Abstract
Glanzmann thrombasthenia, an inherited bleeding disorder, can be caused by a defect or deficiency in platelet integrin αIIbβ3 (GPIIb-IIIa). Studies of thrombasthenia variants have facilitated identification of sites involved in the functions of αIIbβ3 and other integrins. Such sites include those that bind ligand and those that participate in the “activation” of αIIbβ3 required for high affinity binding of ligands such as fibrinogen or PAC1, a monoclonal antibody. Here we describe the isolation of such variants, created in vitro with Chinese hamster ovary cells that express an activated form of αIIbβ3. These cells were exposed to a mutagen, ethyl methane sulfonate, and variants that lost the capacity to bind PAC1 were isolated by fluorescence-activated cell sorting. These variants were grouped into three phenotypic classes. One comprised integrin mutations that disrupt ligand binding function; a second comprised mutations that interfere with the capacity of cells to activate the integrin. Most of these activation-defective mutations were in the integrin cytoplasmic domain, but surprisingly, several were caused by mutations affecting three closely spaced residues in the β3 extracellular domain. A third class of mutants exhibited a defect in integrin activation not ascribable to changes in the integrin sequence. Thus, these may represent mutated signaling molecules required for integrin activation. This unbiased genetic approach provides new insights into the structural basis of integrin function and may assist in identifying the cellular events that regulate integrin function.
Cell adhesion receptors of the integrin family mediate cell–cell and cell–matrix interactions critical to development, immune cell function, and hemostasis (1). Cells can regulate these processes by changing the affinity of integrins between two different states. For example, interactions at the cytoplasmic face of integrins may lead to a conformational change in the extracellular domain, resulting in conversion of the receptor from a low-affinity state to a high-affinity state. Furthermore, the binding of an adhesive ligand to an integrin can result in signals that are transduced to the cytoplasmic face, resulting in intracellular responses, such as protein phosphorylation and cytoskeletal rearrangements (1).
Platelet aggregation is essential for normal hemostasis and is dependent on integrin αIIbβ3 (GPIIb-IIIa; ref. 1). Aggregation depends on the capacity of αIIbβ3 to become activated and thus bind soluble fibrinogen with high affinity. Glanzmann thrombasthenia is an inherited bleeding disorder usually due to reduced or absent αIIbβ3. However, in rare cases, αIIbβ3 is expressed but fails to function due to the presence of mutations or deletions (2). Sequence analysis of the β3 integrin subunit from four such thrombasthenic variants has pinpointed two functionally important regions of the β3 subunit: the ligand binding region (D119Y, R214Q, R214W) and the cytoplasmic domain (S752P). Theoretically, there are two additional sites at which genetic alterations could cause thrombasthenia: within an intracellular signaling molecule that regulates integrin affinity state or between the ligand binding and cytoplasmic regions of the integrin necessary for propagation of the activation response.
Previously, the only method of obtaining functional variants of αIIbβ3 caused by random mutations was to study individuals with Glanzmann thrombasthenia. To overcome the inefficiency inherent in the rarity of this syndrome, we developed a novel method of identifying induced mutations that affect αIIbβ3 integrin function. First, cells expressing a chimeric constitutively active form of αIIbβ3 were chemically mutagenized. Then cells incapable of binding the fibrinogen-mimetic PAC1 antibody were isolated as clonal populations using fluorescence-activated cell sorting (FACS). Using this approach, we have identified mutations in the integrin extracellular domain that result in loss of ligand binding function and mutations in the cytoplasmic domain that perturb integrin signaling. In addition, mutations within a contiguous 24-residue sequence in the β3 extracellular domain that result in defective activation have been identified. Finally, cell lines with defects in integrin activation not ascribable to mutations in the integrin itself have been isolated, implying mutations in putative integrin regulatory proteins.
MATERIALS AND METHODS
DNAs, Cell Lines, and Transfection.
Chinese hamster ovary cells (CHO-K1; American Type Culture Collection) were used because they are hypodiploid (3) and random loss of chromosomes should facilitate the recovery of recessive alleles. αβPy is a stable cell line created by transfecting Chinese hamster ovary cells with the three plasmids: β3β1, αIIbα6A, and a plasmid encoding a neomycin resistance gene, as described (4). The construction of chimeric plasmids containing the extracellular and transmembrane domains of human β3 or αIIb fused to the cytoplasmic domains of β1 (β3β1) or α6A (αIIbα6A) has been described previously (5).
All transfections used Lipofectamine (GIBCO/BRL) according to the manufacturer’s instructions as described (5).
Chemical Mutagenesis.
Approximately 2 × 106 nonconfluent αβPy cells were treated with 200–400 μg/ml ethyl methane sulfonate (EMS; Sigma) for 15–19 hr. After 1 week, ≈1 × 108 cells were harvested for FACS sorting. In the first round of EMS treatment (EMS-1), cells were exposed to 350 μg/ml EMS for 18 hr, and the mutagenic frequency was determined by positive selection for loss of the X-linked hypoxanthine-guanosine phosphoribosyl transferase gene as described (6), using 6-thioguanine. To determine the mutagenesis rate the number of 6-thioguanine-treated clones was divided by the starting number of cells (1 × 106) and corrected for the survival of untreated cells (≈50%).
Flow Cytometry.
Cells were individually sorted on a FACStar Plus (Becton Dickinson) using two-color flow cytometry. The monoclonal antibody, D57, was used to detect expression of αIIbβ3 (5), while PAC1, an activation-specific, fibrinogen-mimetic monoclonal IgM antibody was used to assess the activation state of the integrin (7). In some cases, antibodies were biotinylated according to the manufacturer’s instructions using the reagent biotin-N-hydroxy-succinimide (Sigma). For cell sorting, confluent cells were resuspended by treatment with trypsin and double-stained as described (5). Rare cells that exhibited bright phycoerythrin staining (D57), but weak fluorescein isothiocyanate (FITC) staining (PAC1), were individually sorted into 96-well dishes. Approximately 1 × 107 cells were sorted with 300–500 cells collected (or one cell per 104-105 cells). In two experiments, EMS-2 and EMS-6, cells were mass-sorted before the single-cell sort.
FACS analysis of isolated clones was performed on a FACScan as described (5). PAC1 staining in the presence of the competitive inhibitor Ro43-5054 (2 μM; provided by Beat Steiner, Hoffmann–La Roche, Basel; ref. 8) was used to estimate nonspecific binding. In addition, in some tubes, 2% anti-LIBS6 ascites was added to the cells during the incubation with PAC1. Anti-LIBS6 directly induces αIIbβ3 binding to PAC1 regardless of the status of cellular activation mechanisms and so was used to estimate maximal PAC1 binding (9).
For retransfection analysis, mutant cell lines were transfected with nonmutant αIIbα6Aβ3β1, along with a marker plasmid expressing Tac-α5, a chimeric protein composed of the extracellular and transmembrane regions of the Tac subunit of the interleukin 2 receptor and the integrin α5 cytoplasmic domain (10). Transfected cells were stained with PAC1 and FITC–anti-IgM, as above, along with a biotinylated anti-Tac antibody, 7G7B6 (American Type Culture Collection), followed by phycoerythrin–streptavidin. PAC1 staining was measured in only the subset of cells stained with phycoerythrin, indicating cells that had taken up the transfected DNA.
In some cell lines, binding of the activation-independent αIIbβ3 monoclonal antibody, OPG2, was also analyzed using flow cytometry (11), as described (12).
Sequencing of Recombinant Integrins.
Total cellular RNA was isolated using TRIzol (GIBCO/BRL); cDNA was synthesized using oligo(dT) primers and the cDNA Cycle kit (Invitrogen); and PCR was carried out using the Ampliwax “hot start” method (Perkin–Elmer). PCR products were either subcloned into the TA vector (Invitrogen) or sequenced directly using fluorescent automated sequencing (Applied Biosystems). To eliminate the possibility of PCR errors, two TA vector clones were analyzed per mutant. In the case of direct PCR sequencing, two separate tubes of PCR product per mutant were pooled before sequencing. To confirm the functional significance of specific mutations, PCR products were subcloned into full-length wild-type β3 (in pcDNA1; Invitrogen) and transfected into Chinese hamster ovary cells, along with αIIbα6A DNA. For N-terminal (ligand binding) mutations, the subcloned DNA was a 500-bp KpnI–BstXI fragment; for the central mutants, a 500-bp BstXI–BamHI fragment; and for C-terminal mutations, a 500-bp BamHI–XbaI fragment. C-terminal constructions included the transfer of β1 DNA, thus reconstructing the β3β1 chimera. All other constructions produced a full-length β3. All the mutants transferred into β3 behaved like the parental β3β1 constructs in the original mutant cell lines, as expected (5).
RESULTS AND DISCUSSION
Selection and Characterization of Cell Lines.
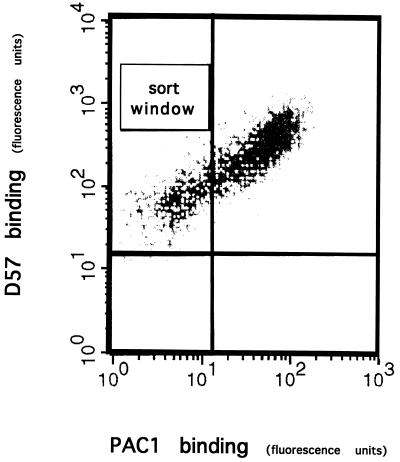
To isolate cells with defective integrin function, cells were exposed to a mutagen, EMS, and then sorted for loss of PAC1 binding activity. The antibody, PAC1, is both a ligand-mimetic and an activation-specific monoclonal antibody (7), allowing the detection of cells with both ligand binding and activation defects. In one experiment, the mutagenic frequency, or probability of inactivating a single gene, was 5 × 10−5 (see Materials and Methods). Two-color FACS was used to isolate the rare cells that failed to bind PAC1, yet continued to express αIIbβ3, as shown by the binding of the D57 monoclonal antibody (Fig. 1). D57 binds to the αIIbβ3 complex regardless of the activation state of the integrin. Using this scheme, one cell per 104-105 was isolated that expressed a recombinant integrin but in an inactive form, as manifest by its lack of PAC1 binding. This procedure was performed on eight separate occasions, to yield multiple independent mutational events. In total, 106 cell lines were isolated that expressed the recombinant integrin, but failed to bind PAC1.
Figure 1.
Isolation of mutants. FACS was used to isolate cells that express recombinant αIIbα6Aβ3β1 integrin, but in an inactive form. Cells were double-stained for expression of αIIbβ3 with D57 (y axis) and with the activation-specific antibody, PAC1 (x axis), as described. Rare cells that expressed the recombinant integrin, but failed to bind PAC1, were isolated (sort window).
Additional FACS analysis showed that the mutant cell lines bound a similar quantity of the anti-αIIbβ3 antibody, D57, as the parent cell line, confirming that the expression of the recombinant integrin was not reduced (data not shown). To distinguish between activation-defective and ligand binding-defective phenotypes, cell lines were tested for their ability to bind PAC1 in the presence of anti-LIBS6, a monoclonal antibody that converts αIIbβ3 to a high-affinity state (9). Cell lines in which anti-LIBS6 failed to restore PAC1 binding were initially classified as ligand binding-defective. When anti-LIBS6 restored PAC1 binding, cell lines were classified as activation-defective.
An activation-defective phenotype could arise from either mutations in the recombinant integrin or in cellular pathways that regulate affinity. To distinguish between these two possibilities, cell lines were retransfected with the wild-type constitutively active chimeric integrin. As these cells already expressed chimeric integrin, a marker plasmid, Tac-α5, was included to identify the population of cells that had taken up wild-type integrin during transfection. If the original defect was intrinsic to the integrin, then reintroduction of the wild-type receptor should restore PAC1 binding. However, if the defect was in an integrin-regulatory protein, then reintroduction of wild-type subunits should have no effect. To identify which subunit contained the defect, wild-type α or β subunits were transfected individually.
PAC1 binding in the presence of the activating anti-LIBS6 was assayed in all 106 cell lines (Figs. 2 and 3). In 34 of these cell lines, there was no PAC1 binding in the presence of anti-LIBS6, indicating a ligand binding-defective phenotype. Subsequent DNA sequence analysis (see below) identified a causative mutation in the ligand binding region of the β subunit in 30 cases. Thus, failure to bind PAC1 in the presence of anti-LIBS6 reliably identified mutations in this region.
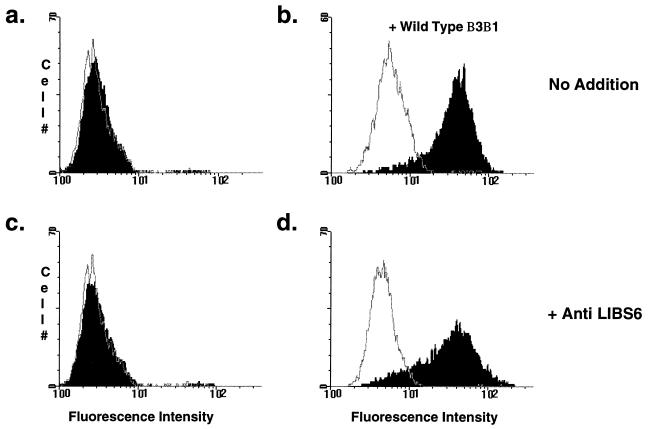
Figure 2.
Analysis of mutants. Cell lines were stained with PAC1 in the presence (open histograms) or absence (filled histograms) of 2 μM Ro43-5054. (a) All mutant cell lines failed to bind PAC1 (filled). Note that in the example shown, PAC1 fluorescence is superimposable on PAC1 fluorescence in the presence of the competitive inhibitor, Ro43-5054 (open). (b) Mutant cell lines were retransfected with wild-type αIIbα6Aβ3β1 and Tac-α5. Cells were double-stained with PAC1 and anti-Tac 48 hr later. Depicted is PAC1 staining on the gated subset of Tac-positive cells. In some transfected mutant cell lines, PAC1 binding was restored, indicating that the defect was in the integrin. (c and d) Mutant cell lines were stained with PAC1 as described in a. In these assays, anti-LIBS6 was added at the same time as PAC1. With the addition of anti-LIBS6, some mutant cell lines still failed to bind PAC1 (c), indicating a ligand binding-defective phenotype. In others, anti-LIBS6 restored PAC1 binding (d), indicating an activation defect.
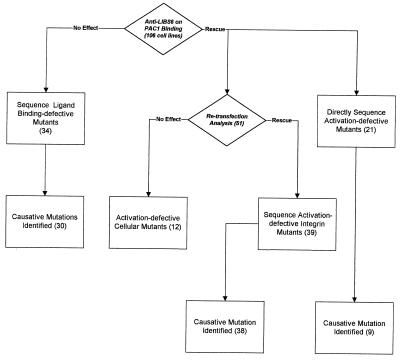
Figure 3.
Characterization of mutants. A total of 106 cell lines was isolated in this study. Of these, 34 failed to bind PAC1 in the presence of anti-LIBS6 and were therefore categorized as ligand binding-defective. Sequence analysis identified a causative mutation in the ligand binding region of the β subunit in 30 cases. Seventy-two cell lines bound PAC1 in the presence of anti-LIBS6 and were therefore categorized as activation-defective. Of these, 51 were retransfected with wild-type chimeric integrin. In 39 cell lines, PAC1 binding was restored, indicating the original defect was in the integrin; 12 cell lines failed to increase PAC1 binding, indicating a possible defect in a signaling molecule responsible for integrin activation. Sequence analysis of an additional 21 activation-defective cell lines identified nine more mutations. Thus, a total of 47 β subunit mutations that cause an activation-defective phenotype were found.
In the remaining 72 cell lines, PAC1 binding was restored in the presence of anti-LIBS6, indicating an activation defect. Of these cell lines, 51 were retransfected with the wild-type integrin, resulting in reconstitution of PAC1 binding in 39 of them. In each case, retransfection with the β subunit alone was sufficient to restore binding function (Fig. 2). Sequence analysis identified causative mutations in the β subunit of 38 of the cell lines. Thus, the retransfection analysis was successful in identifying integrin mutations that disrupt activation.
The use of these two assays, PAC1 binding in the presence of anti-LIBS6 and retransfection analysis, identified three broad classes of mutants. Two classes contained presumptive mutations within the integrin itself, resulting in either a ligand binding-defective or activation-defective phenotype. All identified mutations were within the β subunit. This could be because α subunit mutations may disrupt integrin expression more frequently or because the parent cell line may have had multiple integrated functional α subunit cDNAs. The third class had an activation-defective phenotype, presumably due to disruption of a cellular signaling pathway.
Characterization of Ligand Binding Defective Mutants.
The β subunit was sequenced from reverse transcriptase-PCR products obtained from cDNA. Each cell line appeared to have only one functional copy of the β subunit, because the same mutation was found in multiple cDNA clones. Furthermore, in one case a mutation (GAA TTC → AAA TTC) eliminated an EcoRI site present in β3. All of the PCR products from this cell line were resistant to EcoRI digestion, proving that this mutation was present in all of the expressed β subunit mRNA.
The functional effects of all mutations described were confirmed by introducing the mutation into a plasmid encoding the wild-type β subunit. Chinese hamster ovary cells transiently transfected with cDNA encoding mutant β, along with wild-type chimeric αIIbα6A, were tested for PAC1 binding in the presence and absence of anti-LIBS6. In each case, the transiently transfected mutant integrin failed to bind PAC1, and bound (or failed to bind) PAC1 in the presence of anti-LIBS6, consistent with the original mutant cell line. These results establish that each mutation was responsible for the functional defect.
Sequence analysis of 30 cell lines classified as ligand binding-defective yielded seven mutations within or near the presumptive ligand binding region mapped to Asp109–Glu171 and Ser211–Gly222 (13). There are multiple examples of several of these mutations arising from independent mutational events (Table 1). Two of these changes were at residues affected by naturally occurring mutations in thrombasthenic patients: R214W and D119N. Three other alterations affected oxygenated residues in the ligand binding region (D217N, E220K, and E220Q), and a fourth affected a proline in this region (P219S). In one cell line, a substitution outside the putative ligand binding domain, A252V, was responsible for the ligand binding-defective phenotype. Thus this approach was able to identify multiple mutations that disrupt integrin ligand binding function.
Table 1.
Mutations affecting integrin function
| Ligand binding | Activation, integrin
|
Activation, cellular | |
|---|---|---|---|
| Extracellular | Cytoplasmic | ||
| D119N (1) | E312K (2) | R740X (1) | (19) |
| R214W (2) | G331E (1) | E749K (1) | |
| D217N (14) | S334F (2) | W755X (2) | |
| P219S (1) | P761S (28) | ||
| E220K (8) | 761 dup (1) | ||
| E220Q (1) | Y775X (1) | ||
| A252V (1) | |||
On eight separate occasions (EMS-1–EMS-8), cells were treated with EMS and sorted for loss of PAC1 antibody binding. Each EMS experiment yielded multiple different mutations, which have been classed into four major types. Mutant cell lines that failed to restore PAC1 binding upon retransfection with wild-type integrin were classed as cellular mutations. Mutant cell lines that restored PAC1 binding with retransfection were found to have a mutation in the integrin itself. These mutations were found in the ligand binding region, the cytoplasmic domain, or a discrete extracellular region that may be important in the transmission of signal from the cytoplasm to the ligand binding region. Ligand binding mutants were distinguished from activation mutants, as they failed to bind PAC1 in the presence of anti-LIBS6. Although 106 cell lines were isolated, only those fully characterized are shown here. The residue changes of each integrin mutation are listed, followed in parentheses by the number of independent cell lines isolated that have that mutation. In all cases where a mutation was isolated more than once, those isolates were obtained from at least two different EMS experiments. X, truncation at the indicated residue.
The substitutions identified here provide new insight into the ligand binding function of integrins. All the affected residues, with the exception of Arg214 and Ala252, are highly conserved among different β subunits (Fig. 4). Spontaneous mutations at Asp119 and Arg214 have been previously identified in thrombasthenic patients (2), while the other residues have not yet been observed as naturally occurring mutations. Ligand binding function was impaired by conservative substitutions at Asp217 and Glu220 (D217N and E220Q) indicating that these residues are very sensitive to oxygenated substitutions as well as to substitutions with alanine (12).
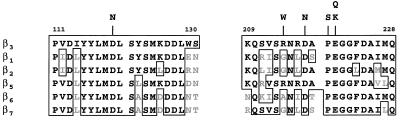
Figure 4.
Mutants of the ligand binding region. An alignment of the ligand binding region of several human integrin β subunits is shown. The boxed areas are conservative residues shared with β3; the gray areas are residues that are not shared. Shown above the alignment are the amino acid changes of the mutants isolated in this study.
Pro219 and Ala252 have also been implicated in ligand binding function by this analysis. Pro219 is a highly conserved residue within the known ligand binding region, while Ala252 is further downstream and is not well conserved among different β subunits. Both mutations eliminated PAC1 binding, both in the presence and absence of anti-LIBS6. Since direct binding of anti-LIBS6 to these cells was not reduced (data not shown), this is consistent with a ligand binding-defective phenotype. However, in contrast to the other mutants, these two bound another ligand-mimetic antibody, OPG2, to the same extent as the wild-type receptor (data not shown). Furthermore, both cell lines adhered to fibrinogen, although significantly more weakly than cells expressing the wild-type chimeric receptor (data not shown). Consequently, P219S and A252V appear to cause a ligand-specific loss of αIIbβ3 function, or block the ability of anti-LIBS6 to activate the integrin.
These data provide support for a recently developed molecular model of the ligand binding domain of β3 (12). Specifically, Asp119, Asp217, and Glu220 were proposed to be part of a metal ion coordination site, similar to the MIDAS motif in the I domain of αM and αL (14, 15). We found that the conservative substitutions of Asn for Asp and Gln for Glu at these positions block ligand binding function. Moreover, the same molecular model predicts that residue Pro219 must be in the cis conformation for Glu220 to efficiently coordinate the divalent cation. It follows that any substitution, such as the mutation P219S, would be in trans. Thus, the adverse effect of this mutation provides independent confirmation of the proposed structure of this putative metal ion binding site.
Characterization of Activation Defective Integrin Mutants.
Retransfection analysis or direct sequencing identified 47 mutations in the β subunit associated with an activation-defective phenotype. Several of these alterations were due to nonsense codons in the cytoplasmic domain, for example, truncations at Arg740 (R740X), Trp755 (W755X), and Tyr775 (Y775X) (Fig. 5A). Also, an insertion of an extra nucleotide base pair between the codons for residues 760 and 761 resulted in a truncation three residues downstream (Asn760-SYLX). Consequently, similar to β3 (16), integrin activation seems to require a near full-length β1 cytoplasmic domain.
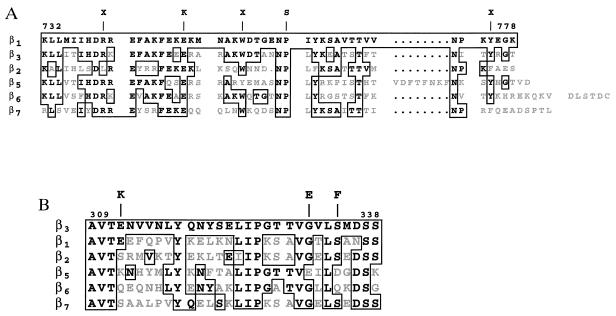
Figure 5.
Integrin activation mutants. Shown is an alignment of the cytoplasmic regions (A) and central extracellular regions (B) of several human β subunits. The boxed areas are conservative residues shared with β1 (A) and β3 (B); the gray areas are residues which are not shared. Shown above the alignment are the amino acid changes of the mutants isolated in this study.
We identified two point mutations that disrupted activation via the β1 cytoplasmic domain. β1(P761S) (Fig. 5A) disrupts the first NPXY motif in the β1 tail. Thus, the Pro in addition to the Asn and Tyr of this motif are required for activation (17). In addition, E749K substitution also blocked activation. This Glu is a completely conserved residue in a motif implicated in binding to α actinin (18). Interestingly, other nonconservative substitutions in this region also disrupt activation (17).
Surprisingly, mutations in a contiguous 23-residue region of the β3 extracellular domain (E312K, G331E, and S334F) also manifested an activation-defective phenotype. Anti-LIBS6 induced integrins containing β3 (E312K) or β3 (G331E) to bind PAC1. In contrast, β3 (S334F) didn’t bind PAC1 in the presence of anti-LIBS6, but did so in the presence of another activating antibody, anti-LIBS1 (data not shown). Glu312 is not a conserved residue, while Gly331 and Ser334 are partially conserved (Fig. 5B). To determine whether these particular mutations also affected the capacity of the integrin to initiate outside-in signaling and regulate cell shape, cell lines expressing β3 containing mutations E312K, G331E, or S334F were plated on fibrinogen. All mutant cell lines exhibited cell spreading (data not shown).
These three mutations eliminated integrin activation but were not located within the cytoplasmic domain. Thus β3 (E312K, G331E, and S334F) define a new class of integrin functional variants. These substitutions appear to interfere with inside-out signaling, that is, the capacity of the ligand binding site to change in response to signals generated at the cytoplasmic face. Alternatively, this region of β3 might be necessary for the binding of a cofactor required for integrin activation. Further analysis of this discrete sequence in β3 may provide the means to identify such a potential cofactor.
Characterization of Activation Defective Cellular Mutants.
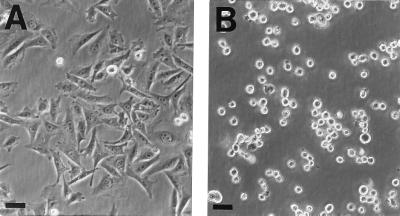
Retransfection of wild-type α and β subunits into 12 cell lines did not reconstitute PAC1 binding (data not shown). Indeed, nine of these lines had an additional phenotype: they failed to spread on substrates such as serum-coated tissue culture plastic (Fig. 6), fibrinogen, or fibronectin (data not shown). Although these cells failed to spread, adherence to saturating concentrations of these substrates (i.e., 10 μg/ml) was not grossly reduced on visual inspection. Cell spreading of these lines on fibrinogen is dependent on the presence of αIIbα6Aβ3β1, while spreading on fibronectin and serum-coated tissue culture plastic is dependent on the presence of endogenous Chinese hamster ovary cell integrins, such as α5β1 and αV integrins. Because the spreading defect was manifest with a variety of substrates, involving different integrins, these cell lines appear to have a defect(s) in the cellular machinery involved in the function of multiple integrins.
Figure 6.
Some activation-defective cellular mutants fail to spread. Equal numbers of cells were plated out on tissue culture plastic in complete medium. After incubation overnight at 37°C, the cells were viewed and photographed using a Nikon Diaphot inverted phase microscope. (A) The parental cell line, expressing αIIbα6Aβ3β1, spread normally on serum-coated tissue culture plastic. The cells had a similar appearance when plated on fibrinogen or fibronectin (data not shown). (B) In contrast, nine of these cell lines failed to spread on any of these substrates. Depicted is the appearance of a representative cell line, plated on tissue culture plastic. (Bars = 20 μm.)
These cell lines, with putative defects in integrin signaling pathways, may provide a means for identification of intermediates in these pathways. Recently, Mobley et al. (19) isolated two Jurkat cell lines that manifested reduced adhesion to fibronectin. In addition, these cells apparently failed to activate β1 and β2 integrins. One of these cells manifested reduced mobility of the mitogen-activated protein kinase, ERK1, on SDS/PAGE, suggesting it might be phosphorylated and activated. Activation of a mitogen-activated protein kinase reduces integrin activation (4), suggesting that mutational activation of an integrin suppressor pathway could account for defective activation in some of the cell lines obtained by us and by Mobley et al. Somatic cell complementation analysis should permit further characterization of these cell lines. This will enable identification of those lines that are suitable to clone cDNAs encoding proteins that complement defects in integrin signaling.
In conclusion, we have developed a method to introduce and characterize cellular mutations that disrupt ligand binding to integrin αIIbβ3. In so doing, new mutations that affect both the ligand binding site and integrin activation have been identified. This in vitro approach should complement the analysis of Glanzmann thrombasthenia in patients and enable a more complete understanding of the molecular basis of the adhesive and signaling functions of this integrin.
Acknowledgments
We thank Virginia Keivens, Jane Forsyth, and Susan Pei for excellent technical assistance. This work was supported in part by grants from the National Institutes of Health and COR Therapeutics. E.K.B. is supported by a fellowship from the Arthritis Foundation. This paper represents Publ. No. 10369VB from Scripps Research Institute.
ABBREVIATIONS
- FACS
fluorescence-activated cell sorting
- EMS
ethyl methane sulfonate
References
- 1.Schwartz M A, Schaller M D, Ginsberg M H. Annu Rev Cell Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 2.Bray P F. Thromb Haemostasis. 1994;72:492–502. [PubMed] [Google Scholar]
- 3.Kao F T, Puck T T. Proc Natl Acad Sci USA. 1968;60:1275–1281. doi: 10.1073/pnas.60.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes, P. E., Renshaw, M. W., Pfaff, M., Forsyth, J., Keivens, V. M., Schwartz, M. A. & Ginsberg, M. H. (1996) Cell, in press. [DOI] [PubMed]
- 5.O’Toole T E, Katagiri Y, Faull R J, Peter K, Tamura R, Quaranta V, Loftus J C, Shattil S J, Ginsberg M H. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Neill J P, Hsie A W. Nature (London) 1977;269:815–817. doi: 10.1038/269815a0. [DOI] [PubMed] [Google Scholar]
- 7.Shattil S J, Hoxie J A, Cunningham M, Brass L F. J Biol Chem. 1985;260:11107–11114. [PubMed] [Google Scholar]
- 8.Alig L, Edenhofer A, Hadvary P, Hurzeler M, Knopp D, Muller M, Steiner B, Trzeciak A, Weller T. J Med Chem. 1992;35:4393–4407. doi: 10.1021/jm00101a017. [DOI] [PubMed] [Google Scholar]
- 9.Frelinger A L, Du X P, Plow E F, Ginsberg M H. J Biol Chem. 1991;266:17106–17111. [PubMed] [Google Scholar]
- 10.LaFlamme S E, Akiyama S K, Yamada K M. J Cell Biol. 1992;117:437–447. doi: 10.1083/jcb.117.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomiyama Y, Tsubakio T, Piotrowicz R S, Kurata Y, Loftus J C, Kunicki T K. Blood. 1992;79:2303–2312. [PubMed] [Google Scholar]
- 12.Collins Tozer E, Liddington R C, Sutcliffe M J, Smeeton A H, Loftus J C. J Biol Chem. 1996;271:21978–21984. doi: 10.1074/jbc.271.36.21978. [DOI] [PubMed] [Google Scholar]
- 13.Loftus J C, Smith J W, Ginsberg M H. J Biol Chem. 1994;269:25235–25238. [PubMed] [Google Scholar]
- 14.Lee J, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 15.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes P, O’Toole T E, Ylanne J, Shattil S J, Ginsberg M H. J Biol Chem. 1995;270:12411–12417. doi: 10.1074/jbc.270.21.12411. [DOI] [PubMed] [Google Scholar]
- 17.O’Toole T E, Ylanne J, Culley B M. J Biol Chem. 1995;270:8553–8558. doi: 10.1074/jbc.270.15.8553. [DOI] [PubMed] [Google Scholar]
- 18.Otey C A, Vasquez G B, Burridge K, Erickson B W. J Biol Chem. 1993;268:21193–21197. [PubMed] [Google Scholar]
- 19.Mobley J L, Ennis E, Shimizu Y. J Immunol. 1996;156:948–956. [PubMed] [Google Scholar]