Abstract
Efficient formation of scrapie isoform of prion protein (PrPSc) requires targeting PrPSc by glycophosphatidyl inositol (GPI) anchors to caveolae-like domains (CLDs). Redirecting the cellular isoform of prion protein (PrPC) to clathrin-coated pits by creating chimeric PrP molecules with four different COOH-terminal transmembrane domains prevented the formation of PrPSc. To determine if these COOH-terminal transmembrane segments prevented PrPC from refolding into PrPSc by altering the structure of the polypeptide, we fused the 28-aa COOH termini from the Qa protein. Two COOH-terminal Qa segments differing by a single residue direct the transmembrane protein to clathrin-coated pits or the GPI form to CLDs; PrPSc was formed from GPI-anchored PrPC but not from transmembrane PrPC. Our findings argue that PrPSc formation is restricted to a specific subcellular compartment and as such, it is likely to involve auxiliary macromolecules found within CLDs.
The posttranslational conversion of the cellular isoform of prion protein (PrPC) into the scrapie isoform of prion protein (PrPSc) is the fundamental process underlying both the transmission and pathogenesis of the prion disease (1, 2). While no difference in the covalent structures of PrPSc and PrPC have been discerned (3), their conformations differ markedly (4). After PrPC is synthesized in the endoplasmic reticulum, it transits through the Golgi to the cell surface where it is bound by a glycophosphatidyl inositol (GPI)-anchor (5, 6). At or near the cell surface, PrPC is either metabolized or converted into PrPSc (7). PrPC seems to reenter cells through caveolae-like domains (CLDs), a subcellular compartment defined biochemically by membranes rich in cholesterol and glycosphingolipids; this compartment also contains many GPI-anchored proteins (7–10). Whether PrPSc formation occurs in the CLDs where PrPC undergoes initial proteolytic cleavage to produce an NH2-terminally truncated protein designated PrPC-II (11, 12) is of considerable interest.
While the results of several studies have established that the formation of PrPSc is a posttranslational process that occurs after PrPC reaches the plasma membrane (13–15), defining the subcellular location where PrPSc is formed has been formidable. Although PrPC like other GPI-anchored proteins is thought to reenter cells through caveolae or CLDs (16), some investigators have argued that PrPC is sorted to clathrin-coated pits where PrPSc formation has been postulated to occur (17). Metabolic labeling studies indicate that PrPSc is formed before PrP transits into acidic endosomes where it is NH2-terminally truncated in cultured cells but not in rodent brain (18–21).
With this background, we sought to determine the role of the GPI anchor in PrPSc formation to gain information about the subcellular trafficking of PrPC and to assess the specificity of PrPSc formation. Our findings argue that PrPSc formation is confined to CLDs and although this is a pathologic process, it occurs within a specific subcellular domain. The apparent restriction PrPSc formation to CLDs would seem to argue that such a process is likely to involve auxiliary macromolecules found within this compartment. Such an auxiliary factor has been implicated in the conversion of PrPC into PrPSc based on the results of transgenetic studies where a chimeric human/mouse (Hu/Mo) PrP molecule but not Hu PrP rendered mice susceptible to Hu prions from patients who died of Creutzfeldt–Jakob disease (22).
MATERIALS AND METHODS
Cultured Cells and Antibodies.
Mouse neuroblastoma (N2a) cells were obtained from American Tissue Culture Collection. Scrapie-infected mouse neuroblastoma (ScN2a) cells are the persistently infected clones as described (23). ScN2a cells expressing the MHM2 PrP were the same as described (24). All the cells were grown and maintained at 37°C in MEM or RPMI 1640 medium supplemented with 10% fetal bovine serum. In some cases, cells were treated with phosphophatidylinositol phospholipase C (PIPLC). Cells grown on 60 mm dishes were rinsed with ice-cold PBS, then incubated with 0.5 unit PIPLC in 1 ml of OptiMEM (GIBCO-BRL) at 37°C for 4 h with swirling.
α-PrP 3F4 is a mAb raised against Syrian hamster (SHa) PrP27–30 (25). To distinguish the signals of MHM2-constructs from endogenous mouse PrPSc, 3F4 mAb was used since this antibody exclusively recognizes SHa as well as MHM2 PrP at Met-109–Met-112 (26). RO73 is an antiserum raised in a rabbit against SDS/PAGE purified SHaPrP 27–30 that reacts with SHaPrP, MoPrP, and MHM2 PrP (13, 27).
Isolation of Triton-Insoluble Complexes.
To test solubility of the Qa-constructs in cold Triton X-100, we used the procedure for the isolation of detergent-insoluble, glycosphingolipid-enriched complexes previously described (28). Briefly, cells grown on 150-mm culture dishes (Corning) were lysed and homogenized in 2 ml of ice-cold Mes-buffered saline (25 mM Mes, pH 6.5/0.15 M NaCl) containing 1% Triton X-100. Homogenates were adjusted to 40% sucrose, then centrifugation was carried out at 39,000 rpm for 16 h at 4°C, and a 5–30% linear sucrose gradient was formed above them. After centrifugation, gradients were fractionated by collecting 1 ml fraction starting from the top of the gradients.
Recombinant Gene Construction and Transfection.
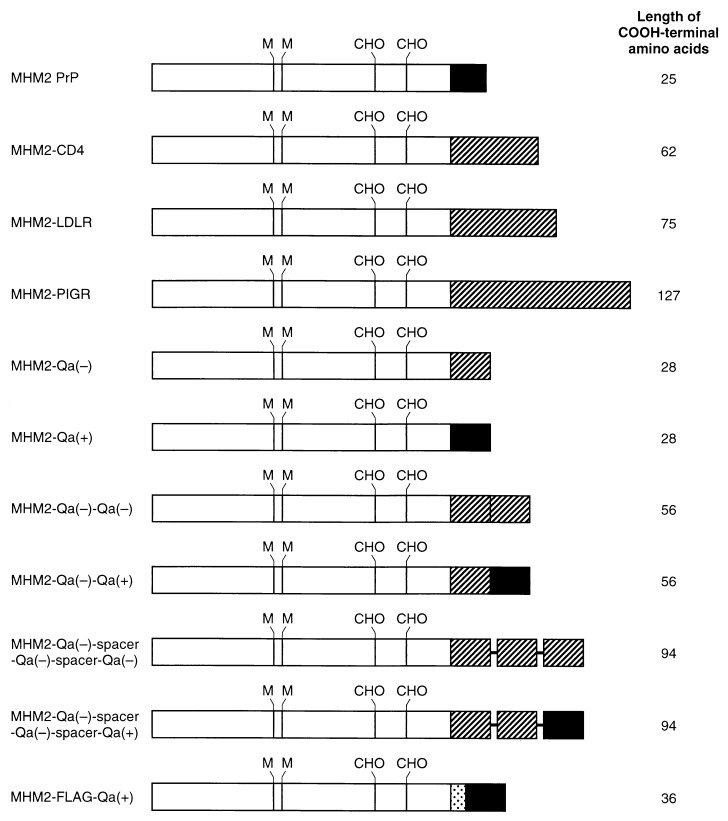
MHM2 PrP with a wild-type GPI anchor was constructed as described (24). PvuI and XhoI restriction sites were introduced in the CD4 by using the mismatched (underlined) CD1 (5′-A GGG GTG AAC CGA T*CG GTG TTC CTG GCT) resulting in the mutations Gln-367 → Arg and Thr-368 → Ser, and CD2 (5′-CTC ATC TGA GGC CTC GAG CCA CCT GCA). BamHI and SalI sites were introduced in the low density lipoprotein receptor (LDLR) by using the mismatched LDL1 (5′-AG AAG CCC GGA TCC GTG AGG GC), resulting in the mutation Ser-704 → Gly, and LDL2 (5′-GGC AGG GGG TCG ACT CCA GGC A). BamHI and XhoI sites were introduced in the polymeric immunoglobulin receptor (PIGR) by using the mismatched PIG1 (5′-AA AGC GGG GGA TCC AAA GTA CT), resulting in the mutations Ser-627 → Gly and Ala-628 → Ser, and PIG2 (5′-GGC GGT GGC TCG AGG TGC CTA G). After the PCR, amplified fragments were digested with restriction enzymes as described above, then ligated into the MHM2 PrP (deletions 231–254) (29), yielding MHM2-CD4, MHM2-LDLR, and MHM2-PIGR (Fig. 1), which were then introduced into the expression plasmid pSPOX (24). The phosphorylated (Ph) Q1 (5′-CG TCT GCC ACC ATC GCT GTC GTG GAT CTT GGA GCC GTC) and unphosphorylated (UPh) Q2 (5′-T CGC GAC GGC TCC AAG AAC CAC GAC AGC GAT GGT GGC AGA CGA T) were ligated into PvuI-digested MHM2 PrP (deletions 231–254), then digested with BglII. The Q1 and Q2 contained a mismatch (underlined) that segregated into two different mutations, MHM2-Qa(−) carrying Val-295 and MHM2-Qa(+) carrying Asp-295. The Ph Q4 (5′-TC GAG CTA GCG GCG ATT CAT GAC GAA GGC GAC AAC AGC TCC GAT GA) and UPh Q3 (5′-GCG ATC ATC GGA GCT GTT GTC GCC TTC GTC ATG AAT CGC CGC TAG C) were ligated into XhoI-cleaved MHM2 PrP, digested with BglII, then ligated to produce a MHM2-Qa(−) or MHM2-Qa(+) (Fig. 1). The Ph Qa-mod12 (5′-CT TCT GCC ACC ATC GCT GTC GTG GAT CTT GGA GCC GTC) and UPh Qa-mod22 (5′-T CGC GAC GGC TCC AAG ATC CAC GAC AGC GAT GGT GGC AGA AGA T) were ligated into PvuI-digested MHM2 PrP (deletions 231–254), then digested with HindIII. The Ph Q4 and UPh Q3 were ligated into XhoI-cleaved MHM2 PrP, digested with HindIII, then ligated together to produce a MHM2-Qa(−)*. The Ph Qa-C12 (5′-CG ATC ATC GGA GCT GTT GTC GCC TTC GTC ATG AAT CGC CGC TAG AT) and UPh Qa-C22 (5′-CTA GCG GCG ATT CAT GAC GAA GGC GAC AAC AGC TCC GAT GAT CG) were ligated into NruI-digested MHM2-Qa(−)*, digested with PvuI, then ligated into PvuI-digested MHM2 Qa(+) or MHM2-Qa(−) to produce a MHM2-Qa(−)-Qa(+) or MHM2-Qa(−)-Qa(−) (Fig. 1).
Figure 1.
Map of recombinant DNA constructs expressed in ScN2a cells using the pSPOX vector. Filled bars, GPI attachment sequences from wild-type MoPrP or Mo Qa(+). Hatched bars: Transmembrane and cytoplasmic sequences from either Mo CD4, rabbit LDLR, human PIGR, or Mo Qa(−). Dotted bar, FLAG sequence of Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys. Bold horizontal lines represent spacer sequence of Ser-Ala-Gly-Ser-Ala. CHO, N-glycosylation sites. M, Met at codon 109 and 112 of SHaPrP.
The Ph Qa-C32 (5′-CT ATC ATC GGA GCT GTT GTC GCC TTC GTC ATG AAT TCG GCC GGG TCG GCC CGC AGA T) and UPh Qa-C42 (5′-CT GCG GGC CGA CCC GGC CGA ATT CAT GAC GAA GGC GAC AAC AGC TCC GAT GAT AG) were ligated into NruI-digested MHM2-Qa(−)*, digested with PvuI, then ligated with PvuI-cleaved MHM2-Qa(−) to produce a MHM2-Qa(−)-spacer-Qa(−) (Fig. 1). Again, the phosphorylated Qa-C32 and unphosphorylated Qa-C42 were ligated into NruI-digested MHM2-Qa(−)-spacer-Qa(−), digested with PvuI, then ligated with PvuI-cleaved MHM2-Qa(−) or MHM2-Qa(+) to produce a MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(+) or MHM2-Qa(−)-spacer-Qa (−)-spacer-Qa(−) (Fig. 1).
The Ph Qa-MOD5FL (5′-CG GAC TAC AAG GAC GAT GAC GAT AAG TCG TCT GCC ACC ATC GCT GTC GTC GTG GAT CTT GGA GCC GTC) and Uph Qa-MOD6FL (5′-T CGC GAC GGC TCC AAG ATC CAC GAC GAC AGC GAT GGT GGC AGA CGA CTT ATC GTC ATC GTC CTT GTA GTC CGA T) were ligated into PvuI-cleaved MHM2 PrP, digested with HindIII. The Ph Q4 and UPh Q3 were ligated into XhoI-cleaved MHM2 PrP, digested with HindIII, then ligated together to produce a MHM2-FLAG-Qa(+) (Fig. 1).
ScN2a cells were either transiently or stably transfected with each construct using a DNA transfection kit (DOTAP, Boehringer Mannheim). In the case of stable transfection, they were then selected and maintained with 1 mg/ml of G418. Cell lysis and Western blot analysis were performed as described (24).
Immunofluorescence.
Immunofluorescence was done according to the methods described, with some modifications (7). Briefly, cells growing on a glass coverslip were incubated with the α-PrP 3F4 mAb (1: 50 of ascitic fluid) for 1 h at 4°C in 1% BSA in PBS. In some cases, cells were chased at 37°C for 45 min to examine whether they were efficiently internalized into a deep compartment. The cells were then examined with a fluorescence microscope (Leitz, Leica) or a Zeiss LSM410 inverse laser scan microscope.
RESULTS
Transmembrane Anchorage Inhibits the PrPSc Formation.
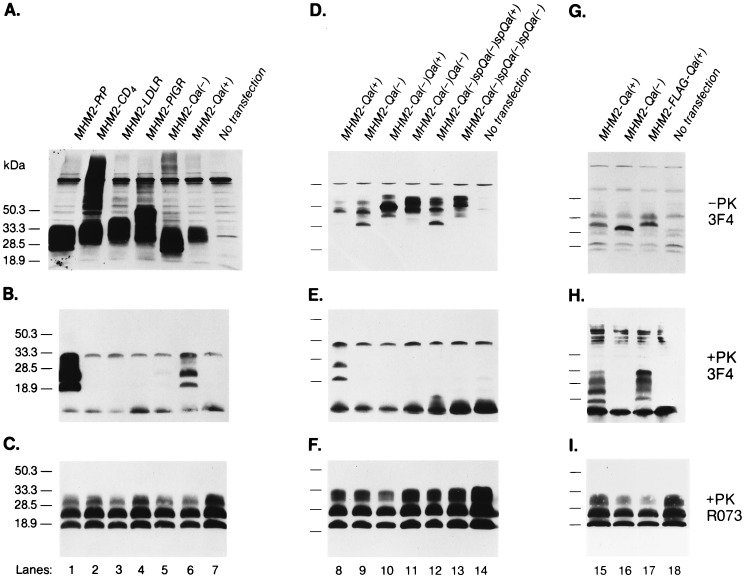
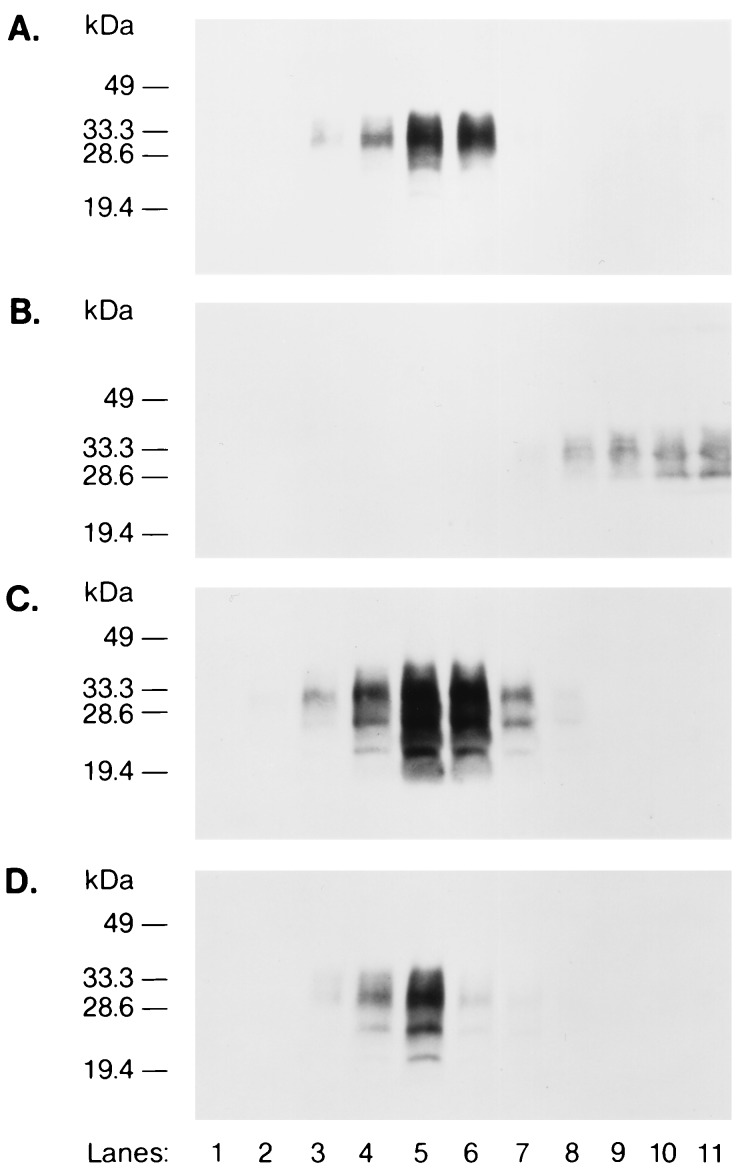
Under conditions where MHM2 PrP permitted maximal levels of MoPrPSc and MHM2 PrPSc to be produced (see Fig. 3 B and C, lane 1), we transfected pSPOX vectors encoding MHM2-CD4, MHM2-LDLR, MHM2-PIGR, and MHM2-Qa(−) (Fig. 1). The CD4, LDLR, PIGR, and Qa(−) COOH termini consisting of 62, 75, 127, and 28 aa, respectively, were fused to residue 231 of MHM2 PrP. All of these constructs produced transmembrane-anchored MHM2 PrP molecules, none of which were transformed into MHM2 PrPSc (Fig. 2 A and B, lanes 2–5). After prolonged exposure with the enhanced chemiluminescence (Amersham) method, faint bands corresponding to the protease-resistant MHM2 PrP were found (data not shown). Since ScN2a cells still formed MoPrPSc after transfection with pSPOX vectors encoding MHM2-CD4, MHM2-LDLR, MHM2-PIGR, and MHM2-Qa(−), we conclude that these chimeric PrP molecules did not inhibit PrPSc formation (Fig. 2C, lanes 2–5).
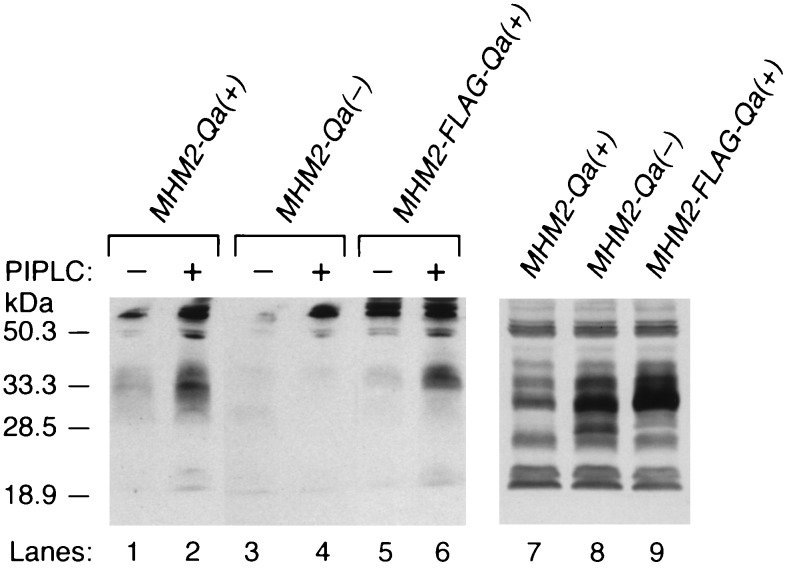
Figure 3.
GPI-anchored chimeric MHM2 PrPC is released by PIPLC digestion but the transmembrane form is not. Chimeric PrP molecules denoted MHM2-Qa(−) for the transmembrane form and MHM2-Qa(+) for the GPI-anchored form were expressed in ScN2a cells. Media (lanes 1–6) and cell lysates (lanes 7–9) were collected from ScN2a cells stably transfected with MHM2-Qa(+) (lanes 1 and 2) or MHM2-Qa(−) (lanes 3, 4, and 8), or MHM2-FLAG-Qa (+) (lanes 5, 6, and 9). Cells were either digested with PIPLC (0.5 U) at 37°C for 4 h (even lanes), or without PIPLC (odd lanes). Western blot analysis was performed with α-PrP 3F4 mAb. Only the GPI-anchored MHM2-Prp was released into the media. Western blot analysis was performed with α-PrP 3F4 mAb.
Figure 2.
GPI-anchored MHM2 PrP is converted into PrPSc but the transmembrane form is not. Western blotting of each MHM2-chimeric construct expressed in ScN2a cells is shown. (A–C) lane 1, MHM2 PrP; lane 2, MHM2-CD4; lane 3, MHM2-LDLR; lane 4, MHM2-PIGR; lane 5, MHM2-Qa(−); lane 6, MHM2-Qa(+); and lane 7, untransfected control ScN2a cells. (D–F) demonstrate that the Qa(−) transmembrane signal predominates in the chimeric constructs with Qa(+) GPI-anchor addition signal. Lane 8, MHM2 -Qa(+); lane 9, MHM2-Qa(−); lane 10, MHM2-Qa(−)-Qa(+); lane 11, MHM2-Qa(−)-Qa(−); lane 12, MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(+); lane 13, MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(−); and lane 14, untransfected control ScN2a cells. (G–I) lane 15, MHM2-Qa(+); lane 16, MHM2-Qa(−); lane 17, MHM2-FLAG-Qa(+); lane 18, untransfected control ScN2a cells. A, D, and G demonstrate the expression of each chimeric MHM2 PrP construct: Forty microliters of undigested cell lysates were applied to each lane and MHM2 PrP was detected by staining with α-PrP 3F4 mAb. B, E, and H demonstrate the conversion of MHM2 PrPC into MHM2 PrPSc and were stained with α-PrP 3F4 mAb. C, F, and I show endogenous MoPrPSc detected with RO73 antiserum. In B and C, E and F, and H and I, 500 μl of cell lysate were digested with proteinase K (20 μl/ml) at 37°C for 1 h followed by centrifugation at 100,000 × g for 1 h at 4°C and the resuspended pellet loaded onto the gels.
Each of the four transmembrane MHM2 PrP molecules has a different COOH-terminal sequence varying in length from 28–127 residues and is joined to a flexible portion of the PrP molecule containing residues 220–231 that recent nuclear magnetic resonance studies document is devoid of organized structure (30). Furthermore, these diverse sequences are unlikely to inhibit PrPSc formation by functionally relevant changes in the structure of these PrP fusion proteins. Instead, it seems more likely that these PrP fusion proteins inhibit PrPSc formation because of their transmembrane anchoring, which targets them to clathrin-coated pits.
GPI-Anchored MHM2-Qa(+) Forms PrPSc.
Mutating codon 295 of the GPI-anchored form of the Qa protein designated Qa(+) produced a transmembrane form of the protein designated Qa(−). In the Qa(+) form of the protein, position 295 encodes an aspartic acid while the Qa(−) form encodes a valine. Although fusion of a 28-aa segment from the COOH terminus of Qa(−) containing a valine prevented PrPSc formation, fusion of the same segment from Qa(+) rendered MHM2-Qa(+) eligible for conversion into PrPSc. While MHM2-Qa(−) is targeted to clathrin-coated pits by its COOH-terminal transmembrane sequence, MHM2-Qa(+) is targeted to CLDs by its GPI anchor addition sequence. Prior to cleavage of the 28 aa fused to the flexible COOH terminus of MHM2-Qa(+) upon addition of the GPI anchor, the MHM2-Qa(+) and MHM2-Qa(−) proteins differ by a single amino acid. Since the COOH-terminal sequences of MHM2-Qa(+) and PrP are different yet both support PrPSc formation, it seems likely that the topology of GPI-anchored PrP molecules is critical for the conversion into PrPSc (Fig. 2B, lanes 1 and 6).
Because the GPI-anchor signal peptide was cleaved and replaced with GPI-anchor moiety, whereas the transmembrane sequence remained fused to the COOH terminus of PrP, we investigated a possibility that the Qa(−) transmembrane sequence itself inhibited PrPSc formation. We examined the behavior of a variety of Qa(−) derivative constructs including MHM2-Qa(−)-Qa(+) and MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(+) (Fig. 1). The conformationally flexible spacer consisted of 5 aa (Ser-Ala-Gly-Ser-Ala). The MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(+) construct was designed to place the GPI anchor on the outer surface of the cell like MHM2-Qa(+) and MHM2 PrP even if the Qa(−) transmembrane segments spanned the plasma membrane. None of the constructs containing a Qa(−) segment was converted into PrPSc as judged by the acquisition of resistance to digestion by proteinase K (Fig. 2E). When the Qa(−) sequence was replaced by hydrophilic FLAG sequence, the formation of PrPSc was restored (Fig. 2H) (31). We chose the FLAG sequence as a control since earlier studies had shown that FLAG fused to the NH2 terminus of PrPC could be converted into PrPSc both in ScN2a cells and Tg mice (G. C. Telling and S.B.P., unpublished work). The eight-residue FLAG sequence does not form a transmembrane segment and thus, does not function as a sorting signal for the clathrin-coated pits. Our data demonstrate that the peptide sequence fused to the COOH terminus of PrP determines whether or not the recombinant PrPC is converted into PrPSc.
Release of MHM2-Qa(+) by Phosphatidylinositol Phospholipase C.
To determine if GPI-anchor addition to MHM2-Qa(+) was properly formed in ScN2a cells, we treated cells expressing either MHM2-Qa(+) or MHM2-Qa(−) with PIPLC. As expected, the GPI-anchored MHM2-Qa(+) as well as the MHM2-FLAG-Qa(+) but not the transmembrane MHM2-Qa(−) was released by PIPLC (Fig. 3, lanes 1–6). From these results, we conclude that MHM2-Qa(+), like MHM2 PrP, is tethered to the external surface of the cell by a GPI anchor. When the cells expressing MHM2-Qa(−)-Qa(+), MHM2-Qa(−)-Qa(−), MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(+), or MHM2-Qa(−)-spacer-Qa(−)-spacer-Qa(−) (Fig. 1) were treated with PIPLC, none of these recombinant molecules were released into the media (data not shown). Thus, none of the PrP molecules containing Qa(−) segments could be released from cells by PIPLC digestion, arguing that none of these molecules were bound to the cell surface by a GPI anchor, and none were converted into PrPSc (Fig. 2 D–F).
MHM2-Qa(−) Is Internalized Through Clathrin-Coated Pits.
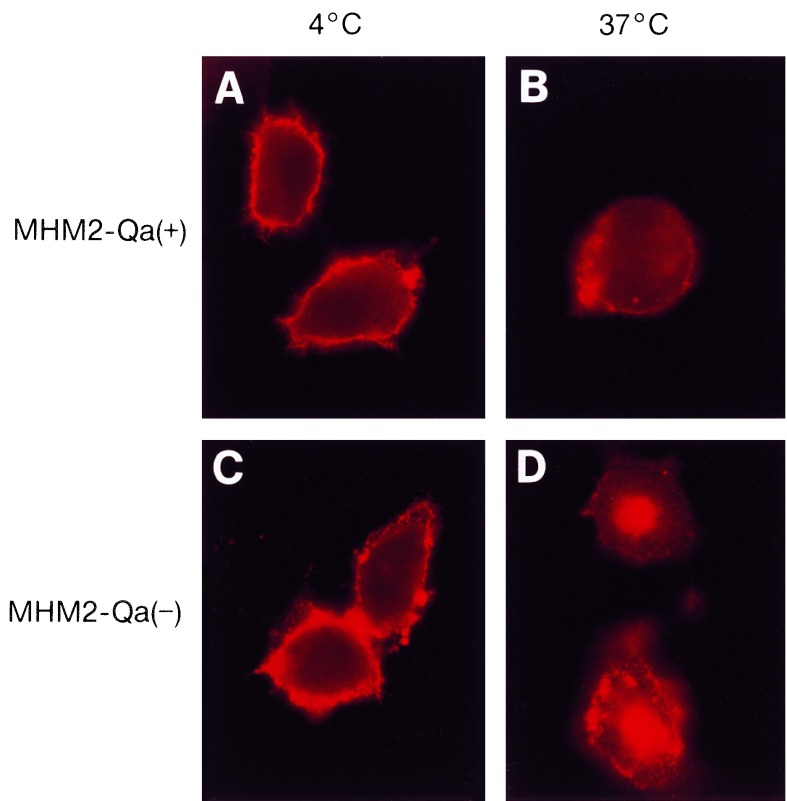
The internalization pattern of GPI-anchored MHM2-Qa(+) was significantly different from that of the transmembrane MHM2-Qa(−). When cells expressing MHM2-Qa(+) or MHM2-Qa(−) were incubated with α-PrP 3F4 mAb for 1 h on ice, both cell lines exhibited a punctate staining pattern confined to the surface (Fig. 4 A and C).
Figure 4.
Internalization of MHM2-Qa(−) and MHM2-Qa(+) expressed in ScN2a cells as measured by indirect immunofluorescence microscope. (A and B) ScN2a cells expressing MHM2-Qa(+). (C and D) ScN2a cells expressing MHM2-Qa(−). All the ScN2a cells were incubated with α-PrP 3F4 mAb at 4°C for 1 h; cells shown in B and D were then chased for 45 min at 37°C. The transmembrane MHM2-Qa(−) accumulated in the perinuclear region after treatment at 37°C, whereas MHM2-Qa(+) remained on or near the cell surface.
When the cells were incubated with α-PrP 3F4 mAb for 45 min at 37°C, the cells transfected with MHM2-Qa(−) cleared the α-PrP 3F4 mAb from the cell surface and delivered it to the perinuclear regions of the cytoplasm (Fig. 4D). In contrast, those cells transfected with MHM2-Qa(+) continued to exhibit a punctate staining pattern on their surface (Fig. 4B). The internalization pattern of MHM2-Qa(−) is similar to that of other transmembrane molecules targeted to the clathrin-coated pits; whereas the pattern for MHM2-Qa(+) is indistinguishable from that of other GPI-anchored molecules (32, 33).
MHM2-Qa(+) Proteins Fractionate with CLDs.
The lysis of cells in cold Triton X-100 allows separation of GPI-anchored proteins from most other proteins by flotation in a sucrose gradient (28, 34). The GPI-anchored proteins in cold Triton X-100 associate with CLDs which contain high levels of cholesterol.
Cells expressing either MHM2-Qa(−) or MHM2-Qa(+) were lysed in cold Triton X-100 and the detergent extracts were fractionated by flotation through sucrose gradients. Because of the low buoyant density of the CLDs, they float in these sucrose gradients. Fractions from these gradients were assayed for MHM2-Qa by immunoblotting with α-PrP 3F4 mAb. MHM2-Qa(−) could only be detected in the lysate fractions (Fig. 5B, lanes 8–11); whereas the majority of the MHM2-Qa(+) molecules were present in the floating fractions (Fig. 5A, lanes 3–6), indicating their association with CLDs. When the fractions were assayed for endogenous MoPrP with α-PrP polyclonal R073 antiserum, MoPrP was found in the floating fractions (Fig. 5 C and D, lanes 3–7). These results show that the GPI anchor of MHM2-Qa(+) rendered the protein insoluble in cold Triton X-100 and caused it to fractionate with the CLDs. In contrast, MHM2-Qa(−) proteins were completely soluble in cold Triton X-100 and did not float with the CLDs.
Figure 5.
MHM2-Qa(+) associates with CLDs. (A) ScN2a cells expressing MHM2-Qa(+) were treated with cold Triton X-100 and the CLDs were separated by flotation into sucrose gradients. Gradients were fractionated and fractions tested for presence of Qa molecules by immunoblotting using 3F4 mAb. Lanes 1–7 represent fractions 1–7 of the gradients (5–30% sucrose), lanes 8–11 represent the lysate fractions (40% sucrose). (B) ScN2a cells expressing MHM2-Qa(−) lysed, fractionated and stained as described for A. C and D were equivalent to A and B, respectively, but were stained with α-PrP polyclonal RO73 antiserum, which recognizes endogenous MoPrP. Although the α-PrP polyclonal RO73 antiserum recognizes both MoPrP and MHM2 PrP, only MoPrP is stained in D. Since MHM2-Qa(−) is expressed at levels lower than MoPrP, the staining reaction was terminated before MHM2-Qa(−) was stained by the α-PrP polyclonal RO73 antiserum.
DISCUSSION
Subcellular Trafficking of PrPC and PrPSc.
The results presented here demonstrate that PrPC targeted to CLDs is converted into PrPSc while chimeric PrPC molecules with transmembrane segments fused to the COOH terminus are not substrates for PrPSc formation as postulated from earlier studies (7). These findings contend that the process of PrPSc formation may be highly restricted topologically. Alternatively, each of the chimeric PrP molecules with different COOH-terminal sequences might be prevented from forming PrPSc because each sequence contains a transmembrane segment. We believe the latter explanation is unlikely because nuclear magnetic resonance studies of PrP fragments containing residues 121–231 suggest that the COOH-terminal 14 residues beyond the helical segment 200–217 provide a flexible tether (30); additionally, the fusion of the hydrophilic FLAG sequence at the COOH terminus of PrPC did not prevent conversion into PrPSc.
The COOH-terminal signal sequence which is cleaved when a GPI anchor is added has been shown to contain at least three elements. At the first and third positions of this sequence, the amino acid residues must have small side chains. Between these residues and a hydrophobic domain of at least 11 aa at the COOH terminus is a spacer domain that has no specific requirements (35, 36). Although no consensus sequence has been recognized on the NH2-terminal side of the cleavage site where the GPI anchors are added (37), our data demonstrate that insertion of the Qa(−) transmembrane segment did predominate GPI-anchor addition. When the Qa(−) transmembrane segment was placed between PrP on the NH2-terminal side and the Qa(+) segment on the COOH-terminal side so that the two Qa sequences were expressed tandem, no release of the recombinant PrPC by PIPLC digestion was found. It is of interest that the mRNAs encoding N-CAM which can exist as either a transmembrane or GPI-anchor protein undergo splicing so that both the transmembrane or GPI anchor addition sequences do not exist in the same mature protein (38). Even though each of the four constructs containing multiple Qa sequences was translated (Fig. 2D), none of the resulting proteins were GPI-anchored as evidenced by a lack of release from the surface of cells by PIPLC digestion (Fig. 3B).
Our inability to create a GPI-anchored PrP molecule containing the Qa(−) transmembrane segment demonstrates that cells regulate the biogenesis of proteins in a manner that prevents the formation of chimeras with the structures that we sought. In view of these results, we substituted the hydrophilic sequence FLAG for Qa(−) and produced the chimeric construct MHM2-FLAG-Qa(+). Unlike the constructs containing the Qa(−) segment, the MHM2-FLAG-Qa(+) protein acquired a GPI anchor and was converted into PrPSc. The results with this construct support our contention that steric hindrance by foreign COOH-terminal sequences is not responsible for the inhibition of PrPSc formation.
Although some investigators found immunostaining of PrP in caveolae in N2a cells (16), others have reported that the endocytosis of chicken (Chk)PrP expressed in Mo N2a cells is mediated by clathrin-coated pits (17). ImmunoGold (Janssen) labeling of N2a cells showed that the concentration of ChkPrP within coated pits was 3–5× higher than over other areas of plasma membrane, and gold particles were also seen within coated vesicles and deeply invaginated coated pits. Furthermore, internalization of ChkPrP was reduced ≈70% after N2a cells were incubated in hypertonic medium, a treatment that inhibits endocytosis by disrupting clathrin lattices. These findings are contrary to the data reported here which show that GPI-anchored PrPC does not reenter cells through clathrin-coated pits. Our results and those reported earlier (7, 9) indicate that the GPI anchor of PrPC directs it to CLDs. Whether the avian PrP sequence, which is only 30% homologous with mammalian PrPs (39), or the overexpression of this protein is responsible for what seem to be aberrant results remains to be determined.
Implications for Prion Propagation.
Our finding that the conversion of PrPC into PrPSc is restricted to a particular subcellular compartment would seem to imply that auxiliary factors localized to that compartment participate in the propagation of prions. Presumably, such auxiliary factors are proteins, but we cannot exclude the possibility that cholesterol, glycosphingolipids, phospholipids, and the membrane surface play a role in PrPSc formation.
This conclusion is consistent with the results of transgenetic studies in which PrPSc formation was inhibited by MoPrPC in mice expressing Hu or chimeric Hu/MoPrP. Such findings seem to be explained most readily by a cellular factor(s), designated protein X, that might function as a molecular chaperone (22). We proposed that protein X facilitates either the unfolding of one or more of the α-helices in PrPC, or their refolding into β-sheets by binding relevant structural intermediates. Protein X appears to bind to the COOH-terminal region of PrPC but not to PrPSc. Whether any of the proteins known to bind to PrP (40–42) function as proposed for protein X remains to be established.
Novel Approaches to Central Nervous System Degeneration Arising From These Studies.
Our investigations argue persuasively that PrPSc formation requires the trafficking of PrPC to a specific subcellular compartment. This finding raises the possibility of designing drugs that divert PrPC away from the specific compartment where PrPSc is formed. If we assume that PrPC performs some as yet unknown function when it resides on the external surface of cells, then modifying its sorting pathway during the degradation phase of its metabolism might provide an effective therapeutic approach.
At present, there are no effective therapies for prion diseases despite numerous studies with DEAE dextran, pentosan sulfate, Congo red, and amphotercin B (43–48). Although ablation of the PrP gene does prevent prion disease (49, 50), such a strategy cannot be implemented since systems to deliver widely PrP antisense polynucleotides to the central nervous system are not currently available. Identification of drugs that modify the trafficking of PrPC might provide a novel approach to treating not only prion diseases but also the more common neurodegenerative illnesses such as Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis, which may also prove to be disorders of protein conformation.
Acknowledgments
We thank Albert Taraboulos and William Welch for their stimulating discussions during the course of these studies. This work was supported by grants from the National Institutes of Health (NS14069, AG08967, AG02132, NS22786, and AG10770) and by gifts from the Leila and Harold Mathers Foundation, Sherman Fairchild Foundation, and Centeon Inc. K.K. was supported in part by Uehara Memorial Foundation and the Japanese Ministry of Education, and M.V. was supported by the Stipendienprogramm Infektionsforschung, Deutsches Krebsforschungszentrum.
ABBREVIATIONS
- PrP
prion protein
- CJD
Creutzfeldt–Jakob disease
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- N2a
mouse neuroblastoma
- ScN2a
scrapie-infected mouse neuroblastoma
- SHa
Syrian hamster
- Mo
mouse
- Hu
human
- Chk
chicken
- GPI
glycophosphatidyl inositol
- CLDs
caveolae-like domains
- Ph
phosphorylated
- UPh
unphosphorylated
References
- 1.Prusiner S B, Bolton D C, Groth D F, Bowman K A, Cochran S P, McKinley M P. Biochemistry. 1982;21:6942–6950. doi: 10.1021/bi00269a050. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Annu Rev Microbiol. 1994;48:655–685. doi: 10.1146/annurev.mi.48.100194.003255. [DOI] [PubMed] [Google Scholar]
- 3.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 4.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl N, Borchelt D R, Hsiao K, Prusiner S B. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B, Race R E, Ernst D, Buchmeier M J, Chesebro B. J Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner S B. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smart E J, Ying Y-S, Mineo C, Anderson R G W. Proc Natl Acad Sci USA. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorodinsky A, Harris D A. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond S J, Smart E J, Anderson R G W, Taraboulos A, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraguchi T, Fisher S, Olofsson S, Endo T, Groth D, Tarantino A, Borchelt D R, Teplow D, Hood L, Burlingame A, Lycke E, Kobata A, Prusiner S B. Arch Biochem Biophys. 1989;274:1–13. doi: 10.1016/0003-9861(89)90409-8. [DOI] [PubMed] [Google Scholar]
- 12.Pan K-M, Stahl N, Prusiner S B. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taraboulos A, Serban D, Prusiner S B. J Cell Biol. 1990;110:2117–2132. doi: 10.1083/jcb.110.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caughey B, Raymond G J. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 15.Borchelt D R, Taraboulos A, Prusiner S B. J Biol Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 16.Ying Y-S, Anderson R G W, Rothberg K G. Cold Spring Harbor Symp Quant Biol. 1992;57:593–604. doi: 10.1101/sqb.1992.057.01.065. [DOI] [PubMed] [Google Scholar]
- 17.Shyng S-L, Heuser J E, Harris D A. J Cell Biol. 1994;125:1239–1250. doi: 10.1083/jcb.125.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinley M P, Taraboulos A, Kenaga L, Serban D, Stieber A, DeArmond S J, Prusiner S B, Gonatas N. Lab Invest. 1991;65:622–630. [PubMed] [Google Scholar]
- 19.Caughey B, Raymond G J, Ernst D, Race R E. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taraboulos A, Raeber A J, Borchelt D R, Serban D, Prusiner S B. Mol Biol Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinley M P, Meyer R, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner S B. J Virol. 1991;65:1440–1449. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 23.Butler D A, Scott M R D, Bockman J M, Borchelt D R, Taraboulos A, Hsiao K K, Kingsbury D T, Prusiner S B. J Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers M, Serban D, Gyuris T, Scott M, Torchia T, Prusiner S B. J Immunol. 1991;147:3568–3574. [PubMed] [Google Scholar]
- 27.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 28.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 29.Rogers M, Yehiely F, Scott M, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 31.Brizzard B L, Chubet R G, Vizard D L. BioTechniques. 1994;16:730–735. [PubMed] [Google Scholar]
- 32.Keller G A, Siegel M W, Caras I W. EMBO J. 1992;11:863–874. doi: 10.1002/j.1460-2075.1992.tb05124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritter T E, Fajardo O, Matsue H, Anderson R G W, Lacey S W. Proc Natl Acad Sci USA. 1995;92:3824–3828. doi: 10.1073/pnas.92.9.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sargiacomo M, Sudol M, Tang Z, Lisanti M P. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodukula K, Gerber L D, Amthauer R, Brink L, Udenfriend S. J Cell Biol. 1993;120:657–664. doi: 10.1083/jcb.120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coyne K E, Crisci A, Lublin D M. J Biol Chem. 1993;268:6689–6693. [PubMed] [Google Scholar]
- 37.Ferguson M A J, Williams A F. Annu Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 38.He H-T, Finne J, Goridis C. J Cell Biol. 1987;105:2489–2500. doi: 10.1083/jcb.105.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris D A, Falls D L, Walsh W, Fischbach G D. Soc Neurosci Abstr. 1989;15:70.7. [Google Scholar]
- 40.Oesch B, Teplow D B, Stahl N, Serban D, Hood L E, Prusiner S B. Biochemistry. 1990;29:5848–5855. doi: 10.1021/bi00476a029. [DOI] [PubMed] [Google Scholar]
- 41.Kurschner C, Morgan J I. Mol Brain Res. 1995;30:165–168. doi: 10.1016/0169-328x(95)00013-i. [DOI] [PubMed] [Google Scholar]
- 42.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker E-L. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehlers B, Diringer H. J Gen Virol. 1984;65:1325–1330. doi: 10.1099/0022-1317-65-8-1325. [DOI] [PubMed] [Google Scholar]
- 44.Caughey B, Race R E. J Neurochem. 1992;59:768–771. doi: 10.1111/j.1471-4159.1992.tb09437.x. [DOI] [PubMed] [Google Scholar]
- 45.Kimberlin R H, Walker C A. Arch Virol. 1983;78:9–18. doi: 10.1007/BF01310854. [DOI] [PubMed] [Google Scholar]
- 46.Xi Y G, Ingrosso L, Ladogana A, Masullo C, Pocchiari M. Nature (London) 1992;356:598–601. doi: 10.1038/356598a0. [DOI] [PubMed] [Google Scholar]
- 47.Ingrosso L, Ladogana A, Pocchiari M. J Virol. 1995;69:506–508. doi: 10.1128/jvi.69.1.506-508.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenzie D, Kaczkowski J, Marsh R, Aiken J. J Virol. 1994;68:7534–7536. doi: 10.1128/jvi.68.11.7534-7536.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Büeler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 50.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S-L, DeArmond S J. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]