suggest that MEL1 function is essential for the completion of normal sporogenesis and meiosis in both male and female organs.
Meiosis Is Arrested at Early Prophase I in mel1 Mutants
In mel1-1 mutant PMCs arrested at early meiosis I, meiotic chromosomes frequently exhibited uncondensed morphology, like in leptotene or zygotene, when wild-type PMCs had already entered into microgametogenesis (Figure 7A). In 0.9- to 1.1-mm anthers in the mutant, ∼5% of meiocytes escaped meiotic arrest to reach meiosis II or the microspore stages (arrows in Figure 7A). We used anther length as a surrogate for stage progression for comparison of phenotypes between the wild type and mutant, because the mel1 mutation had no effect on anther elongation until the end of meiosis (data not shown).
Figure 7.
Meiotic Chromosomes Failed to Undergo Normal Condensation and Rarely Carried Hypomethylated Histone H3K9 at Pericentromeres in PMCs of mel1-1 Mutants.
(A) Frequency of PMCs at various meiotic stages in anthers ranging from 0.5 to 1.1 mm in length. Blue and red bars indicate the frequency of wild-type and mutant PMCs at the various stages. Arrows indicate the frequencies of escape from meiotic arrest in the mutant (each was 5% or less).
(B) to (I) Indirect immunofluorescence of PMCs. In each section, the left and middle panels represent the results using anti-dimethylated H3K9 (green) and anti-Os CenH3 (red) antibodies, respectively, and both images are merged in the right panels with the chromatin image counterstained with TO-PRO-3 (Invitrogen) (blue). Bars = 5 μm.
(B) to (E) In wild-type PMCs, H3K9 was dimethylated extensively, but mainly at pericentromeric regions during leptotene (B), zygotene (C), pachytene (D), and anaphase I (E).
(F) to (H) Of the mel1-1 PMCs arrested at early meiosis I, most were dimethylated to a normal (F) or moderate (G) level, but markedly hypomethylated PMCs appeared at low frequency (H).
(I) All PMCs that had escaped from meiotic arrest in mutants were dimethylated on H3K9 at a wild-type level.
Indirect immunofluorescence using the anti-PAIR2 and anti-phosphorylated histone H3 serine-10 (H3S10) antibodies was performed to ascertain the details of chromosome condensation status. In wild-type PMCs, the PAIR2 protein is known to transiently associate with meiotic chromosome axes during leptotene and zygotene, and at pachytene it has been removed from the axes (Nonomura et al., 2006). The anti-phosphorylated H3S10 antibody is generally used as a mitosis marker. In wild-type rice meiocytes, the PAIR2 associated with meiotic chromosome axes until diakinesis, but the signal was replaced by that of phosphorylated H3S10 on the entire chromatins at and after early metaphase I (see Supplemental Figure 4 online). In all mutant PMCs carrying uncondensed chromosomes, PAIR2 associated with chromosome axes, but H3S10s were phosphorylated only to a very limited extent. This result confirmed that most mutant PMCs became arrested at early meiosis I with respect to chromatin modification. mRNA of another meiotic gene, PAIR1 (Nonomura et al., 2004), was also detected in mel1-1 mutant anthers (Figure 2B). In addition, electron-dense and filamentous structures, generally representing the axial elements of meiotic chromosomes, were observed in the nuclei at leptotene in both wild-type and mutant PMCs (Figures 5I and 5J). These results indicate that the mutant meiocytes achieve the meiotic process.
Chromatin Modification Is Altered in mel1 Mutant Meiocytes
The influence of the mel1 mutation on histone H3 methylation was investigated, because several AGO members regulate H3 lysine-9 (H3K9) methylation and chromosome dynamics, depending on the small RNA–mediated silencing machinery (Volpe et al., 2002; Hall et al., 2003; Pal-Bhadra et al., 2004). In wild-type rice meiocytes, H3K9 was generally dimethylated, but mainly at pericentromeric regions during meiosis (Figures 7B to 7E). In mel1-1 mutant PMCs, H3K9 dimethylation also occurred at pericentromeric regions of both arrested (Figures 7F to 7H) and escaped (Figure 7I) chromosomes. In leptotene and zygotene stages, no remarkable difference was observed in the average signal intensity of dimethylated H3K9 between wild-type and mel1-1 PMCs: 40.3 ± 12.5 (n = 13) and 31.3 ± 10.9 (n = 25), respectively (see Methods for a calculation of signal intensity). However, in at least 2 of 25 mel1-1 PMCs observed, all of which were arrested at leptotene or zygotene, the intensity of H3K9 dimethylation was reduced significantly (13.7 and 18.3; an example of two PMCs is shown in Figure 7H). A relationship between MEL1 function and H3K9 dimethylation could not be ruled out, but it was thought to be weak if it existed at all.
Interestingly, strong PAIR2 association within the nucleolus, which is never observed in wild-type meiocytes (Figure 8A) (Nonomura et al., 2006), frequently occurred in mel1-1 PMCs (Figures 8B and 8C). These intense PAIR2 foci corresponded to the nucleolar-organizing region (NOR) (Figures 8D to 8F), which is composed of ∼850 copies of the rRNA gene repeat unit in a diploid genome (Oono and Sugiura, 1980). During electron microscopic observations of the mel1-1 PMCs, an abnormal and faintly stained structure that was heterogeneous with the electron-dense nucleolar materials was frequently observed (Figure 5J). This may reflect altered chromatin structure in NORs in the mel1 mutant.
Figure 8.
Dramatically Increased Accessibility of the Meiotic Protein PAIR2 Observed in mel1-1 Mutant PMCs.
(A) In wild-type PMCs at leptotene, no PAIR2 signal (green) was observed within the nucleolus (dashed circle).
(B) and (C) In mel1-1 PMCs, strong PAIR2 signals were observed within the nucleolus.
(D) to (F) The aberrant PAIR2 focus within the nucleolus (D) corresponded to the fluorescence in situ hybridization signal of the NOR (25S rDNAs) (E). Images shown in (D) and (E) are pseudocolored in green and red, respectively, and merged in (F). Bar = 5 μm.
DISCUSSION
This study revealed that the AGO family member MEL1 is involved in germ cell development in cultivated rice. The mel1 mutation also affected the development of support cells of SCs. This is likely to be a secondary effect of the mutation in anther wall development, because of tapetum development tightly coupling with SC development, as shown by the Arabidopsis spl study (Yang et al., 1999), while there is another possibility that MEL1 plays some roles in the development of archesporial cells, the origin of both SCs and support cells. In mice, the miwi2 mutant becomes infertile and progression of meiosis becomes defective at early prophase I, as in mel1 in rice, in addition to loss of germ cells (Carmell et al., 2007). Sertoli cells, which are somatic support cells in seminiferous tubules, became increasingly vacuolar in miwi2 mutants, also resembling the rice mel1 mutant. Taken together with the results from various organisms, it is likely that multiple AGOs participate in the regulation of plant germ cell development that is dependent on the RNA-mediated silencing system. The existence of multiple AGO copies in Arabidopsis and rice indicates that the functions of AGO family members differ spatially and temporally in the life cycles of these plants. The MEL1 subgroup comprises a single member in Arabidopsis and five members in rice, whereas other subgroups include multiple copies in both Arabidopsis and rice (Figure 3). This may suggest that the role of At2g27880, the sole Arabidopsis member of the MEL1 subgroup, has come to be shared by multiple rice AGOs during the differentiation of monocots from dicots and that the expression of At2g27880 is not restricted within germ cell development in Arabidopsis.
The AGO1/PNH subgroup was phylogenetically closest to the MEL1 subgroup (Figure 3). At AGO1 acts in miRNA-directed mRNA cleavage, also targeting AGO1 by itself, and in proper plant development (Vaucheret et al., 2004). The existence of miRNA pathways has also been reported in rice reproduction. Os GAMYB, a positive regulator of gibberellin signaling, plays multiple roles in the development of tapetal cells and the aleurone layer (Kaneko et al., 2004) and is regulated by a miRNA, miR159a, during anther development (Tsuji et al., 2006). We plan to investigate the transcriptional level of known miRNAs and reproduction-related genes in the mel1 mutant by means of microarray analysis.
It is also known that several AGO family members mediate heterochromatin assembly. For example, in fission yeast, deletion of AGO1, Dicer, or RNA-dependent RNA polymerase disrupts heterochromatin-mediated silencing, which correlates with the loss of H3K9 methylation and SWI6/HP1 association with heterochromatic loci (Volpe et al., 2002; Hall et al., 2003). In this study, we found that H3K9 dimethylation was somewhat reduced in mel1 meiocytes arrested at early meiosis I (Figure 7). In addition, structural alteration of the nucleolus and NORs was observed in mel1-1 mutant meiocytes (Figures 5J and 8). The nucleolus is an essential nuclear organelle for ribosome assembly, and excessive high recombination within NORs is harmful to organisms (Kobayashi et al., 2004). Thus, it may be expected that organisms have developed systems to regulate recombination within rDNA repeats. In Saccharomyces cerevisiae, the stability of rDNA copy number is controlled by transcription from a noncoding bidirectional promoter within the rDNA spacer (Kobayashi and Ganley, 2005). H3K9 methylation and the RNAi pathway are required for the normal organization of rDNAs, satellite DNAs, and nucleoli in Drosophila (Peng and Karpen, 2007). In this study, dramatically increased accessibility of PAIR2 to mel1 mutant NORs (Figure 8) suggests that the rice MEL1 may insulate rDNA repeats from homologous synapsis and meiotic recombination to maintain the appropriate rDNA copy number.
How does the cytokinesis of PMCs become incomplete, and is meiosis arrested at very early stages in most mel1 meiocytes? Our first hypothesis was that the regulatory pathway that occurs in tapetal cells is ectopically expressed in mel1 PMCs. This idea was guided by several observations: (1) fused aberrant PMCs appear following differentiation of the tapetum (Figure 5H); (2) PMCs with incomplete cytokinesis and multiple nuclei resemble the binucleated tapetal cells that are usual in wild-type rice anthers; (3) mitochondria in mel1 PMCs are converted from the germ cell type into the somatic cell type (Figure 5E); and (4) many vacuoles appear in mel1 PMCs (Figure 5B) and rupture during early meiosis (Figure 5H), similar to the programmed cell death of wild-type tapetum that is marked by vacuolation and vacuole rupture in addition to cell shrinkage (Papini et al., 1999; Wu and Cheung, 2000). We examined the in situ expression of genes relating to tapetum development, that is, MSP1 (Nonomura et al., 2003) and Os GAMYB (Kaneko et al., 2004; Tsuji et al., 2006), but the expression profile of both genes in mel1 mutants seemed identical to that in the wild type (data not shown). Although we have obtained no findings to date in support of our first hypothesis that MEL1 represses the tapetum program in rice PMCs, the observation that the mel1 PMCs carry somatic cell–type mitochondria suggests a possibility that MEL1 represses somatic gene expression in rice germ cells.
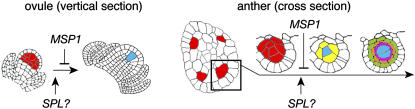
Finally, in situ profiling of MEL1 mRNA expression yielded insights into the process by which archesporial cells initiate and develop in rice. Generally, the female archesporial cell of flowering plants is believed to directly mature into a single MMC in the ovule primordium (Reiser and Fischer, 1993). However, the area stained by the MEL1 probe, which might mark the locations of archesporial initials, was larger in earlier stages (Figures 4C and 4E) than in later stages in the ovule primordium (Figure 4H). We propose a new model in which the archespores are generated in excess of subsequently developed single MMCs (Figure 9). This idea may be supported by our previous finding that in the reproductive organs of rice, hypodermal nucellar cells surrounding the MMC have the potential to become MMCs (Nonomura et al., 2003). The analogy of the female processes is applicable for male processes—that is, the interior cells of the excess archespores in microsporangia might differentiate into SCs, probably dependent on the function of the rice ortholog of Arabidopsis SPL, and the archespores adhered to the SCs might be determined by their identity as parietal cells, probably by the signal from the SCs (Figure 9). This model well explains the rice msp1 phenotype, in which the anther generates excessive SCs and fails inner wall development (Nonomura et al., 2003). In addition, hypodermal MEL1 signals appeared in founder regions of microsporangia as stepping stones along the transverse stamen axes (Figure 4B). Although Raghavan (1988) proposes that in rice stamens a single row of hypodermal archespores is initiated over the entire length of each microsporangium, the results of our study suggest the possibility that archesporial initials develop discontinuously in stamen primordia and subsequently proliferate to occupy the microsporangia.
Figure 9.
New Models for Plant Archesporial Initiation in the Ovule and Anther.
Cells colored in red, blue, yellow, magenta, orange, and green indicate archesporial cells, SCs, primary parietal cells, tapetal cells, middle layer cells, and endothecium cells, respectively. Excess female archespores are generated in the ovule, and one of the archespores only develops in an SC, probably by means of SPL-like gene function to promote SC generation and MSP1 function to repress support cells of SCs developing into SCs; no rice ortholog of the SPL gene has been identified yet. Analogous with ovule development, male archespores might also be generated in excess of SCs, and SPL and MSP1 might determine the fate of SCs and parietal cells in microsporangia.
In conclusion, rice MEL1 is a key gene involved in meiosis in plant germ cells. Phylogenetic, cytological, and mRNA expression analyses suggest that MEL1 is a novel plant AGO that functions to maintain germ cell identity, possibly by repressing the ectopic expression of somatic genes and organizing the structure and modification of heterochromatin or NORs. To elucidate the relationship between MEL1 and other AGOs will reveal a novel view of the gene regulatory system via small RNA–mediated gene silencing in plant sexual reproduction.
METHODS
Plant Materials
Rice (Oryza sativa) calli derived from the japonica cv Nipponbare were cultured in a suspension for 5 months to activate a transposition of Tos17 and regenerated into mature plants as described by Hirochika et al. (1996). From 27,998 regenerated plants, reduction of seed fertility was observed in 4032 lines, in both the first generation of regenerated plants (R1 generation) and in their first and second generation progeny (R2 and R3 generations). From those sterile lines, 600 lines segregating sterile plants in the R2 generation were randomly chosen and used for further analyses. The mel1-1 homozygous mutants and their wild-type siblings used for genetic analyses in this study were mainly R4 generation individuals, and those used for histological and molecular analyses were mainly BC4F2 generation individuals. All materials were grown in a field in the city of Mishima, Shizuoka, Japan, or in a greenhouse at temperatures of 30°C during the day and 24°C at night.
Genetic Analysis
The linkage relationship between the sterile phenotype and transposed Tos17 fragments was analyzed by DNA gel blot hybridization and PCR using 295 R3 plants. DNA extraction, DNA gel blotting, cloning of the Tos17-tagged sequence, and isolation of the flanking sequence of the Tos17-tagged MEL1 gene were performed as described by Nonomura et al. (2003).
PCR genotyping for the mel1-1 population was performed using a mixture of three primers: 494, 5′-GCTTTTGGATTTTTGCTCAC-3′; 606, 5′-GCATTGTCTCAAGCAGAGTTAAGGC-3′; and T17LTR2MR, 5′-GCTAATACTATTGTTAGGTTGCAAG-3′. We confirmed that the 494 and 606 primers amplified only the MEL1 locus of numerous rice AGO loci (see Supplemental Figure 2C online). PCR conditions were as follows: initial incubation at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 4 min. The 4.0-kb product from the wild-type allele and the 3.6-kb product from the mutant allele were divided on 0.8% agarose gels by electrophoresis at 100 V for 1 h.
Molecular Cloning of the MEL1 cDNA
Full-length cDNA was obtained from young rice panicles by both screening a Uni-ZAP XR (Stratagene) cDNA library and RACE technology using the Marathon cDNA amplification kit (BD Biosciences). First, the Tos17-flanking sequence of MEL1 was used as a probe to screen ∼35,000 plaques of the Uni-ZAP library. Of 20 randomly selected positive clones, the Uni-ZAP clone with the largest insert was used for the synthesis of a full-length cDNA. For 5′ RACE, total RNAs were extracted from 10- to 30-mm-long young panicles using the RNeasy plant mini kit (Qiagen), and poly(A)+ RNAs were purified using Oligotex-dT30 Super (Takara Bio). The specific antisense 579 primer, 5′-TGTCTCGCCTCCCATGAACCTCTG-3′, and the AP1 primer were used for the first RACE PCR, and the 579 and AP2 primers were used for the nested PCR (the AP primers were supplied in the Marathon kit). The RACE product was cloned into the pCR-BluntII-TOPO vector (Invitrogen) to add an EcoRI site to the 5′ end, excised using EcoRI and NheI, and inserted upstream of the Uni-ZAP insert. The resultant full-length MEL1 cDNA was inserted between the EcoRI and XhoI sites in pBluescript SK− phagemid (Stratagene). The cDNA sequence was determined using an ABI3100 sequencer (Applied Biosystems) and GENETYX-MAC 12.0 software (Genetyx).
Complementation Test for the mel1 Mutant Phenotype
The 5′ terminus of the MEL1 coding region was highly GC-rich, and the upstream cis region was highly AT-rich (data not shown). Thus, the genomic MEL1 sequence was difficult to subclone. We succeeded in subcloning this region by extracting an 18-kb fragment twofold larger than the MEL1 gene. First, a 26-kb XhoI fragment including the MEL1 gene was isolated from the rice BAC clone OSJNBa0052F07 using the CHEF Mapper pulsed-field gel electrophoresis system (Bio-Rad). Next, both 10- and 8-kb BamHI fragments were isolated from the 26-kb fragment, and the 18-kb fragment including the MEL1 gene was reconstructed within the pPZP2H-lac binary vector (Fuse et al., 2001), named pKN16. The pKN16 and empty vectors were introduced into mel1-1/mel1-1 homozygous and +/mel1-1 heterozygous calli in accordance with the method of Hiei et al. (1994).
Phylogenetic Analysis
The peptide sequences of the PAZ and PIWI domains were aligned using ClustalW on the DNA Data Bank of Japan website (http://www.ddbj.nig.ac.jp/). After insertions and deletions were trimmed, the sequences were again aligned and used to construct a phylogenetic tree using the neighbor-joining method (Saitou and Nei, 1987) and TreeView1.6.6 software (Rod Page; http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
Expression Analysis of mRNAs
In situ hybridization and RT-PCR were performed in accordance with the method described by Nonomura et al. (2003). For the synthesis of RNA probes, the downstream half of the MEL1 cDNA was amplified by PCR and cloned into the pCRII-TOPO vector (Invitrogen) using the primers 606 and 795, 5′-CCTGAAATCACCAAATACCG-3′. The antisense and sense RNA probes were transcribed in vitro from the SP6 or T7 promoters using the DIG RNA labeling kit (Roche). The 795 and 606 primers were also used for RT-PCR.
Histological Analysis of Reproductive Organs
For TEM observations, young rice flowers ranging from 0.6 to 2.0 mm in length were fixed with 4% (w/v) paraformaldehyde in PMEG buffer (25 mM PIPES, 5 mM EGTA, 2.5 mM MgSO4, 4% glycerol, and 0.2% DMSO, pH 6.8) for 3 h, washed six times with PMEG for 20 min each, and stored at 4°C. After the first fixation, samples were refixed with 2.5% glutaraldehyde in PBS at 4°C for 2 h. Ultrathin sectioning, staining with saturated uranyl acetate/lead citrate, and TEM observations were performed in accordance with the method described by Nonomura et al. (2006). For optical tissue sectioning, young anthers were stained with propidium iodide, whole-mounted on a glass slide, and observed using the FluoView CLSM system (Olympus) in accordance with the method of Nonomura et al. (2003).
Indirect Immunofluorescence and Fluorescence in Situ Hybridization
Indirect immunofluorescence and meiotic chromosome observation of rice PMCs isolated by the enzymatic digestion of anthers were performed. For whole-mount immunofluorescence, the tip of an anther from a 1.0-cm rice flower was cut off with forceps, incubated in an enzyme cocktail containing 2% cellulase Onozuka-RS (Yakult Honsha), 0.3% pectolyase Y-23 (Kikkoman), and 0.5% Macerozyme-R10 (Yakult Honsha) in PMEG buffer at 37°C for 30 min, and supplied for immunological staining steps in accordance with the method of Nonomura et al. (2006). Rabbit polyclonal IgG of anti-dimethyl-histone H3 (Lys-9) (No. 07-441; used at 1:300) and anti-phospho-histone H3 (Ser-10) (No. 06-570; used at 1:100) (both from Upstate Biotechnology) were used to stain dimethylated H3K9 and phosphorylated H3S10, respectively. To stain spindle microtubules, monoclonal antibody against rat α-tubulin subunit OBT0614S (AbD Serotec) was used at a 1:100 dilution. The centromere and meiotic chromosomal axes were stained with anti-Os CenH3, guinea pig polyclonal IgG (used at 1:2000) and anti-PAIR2, mouse polyclonal IgG (used at 1:3000) (Nonomura et al., 2006). The chromosomes were counterstained with 40 μg/mL TO-PRO-3 iodide (Invitrogen)/Vectashield (Vector).
The signal intensity of dimethylated H3K9 was estimated after normalization against the intensity of counterstaining. After the images obtained above were converted into TIFF format, the gray value of the chromatin area counterstained with the TO-PRO-3 was measured using ImageJ 1.36b software (Abramoff et al., 2004). The gray value of the dimethylated H3K9 signal was also measured in the same area with TO-PRO-3 and then divided by the gray value of the TO-PRO-3 signal for normalization (i.e., the H3K9 intensity was indicated by the percentage against TO-PRO-3 intensity in the chromatin area in this study).
For double staining of rice PMCs with indirect immunofluorescence and fluorescence in situ hybridization, digoxigenin-labeled 25S rDNA was amplified from cv Nipponbare genomic DNAs by PCR and using the DIG DNA labeling kit (Roche) as described by Nonomura et al. (2004). Fixation of anthers, enzymatic digestion, and slide preparation were performed in accordance with the standard protocol for indirect immunofluorescence described above. PAIR2 detection and CLSM observations were performed in accordance with the standard protocol for indirect immunofluorescence (Nonomura et al., 2006).
Accession Numbers
The DNA Data Bank of Japan accession number for the MEL1 cDNA is AB297928. The Rice Annotation Project Database (http://rapdb.lab.nig.ac.jp/index.html) locus identifier for MEL1 is Os03g0800200.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genetic Linkage Analysis for the mel1-1 Sterile Phenotype and Tos17 Insertion.
Supplemental Figure 2. Genomic Structure of the MEL1 Gene.
Supplemental Figure 3. Histological Analysis of Wild-Type and mel1-1 Gametogenesis in Anthers by Optical Sectioning.
Supplemental Figure 4. PMCs of the mel1-1 Mutant Were Arrested at Early Meiosis I.
Supplementary Material
Acknowledgments
We thank E. Suzuki (National Institute of Genetics, Japan), N. Nagata (Japan Women's University), K. Kusumi (Kyushu University), and S. Arimura and M. Nakazono (University of Tokyo) for assistance with the TEM observations of mitochondria. This work was supported partly by the Green Techno Project “Comparative Genomics for Understanding the Diversity of Cereal Crops” (Grant GD2002) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (to K.-I.N.) and partly by Grants-in-Aid for Scientific Research on the 2006–2010 Specified Study “Genome Barriers in Plant Reproduction” from the Ministry of Education and Science of Japan (to N.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ken-Ichi Nonomura (knonomur@lab.nig.ac.jp).
Online version contains Web-only data.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophoto. Int. 11 36–42. [Google Scholar]
- Aravin, A., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 203–207. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12 1718–1727. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Girard, A., van de Kant, H.J., Bourc'his, D., Bestor, T.H., de Rooij, D.G., and Hannon, G.J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12 503–514. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. (2002). The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16 2733–2742. [DOI] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, G.L. (1966). Systematic Embryology of the Angiosperms. (New York: John Wiley & Sons).
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18 219–222. [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–51. [DOI] [PubMed] [Google Scholar]
- Grivna, S.T., Pyhtila, B., and Lin, H. (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 103 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M., Noma, K., and Grewal, S.I. (2003). RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Sun, H., and Poethig, R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. [DOI] [PubMed] [Google Scholar]
- Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., Riechmann, J.L., and Meyerowitz, E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 356–360. [DOI] [PubMed] [Google Scholar]
- Itoh, J., Nonomura, K.I., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., Hattori, T., Miyao, A., Hirochika, H., Ashikari, M., and Matsuoka, M. (2004). Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, A., Oliva, M., Mosquna, A., Hakim, O., and Ohad, N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37 707–719. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., Yamaguchi, S., and Carthew, R.W. (2002). RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 16 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2005). The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev. Biol. 280 504–517. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., and Ganley, A.R. (2005). Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Horiuchi, T., Tongaonkar, P., Vu, L., and Nomura, M. (2004). SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117 441–453. [DOI] [PubMed] [Google Scholar]
- Lin, H., and Spradling, A.C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124 2463–2476. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126 469–481. [DOI] [PubMed] [Google Scholar]
- Mamun, E.A., Cantrill, L.C., Overall, R.L., and Sutton, B.G. (2005). Cellular organisation and differentiation of organelles in pre-meiotic rice anthers. Cell Biol. Int. 29 792–802. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1997). Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A., Ito, M., Kamiya, N., Sato, Y., and Matsuoka, M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30 189–201. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.I., Miyoshi, K., Eiguchi, M., Suzuki, T., Miyao, A., Hirochika, H., and Kurata, N. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura, K.I., Nakano, M., Eiguchi, M., Suzuki, T., and Kurata, N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119 217–225. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.I., Nakano, M., Fukuda, T., Eiguchi, M., Miyao, A., Hirochika, H., and Kurata, N. (2004). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono, K., and Sugiura, M. (1980). Heterogeneity of the ribosomal RNA gene clusters in rice. Chromosoma 76 85–89. [Google Scholar]
- Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. (2004). Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669–672. [DOI] [PubMed] [Google Scholar]
- Papini, A., Mosti, S., and Brighigna, L. (1999). Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 207 213–221. [Google Scholar]
- Peng, J.C., and Karpen, G.H. (2007). H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan, V. (1988). Anther and pollen development in rice (Oryza sativa). Am. J. Bot. 75 183–196. [Google Scholar]
- Reiser, L., and Fischer, R.L. (1993). The ovule and the embryo sac. Plant Cell 5 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Nishida, K.M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., and Siomi, M.C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., and Dickinson, H.G. (2004). Stamen structure and function. Plant Cell 16 (suppl.): 46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A., Guerineau, F., Canales-Holzeis, C., Dickinson, H.G., and Scott, R.J. (2002). A novel extinction screen in Arabidopsis thaliana identifies mutant plants defective in early microsporangial development. Plant J. 29 581–594. [DOI] [PubMed] [Google Scholar]
- Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., Nishizawa, N.K., Gomi, K., Shimada, A., Kitano, H., Ashikari, M., and Matsuoka, M. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47 427–444. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, D.P. (2001). Cytokinesis and building of the cell plate in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 751–784. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wu, H.M., and Cheung, A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44 267–281. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M., Tsugawa, H., Miyao, A., Yano, M., Wu, J., Yamamoto, S., Matsumoto, T., Sasaki, T., and Hirochika, H. (2001). The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol. Genet. Genomics 265 336–344. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit, E., Batista, P.J., Bei, Y., Pang, K.M., Chen, C.C., Tolia, N.H., Joshua-Tor, L., Mitani, S., Simard, M.J., and Mello, C.C. (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747–757. [DOI] [PubMed] [Google Scholar]
- Zhao, D.Z., Wang, G.F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.