Abstract
Parsley (Petroselinum crispum) plants and suspension-cultured cells have been used extensively for studies of non-host-resistance mechanisms in plant/pathogen interactions. We now show that treatment of cultured parsley cells with a defined peptide elicitor of fungal origin causes rapid and large changes in the levels of various unsaturated fatty acids. While linoleic acid decreased and linolenic acid increased steadily for several hours, comparatively sharp increases in oleic acid followed a biphasic time course. In contrast, the overall level of stearic acid remained unaffected. Using a PCR-based approach, a parsley cDNA was isolated sharing high sequence similarity with ω-3 fatty acid desaturases. Subsequent isolation and characterization of a full-length cDNA enabled its functional identification as a plastid-localized ω-3 fatty acid desaturase by complementation of the Arabidopsis thaliana fad7/8 double mutant which is low in trienoic fatty acids. ω-3 Fatty acid desaturase mRNA accumulated rapidly and transiently in elicitor-treated cultured parsley cells, protoplasts, and leaves, as well as highly localized around fungal infection sites in parsley leaf buds. These results indicate that unsaturated fatty acid metabolism is yet another component of the highly complex, transcriptionally regulated pathogen defense response in plants.
Keywords: cultured parsley cells, fungal elicitor, Phytophthora sojae, unsaturated fatty acids
Plant defense against pathogens encompasses a wide array of biochemical reactions resulting in extensive metabolic changes within the affected tissue (1, 2). Many of these reactions are triggered upon perception of the pathogen or pathogen-derived elicitors and involve the de novo synthesis of proteins via transcriptional activation of defense-related genes (3, 4). Several defense-related genes have been identified using various plant pathosystems, and many of their products were found to represent diverse enzymes involved in many different metabolic routes. For example, all three enzymes of the general phenylpropanoid pathway, phenylalanine ammonia-lyase, cinnamate 4-hydroxylase, and 4-coumarate:CoA ligase, which link primary with secondary metabolism, are transcriptionally induced by elicitor (5), as well as many enzymes involved in the subsequent branch pathways that lead to the production of antimicrobial phytoalexins, lignin, and other soluble or cell wall-bound phenolics (6–8). Similarly, genes encoding enzymes of the shikimate pathway have been shown to be activated by elicitor (9, 10).
Challenge of parsley leaves with the fungus Phytophthora sojae results in a non-host-resistance reponse that very effectively limits pathogen ingress (11). Fungal growth is restricted to a few invaded cells directly adjacent to the penetration site, which then rapidly undergo cell death (hypersensitive response). Elucidation of the molecular and biochemical events associated with this response have been aided greatly by the use of suspension-cultured parsley cells that, when treated with a defined fungal peptide elicitor molecule (Pep25), closely mimic various facets of the infection-induced plant defense response (12, 13). Using this simplified system, we have isolated and characterized numerous parsley genes whose transcription is rapidly activated both in elicitor-treated cells and in fungus-infected leaf tissue (14, 15). These genes encode not only various enzymes of secondary metabolism but also enzymes of primary metabolism, such as S-adenosyl-methionine synthetase and S-adenosyl-homocyteine hydrolase, two enzymes of the activated methyl cycle (16). Recently, sequence analysis suggested that some of the elicitor-stimulated genes also encode enzymes involved in fatty acid biosynthesis (17).
Here we report that treatment of parsley cells with Pep25 elicitor leads to large changes in the levels of unsaturated fatty acids. Furthermore, we show by complementation experiments that an isolated parsley cDNA encodes ω-3 fatty acid desaturase (ω-3FAD) and demonstrate that expression of the corresponding gene is strongly enhanced in elicitor-treated cells and in fungus-infected leaf buds of parsley. Various possible functional implications, including a role in defense-related signaling, will be discussed.
MATERIALS AND METHODS
Plant/Fungal Material and Elicitor Treatment.
Cultured parsley (Petroselinum crispum L.) cells were grown in the dark for 6 days under previously described conditions (18) and used for the isolation of protoplasts (19). Parsley leaf RNA was obtained from 5-month-old greenhouse-grown plants. Oligopeptide elicitor (Pep25) treatment was performed by direct addition to the suspension cultures/protoplasts (final concentration, 0.5 μg/ml) or by pressure infiltration via the stomata on nondetached leaves using a 1-ml syringe (1 μg/ml). Conditions for growth of parsley seedlings and generation of zoospores from the fungus P. sojae for in situ RNA hybridization experiments were as described (20). Arabidopsis thaliana fatty acid double mutant, fad7-1/8-1 (21), seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH; stock no. CS8036). For the generation of transgenic lines, the A. thaliana plants were grown in phytochambers under short-day light (8 h) or for flower induction under long-day light (12–16 h) conditions. Selection of surface-sterilized seeds was carried out on plates containing 0.5× Murashige–Skoog salt mixture, 250 mg/l claforan, and 100 mg/l kanamycin. After 7-8 days plants were transferred to soil.
Fatty Acid Analysis.
Fatty acid methyl esters were prepared according to the method of Browse et al. (22). Briefly, 1 ml of freshly prepared methanolic HCl (143 μl acetyl chloride in 1 ml methanol) was added to 50 mg of plant material in Teflon-sealed glass vials, heated for 1 h at 80°C, and then mixed with 1 ml of 150 mM NaCl. Fatty acid methyl esters were extracted into 300 μl of isooctane, and 1–5 μl samples were analyzed in a 10-m, 0.2-mm inner diameter carbowax gas chromatography column (26 cm·s−1 H2, 140–230°C temperature gradient). The fatty acid methyl esters were identified by comparison of their retention times to authentic standards.
RNA Isolation and Analysis.
Total RNA was isolated from leaves of transgenic A. thaliana plants according to the procedure of Verwoerd et al. (23). RNA was isolated from cultured parsley cells, protoplasts and leaves using the total RNA kit (Qiagen, Hilden, Germany). Approximately 20 μg of RNA per lane was denatured and separated in 1.2% (wt/vol) formaldehyde-agarose gels. The RNA was transferred to Hybond-N nylon membranes (Amersham) and cross-linked by UV irradiation. Prehybridization and hybridization conditions were as described (16).
Cloning of ω-3FAD cDNAs.
Two degenerate primers, 5′-CGGAATTCAT(A/C/T)CC(A/T)AA(A/G)CA(C/T)TG(C/T)TGG-3′ and 5′-CGGGATCCA(C/T)TC(C/T)TT(G/C)CC(G/T)C(G/T)(A/G)TACCA-3′, encompassing two conserved regions within published plant ω-3FADs and containing convenient EcoRI/BamHI cloning sites were synthesized and used for reverse transcription–PCR amplification of specific cDNAs employing RNA from elicitor-treated parsley cells. The PCR-amplified product was digested with EcoRI and BamHI, cloned into the pBluescript II SK vector (Stratagene), and sequenced. The excised EcoRI/BamHI PCR fragment was labeled with [32P]dCTP by random priming (24) and used to screen 250,000 recombinant phage clones from a parsley λgt11 cDNA library (16). Three positive clones differing in insert size (clone 1, 1465 bp; clone 2, 1325 bp; and clone 3, 743 bp) were plaque purified and subcloned into the pBluescript II SK vector. The dideoxy chain-termination method was used for sequencing (25). All three clones were sequence identical within their overlapping regions and to the original PCR product.
Plant Transformation and Regeneration.
For functional complementation of the A. thaliana fad7-1/8-1 double mutant (21), a chimeric 1556-bp parsley cDNA was constructed. The cDNA contained the first 445 bp of clone 2, including 95 bp of 5′ untranslated region and 1111 bp of the remaining sequence from clone 1. These two cDNA clones were identical within their coding regions with the exception that clone 1 contained only 4 bp of 5′ untranslated sequence whereas clone 2 was shorter at the 3′ end. The chimeric insert was fused to the cauliflower mosaic virus 35S (−417) promoter thus replacing the GUS reporter gene cassette in the binary vector pGPTV-Kan (26). The construct was introduced into the Agrobacterium tumefaciens strain GV3101 (pMP90) (27) by the freeze/thaw transformation procedure.
A modified version of the in planta Agrobacterium-mediated gene transformation method (28) was employed for the generation of transgenic plants. Transformants containing the chimeric vector construct were transferred to soil and allowed to set seed. Seeds were plated on selective media and the ratio of survivors to nonsurvivors counted. Survivors of each line were planted in soil and pools of each line were analyzed.
In Situ Hybridization.
Primary leaf buds obtained from 7-day-old parsley seedling were harvested 6–10 h postinoculation with fungal zoospores. Fixation of the tissue, paraffin embedding, sectioning, and the procedure for RNA/RNA hybridization in situ were performed as described (20).
RESULTS
Elicitor-Induced Changes in Fatty Acid Composition.
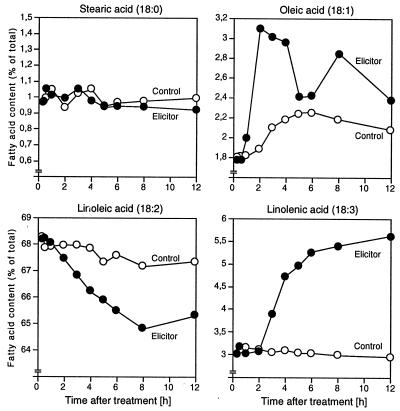
Treatment of parsley cells with Pep25 elicitor resulted in large changes in the levels of unsaturated fatty acids (Fig. 1). A rapid and sharp increase of oleic acid (18:1) within the first 2 h was followed by a transient decrease and by a second peak around 8 h. The biphasic nature of these changes was consistently observed in four independent experiments. In contrast, linolenic acid (18:3) increased steadily and reached nearly twice the initial value within 12 h, whereas linoleic acid (18:2) decreased to a similar extent, suggesting a direct metabolic flux. During the time period tested, the level of stearic acid (18:0) remained unchanged. Palmitic acid (16:0), which constituted about 19% of the total fatty acids in cultured parsley cells, decreased by about 1% (data not shown). The corresponding desaturated 16:1, 16:2, and 16:3 fatty acids together comprised less than 1% of the total fatty acids and were not analyzed in detail.
Figure 1.
Effects of Pep25 elicitor on four selected fatty acids in cultured parsley cells. Relative contents of the acids in control (○) and elicitor-treated cells (•) were measured at the indicated times by gas chromatography.
Cloning of a Parsley ω-3FAD.
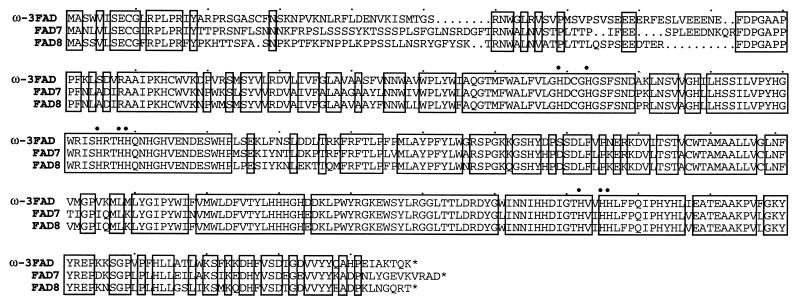
The observed increase in the 18:3 fatty acid level suggested an induction of the immediately responsible enzyme, ω-3FAD. To investigate this possibility further, degenerate primers, corresponding to two conserved regions found in various plant ω-3FADs, were synthesized and used for cDNA cloning by reverse transcription–PCR. A 586-bp PCR product was obtained, subcloned, sequenced, and used to isolate a near full-length cDNA from a random-primed parsley cDNA library (16). Three independent recombinant phage clones harboring inserts of 1465, 1325, and 743 bp, respectively, were isolated. The inserts were identical in sequence within their overlapping regions as well as with the original PCR fragment. The largest insert contained a 1317-bp ORF corresponding to 439 amino acids with a predicted Mr of 50,391 and a pI of 8.4. Sequence comparison with proteins in the database revealed highest identity/similarity to several plastid-localized ω-3FADs from A. thaliana (Fig. 2), castor bean and tobacco (21, 29–31), and to a somewhat lesser extent to other putative plastid-localized ω-3FADs from diverse plant species. Computer-assisted identification of a potential transit peptide at the N terminus of the deduced parsley protein indicates that it is targeted to plastids (32). Furthermore, the protein contains a putative membrane-spanning domain and the eight highly conserved histidine residues shown to be necessary for fatty acid desaturase function (ref. 33; Fig. 2).
Figure 2.
Comparison of the amino acid sequences deduced from the parsley cDNA and for two previously established A. thaliana plastidial ω-3FADs. Identical positions are boxed. The eight invariant histidines are indicated by dots.
Complementation of the Arabidopsis fad7/8 Double Mutant.
To functionally verify that the parsley cDNA codes for a plastid-localized ω-3FAD, genetic complementation experiments were conducted. The parsley cDNA containing 95 bp of 5′ untranslated region and the entire coding region was inserted into the binary Ti plasmid pGPTV-Kan under the control of the cauliflower mosaic virus 35S promoter (26). The construct was used to transform the A. thaliana fatty acid desaturation double mutant line fad7-1/fad8-1 (21) via in planta Ag. tumefaciens-mediated gene transfer.
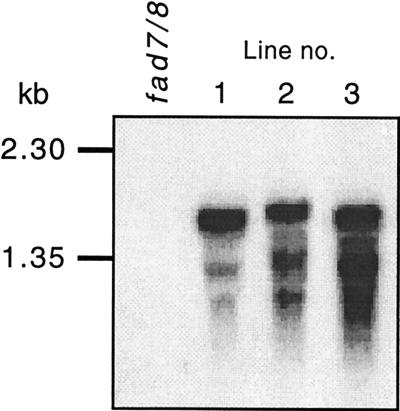
The trienoic (16:3, 18:3) fatty acid content in the fad7/8 double mutant is strongly reduced compared to both the wild-type and the respective single mutant lines (34). The presence of the parsley cDNA within the genome of T2 plants was verified by PCR amplification using specific primers. Descendents of the primary transformants segregated on selective media plates as expected for single, or in one case double, integrations of the parsley cDNA. Three lines showing strong expression of the transgene in leaves (Fig. 3) were selected for further analysis.
Figure 3.
mRNA levels of parsley ω-3FAD in three selected transgenic A. thaliana fad7/8 double mutant lines as compared with the untransformed line. The parsley 586-bp PCR product was used as probe for RNA blot hybridization.
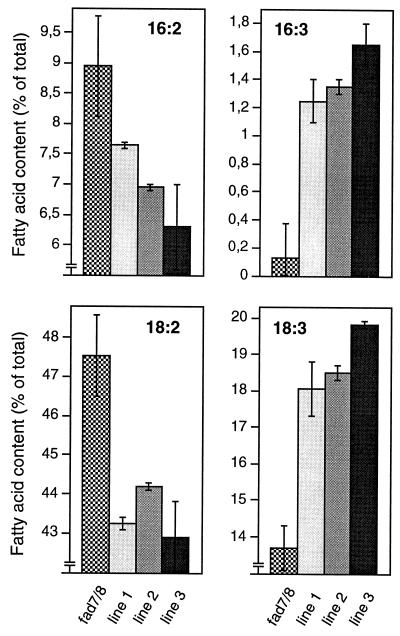
Gas chromatography analysis of the leaf fatty acid composition of the individual transformed lines demonstrated a clear increase in trienoic and a corresponding decrease in dienoic (16:2, 18:2) fatty acids in all three transgenic lines, as compared to that of the double mutant (Fig. 4), thus confirming that the parsley cDNA encodes an ω-3FAD.
Figure 4.
Functional complementation of the A. thaliana fad7/8 fatty acid double mutant with the parsley ω-3FAD cDNA. Polyunsaturated fatty acid contents of three transgenic lines were compared to those of the untransformed mutant.
Effects of Elicitor Treatment and Fungal Infection on ω-3FAD mRNA Levels.
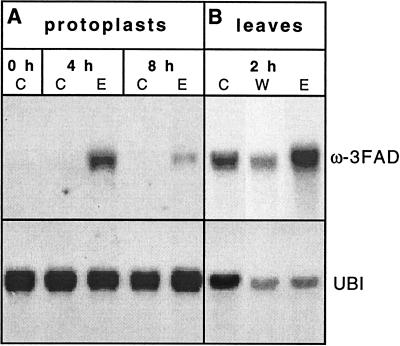
RNA blot analysis was used to measure the effect of Pep25 treatment on the ω-3FAD mRNA levels in cultured parsley cells, protoplasts, and leaves. In cultured cells, ω-3FAD mRNA accumulated very rapidly, strongly, and transiently upon elicitor treatment, with highest levels occurring within 1–2 h (Fig. 5). In accordance with the specific binding of Pep25 elicitor to sites on the plasma membrane and with the efficient triggering of various selected defense responses in the absence of the cell wall (13), Pep25 also induced the accumulation of ω-3FAD mRNA in parsley protoplasts (Fig. 6A). Pep25 infiltration into parsley leaves via the stomata likewise resulted in an increase in ω-3FAD mRNA, albeit from a higher basal level than in cultured cells (Fig. 6B). Control experiments using a ubiquitin cDNA (Upstate Biotechnology) as probe indicated that the amounts of loaded RNA from water (lane W) and elicitor (lane E)-infiltrated parsley leaves were comparable but significantly lower than from untreated leaves (lane C). And yet, the highest level of ω-3FAD mRNA accumulated in elicitor-infiltrated leaves.
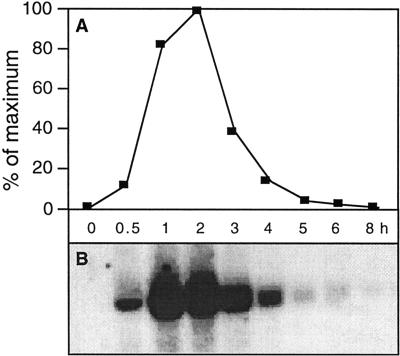
Figure 5.
Time course of changes in the ω-3FAD mRNA level in cultured parsley cells treated with Pep25 elicitor. Relative mRNA amounts (A) were determined from an RNA blot (B) using a PhosporImager. The blot was probed with a 350-bp fragment derived from the 5′ end of the parsley ω-3FAD cDNA.
Figure 6.
Changes in the ω-3FAD mRNA levels in parsley protoplasts and leaves upon treatment with Pep25 elicitor. RNA was isolated at the times indicated either (A) from control (lane C) or elicitor-treated (lane E) protoplasts or (B) from control (lane C), water-infiltrated (lane W), or elicitor-infiltrated (lane E) leaves. The RNA blots were first probed with a 350-bp fragment derived from the 5′ end of the parsley ω-3FAD cDNA and then reprobed with a ubiquitin cDNA (Upstate Biotechnology, Lake Placid, NY) to monitor variations in RNA loading.
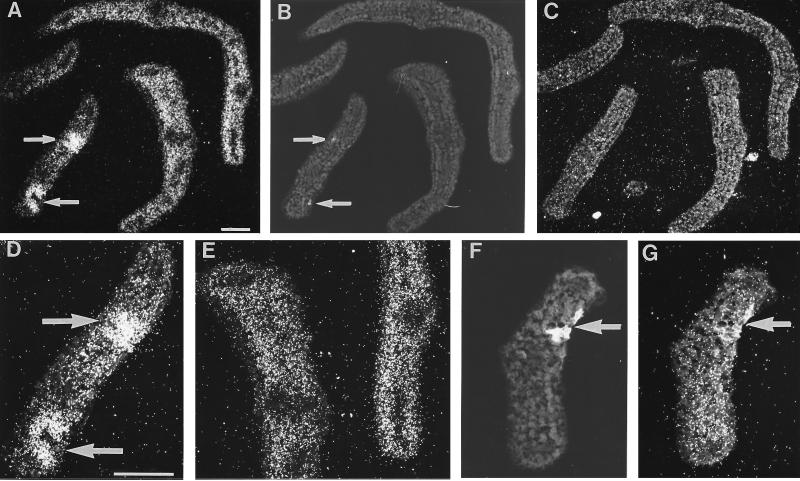
In situ RNA/RNA hybridization in parsley leaf buds (Fig. 7) demonstrated the predominant occurrence of ω-3FAD mRNA in the photosynthetically active mesophyll cells of uninfected tissue, whereas much lower signal intensities were detected in vascular tissue and in the epidermis (Fig. 7 A and E). In tissue infected with P. sojae, ω-3FAD mRNA strongly accumulated in sharply confined areas around fungal penetration sites (arrows in Fig. 7 A, B, and D). Concomitant with the induction at infection sites, the level of ω-3FAD mRNA appeared to decrease in the adjacent mesophyll (Fig. 7 A and D). The induction by fungal infection was strong as early as 6 h postinoculation with fungal spores. However, already 10 h postinoculation, the hybridization signal at the infection site was no longer discernible from that in the surrounding tissue (Fig. 7G), indicating a very rapid and transient accumulation of the transcript.
Figure 7.
In situ localization of ω-3FAD mRNA in infected (arrows) and uninfected areas of parsley leaf buds. Two adjacent cross sections were hybridized with antisense (A) or sense (C) ω-3FAD RNA 6 h postinoculation with P. sojae spores. Infection sites (arrows) were detected by UV fluorescence of necrotic spots (B) in the same section as shown in A. Larger magnifications of selected areas in A are shown in D and E. Another cross-section through a separate leaf bud was taken 10 h postinoculation for UV light exposure (F) and hybridization with ω-3FAD antisense RNA (G). (Bars = 100 μm.)
DISCUSSION
These results demonstrate that the previously reported large complexity of the response of cultured parsley cells to fungal elicitor also includes changes in the metabolism of unsaturated fatty acids. More specifically, we showed that one of the strongly elicitor-responsive genes encodes ω-3FAD and that expression of this gene occurs rapidly and highly locally around fungal penetration sites. The combination of all three aspects, the functional involvement in the biosynthesis of unsaturated fatty acids, the extremely rapid and transient activation, and the sharply localized expression at infection sites, makes this gene particularly interesting and sets it apart from all other elicitor- and pathogen-responsive genes so far analyzed in parsley.
In terms of speed of mRNA appearance and disappearance in elicitor-treated parsley cells, ω-3FAD is second only to two recently identified genes encoding the putative transcription factors WRKY1 and WRKY3 (35), but is unique among all elicitor-responsive enzyme-encoding genes. It is remarkable that all of the genes tested so far with respect to their expression behavior at fungal infection sites in whole-plant tissue exhibited similar relative time courses of transcriptional activation as in elicitor-treated cell cultures. This includes such extreme cases as the particularly late response of a gene encoding the phytoalexin-biosynthetic enzyme bergaptol O-methyltransferase (14) and the biphasic, coordinated responses of the genes encoding phenylalanine ammonia-lyase and 4-coumarate:CoA ligase (5, 20). ω-3FAD represents a different type of extreme, where an exceptionally early and transient mRNA induction in elicitor-treated cell cultures again mimics a very similar behavior at true infection sites. In fact, to our knowledge ω-3FAD is the first infection-responsive gene analyzed whose transcript has returned to background level as early as 10 h postinoculation of parsley leaves with fungal spores. Moreover, no other gene activation has so far been so sharply confined around fungal infection sites, with the possible exception of a peroxidase gene that, however, responded much more slowly (36).
Earlier biochemical studies using Phaseolus vulgaris and Poa pratensis had already indicated alterations in lipid metabolism upon fungal infection of whole plant tissue (37, 38). However, research in this area has remained rather limited. Recently, Mueller et al. (39) demonstrated a rapid increase in free 18:3 fatty acids in Eschscholtzia californica cell suspension cultures treated with a yeast elicitor. Treatment of tobacco cells with a fungal elicitor derived from Phytophthora parasitica var nicotianae resulted in a decrease in phospholipids concomitant with an increase in phospholipase A activity (40). Furthermore, expression of a putative ω-6 fatty acid desaturase gene was strongly induced in tomato plants infected with the citrus exocortis viroid (41). In parsley, we have previously identified numerous fungal elicitor-responsive genes (15), two of which, ELI7 and ELI12, showed high sequence similarity with microsomal fatty acid desaturases/hydroxylases (17, 42). Subsequent screening of an A. thaliana genomic library with the parsley ELI12 probe resulted in the isolation of the microsomal ω-6 fatty acid desaturase gene locus, fad2, previously identified by Browse and coworkers (43). Expression of this fad2 gene was shown to be strongly induced in cultured A. thaliana cells by fungal elicitor (44). In contrast, no increase in the level of the microsomal fatty acid desaturase 3 (FAD3) mRNA was observed in A. thaliana ecotypes Columbia or Pr-0 leaves inoculated with the bacterial pathogen Xanthomonas campestris pv campestris (45). Whether this difference is due to differential regulation of the genes tested or to the different conditions used remains to be elucidated.
The parsley cDNA described here most likely encodes a plastid-localized ω-3FAD, as concluded from both the presence of a putative N-terminal transit peptide (32) and the efficient complemention of an A. thaliana double mutant line deficient in plastid desaturase activity. Results from DNA blot analysis suggest that ω-3fad is a member of a small family of two or three genes per haploid parsley genome (data not shown). In A. thaliana, two members, fad7 and fad8, have been isolated that can be distinguished on the basis of their differential expression behavior at various temperatures (21). Whether a similar situation exists in parsley remains to be elucidated.
The most intriguing question arising from our present data concerns the functional significance of such rapid changes in fatty acid metabolism in response to pathogen attack. A surprising finding was recently reported for garden geranium where formation of ω5 anacardic acid via expression of a specific Δ9 14:0-acyl carrier protein fatty acid desaturase gene appeared to directly confer pest resistance (46). On the other hand, polyunsaturated fatty acids, particularly 18:2 and 18:3, have been proposed to be important precursors in the generation of specific cellular signals such as jasmonate (47, 48). Endogenous jasmonate and its methyl ester have been shown to increase rapidly in diverse plants upon wounding or pathogen attack and to activate defense-related genes (49). Recent studies imply that the chloroplast membrane plays an important role in jasmonate formation both by releasing the linolenic acid substrate and as the site of two enzymes of jasmonate biosynthesis, allene oxide synthase, and lipoxygenase (50–54). In parsley cells, addition of Pep25 elicitor led to a rapid and massive increase in jasmonate production, concurrent with the release of hydrogen peroxide (oxidative burst) and before the induction of numerous defense-related genes (2, 55, 56). Thus, activation of the ω-3fad gene may serve to indirectly supply the substrate for jasmonate synthesis and replenish depleted membrane lipids of the chloroplast and/or to restore membranes damaged through the generation of free radicals and lipid hydroperoxides during the oxidative burst. Alternatively, ω-3fad activation may be involved in the generation of other fatty acid derivatives that either directly act as endogenous signal molecules or indirectly serve as precursors for the synthesis of polymers associated with local suberization of cells or for reinforcement of cell walls.
Apart from these functional considerations, the rapid, highly localized response of the ω-3fad gene at fungal infection sites renders its promoter particularly interesting with regard to the pursuit of novel strategies for disease resistance breeding (57).
Acknowledgments
This study is dedicated to Prof. Benno Parthier (Halle, Germany) on the occasion of his 65th birthday. We thank Dr. Olaf Batz for the RNA blot hybridization depicted in Fig. 5, Dr. Nobert Martini for help with the fatty acid analysis, and Dr. Robert Cormack (all of this institute) for critical reading of the manuscript. M.T.-K. was recipient of a Deutscher Akademischer Austauschdienst fellowship.
ABBREVIATIONS
- 18:2
linoleic acid
- 18:3
linolenic acid
- ω-3FAD
ω-3 fatty acid desaturase
Footnotes
References
- 1.Bowles D J. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 2.Kombrink E, Somssich I E. In: Defense Responses of Plants to Pathogens. Andrews J H, Tommerup I C, editors. Vol. 21. London: Academic; 1995. pp. 1–34. [Google Scholar]
- 3.Dixon R A, Harrison M J. Adv Genet. 1990;28:165–234. doi: 10.1016/s0065-2660(08)60527-1. [DOI] [PubMed] [Google Scholar]
- 4.Somssich I E. In: Regulatory Elements Governing Pathogenesis-Related (PR) Gene Expression. Nover L, editor. Vol. 20. Berlin: Springer; 1994. pp. 163–179. [DOI] [PubMed] [Google Scholar]
- 5.Logemann E, Parniske M, Hahlbrock K. Proc Natl Acad Sci USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudet A M. New Phytol. 1995;129:203–236. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 7.Smith C J. New Phytol. 1996;132:1–45. doi: 10.1111/j.1469-8137.1996.tb04506.x. [DOI] [PubMed] [Google Scholar]
- 8.Hahlbrock K, Scheel D. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- 9.Herrmann K M. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmid J, Amrhein N. Phytochemistry. 1995;39:737–749. [Google Scholar]
- 11.Jahnen W, Hahlbrock K. Planta. 1988;173:197–204. doi: 10.1007/BF00403011. [DOI] [PubMed] [Google Scholar]
- 12.Hahlbrock K, Scheel D, Logemann E, Nürnberger T, Parniske M, Reinold S, Sachs W R, Schmelzer E. Proc Natl Acad Sci USA. 1995;92:4150–4157. doi: 10.1073/pnas.92.10.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nürnberger T, Nennstiel D, Jabs T, Sacks W, Hahlbrock K, Scheel D. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- 14.Schmelzer E, Krüger-Lebus S, Hahlbrock K. Plant Cell. 1989;1:993–1001. doi: 10.1105/tpc.1.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somssich I E, Bollmann J, Hahlbrock K, Kombrink E, Schulz W. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- 16.Kawalleck P, Plesch G, Hahlbrock K, Somssich I E. Proc Natl Acad Sci USA. 1992;89:4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamiya-Wik M. Ph.D. thesis. Cologne, Germany: Univ. of Cologne; 1995. [Google Scholar]
- 18.Kombrink E, Hahlbrock K. Plant Physiol. 1986;81:216–221. doi: 10.1104/pp.81.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dangl J L, Hauffe K-D, Lipphardt S, Hahlbrock K, Scheel D. EMBO J. 1987;6:2551–2556. doi: 10.1002/j.1460-2075.1987.tb02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinold S, Hahlbrock K. Plant Physiol. 1996;112:131–140. doi: 10.1104/pp.112.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson S, Arondel V, Iba K, Somerville C. Plant Physiol. 1994;106:1615–1621. doi: 10.1104/pp.106.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browse J, McCourt P J, Somerville C R. Anal Biochem. 1986;152:141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]
- 23.Verwoerd T C, Dekker B M M, Hoekema A. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg A P, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen E Y, Seeburg P H. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 26.Becker D, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 27.Koncz C, Schell J. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 28.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 29.Yadav N S, Wierzbicki A, Aegerter M, Caster C S, Pérez-Grau L, Kinney A J, Hitz W D, Booth R, Jr, Schweiger B, Stecca K L, Allen S M, Blackwell M, Reiter R S, Carlson T J, Russel S H, Feldmann K A, Pierce J, Browse J. Plant Physiol. 1993;103:467–476. doi: 10.1104/pp.103.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Loo F J, Somerville C. Plant Physiol. 1994;105:443–444. doi: 10.1104/pp.105.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada T, Nishiuchi T, Kodama H, Nishimura M, Iba K. Plant Cell Physiol. 1996;37:606–611. doi: 10.1093/oxfordjournals.pcp.a028988. [DOI] [PubMed] [Google Scholar]
- 32.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 34.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rushton P J, Torres J T, Parniske M, Wernert P, Hahlbrock K, Somssich I E. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 36.Kawalleck P, Schmelzer E, Hahlbrock K, Somssich I E. Mol Gen Genet. 1995;247:444–452. doi: 10.1007/BF00293146. [DOI] [PubMed] [Google Scholar]
- 37.Hoppe H H, Heitefuss R. Physiol Plant Pathol. 1974;4:11–24. [Google Scholar]
- 38.Lösel D M. New Phytol. 1978;80:167–174. [Google Scholar]
- 39.Mueller M J, Brodschelm W, Spannagl E, Zenk M H. Proc Natl Acad Sci USA. 1993;90:7490–7494. doi: 10.1073/pnas.90.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy S, Pouénat M-L, Caumont C, Cariven C, Prévost M-C, Esquerré-Tugayé M-T. Plant Sci. 1995;107:17–25. [Google Scholar]
- 41.Gadea J, Mayda M E, Conejero V, Vera P. Mol Plant-Microbe Interact. 1996;9:409–415. doi: 10.1094/mpmi-9-0409. [DOI] [PubMed] [Google Scholar]
- 42.van de Loo F J, Braun P, Turner S, Somerville C. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trezzini G F, Horrichs A, Somssich I E. Plant Mol Biol. 1993;21:385–389. doi: 10.1007/BF00019954. [DOI] [PubMed] [Google Scholar]
- 45.Buell C R, Somerville S C. Mol Plant-Microbe Interact. 1995;8:435–443. [Google Scholar]
- 46.Schultz D J, Cahoon E B, Shanklin J, Craig R, Cox-Foster D L, Mumma R O, Medford J I. Proc Natl Acad Sci USA. 1996;93:8771–8775. doi: 10.1073/pnas.93.16.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creelman R A, Mullet J E. Proc Natl Acad Sci USA. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doares S H, Syrovets T, Weiler E W, Ryan C A. Proc Natl Acad Sci USA. 1995;92:4095–4098. doi: 10.1073/pnas.92.10.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conconi A, Miquel M, Browse J A, Ryan C A. Plant Physiol. 1996;111:797–803. doi: 10.1104/pp.111.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song W-C, Funk C D, Brash A R. Proc Natl Acad Sci USA. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y-L, Shirano Y, Ohta H, Hibino T, Tanaka K, Shibata D. J Biol Chem. 1994;269:3755–3761. [PubMed] [Google Scholar]
- 53.Bell E, Creelman R A, Mullet J E. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler E W. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- 55.Ellard-Ivey M, Douglas C J. Plant Physiol. 1996;112:183–192. doi: 10.1104/pp.112.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dittrich H, Kutchan T M, Zenk M H. FEBS Lett. 1992;309:33–36. doi: 10.1016/0014-5793(92)80733-w. [DOI] [PubMed] [Google Scholar]
- 57.Strittmatter G, Janssens J, Opsomer C, Botterman J. Bio/Technology. 1995;13:1085–1089. [Google Scholar]