Abstract
The proper guidance of the Caenorhabditis elegans hermaphrodite sex myoblasts (SMs) requires the genes egl-15 and egl-17. egl-15 has been shown to encode the C. elegans orthologue of the fibroblast growth factor receptor (FGFR). Here we clone egl-17 and show it to be a member of the fibroblast growth factor (FGF) family, one of the first functional invertebrate FGFs known. egl-17 shares homology with other FGF members, conserving the key residues required to form the distinctive tertiary structure common to FGFs. Genetic and molecular evidence demonstrates that the SM migration defect seen in egl-17 mutant animals represents complete loss of egl-17 function. While mutations in egl-17 affect only SM migration, mutations in egl-15 can result in larval arrest, scrawny body morphology, and the ability to suppress mutations in clr-1. We propose that EGL-17 (FGF) acts as a ligand for EGL-15 (FGFR) specifically during SM migration and that another ligand(s) activates EGL-15 for its other functions.
Egg laying in Caenorhabditis elegans hermaphrodites requires the proper positioning, attachment, and functioning of a set of 16 vulval and uterine muscles. These muscles, collectively called the sex muscles, arise from a pair of bilaterally symmetrical sex myoblast (SM) cells that are born in the posterior body region of C. elegans larvae at the end of the first larval stage. During the second and the early portions of the third larval stage, the SMs migrate anteriorly approximately 65 μm to reach the precise center of the developing gonad, where they divide and give rise to the sex muscles (1). The migrations of the SMs are critical for normal egg laying, as SMs that fail to migrate anteriorly result in mispositioned sex muscles and an egg-laying-defective (Egl) phenotype (2).
The migrations of the SMs are controlled by two mechanisms (3). A gonad-independent mechanism, revealed by laser removal of the gonad, allows the SMs to migrate anteriorly to a broad range of final positions that span the central region of the animal. The second mechanism allows for the precise positioning of the SMs flanking the center of the gonad. In this gonad-dependent mechanism, somatic cells in the gonad are postulated to send attractive signals to the SMs, resulting in their precise positioning (3).
Mutations in two genes, egl-15 and egl-17, alter the interaction between the SMs and the gonad (2). The SMs, normally attracted to the gonad, are actively repelled by the gonad in egl-15 and egl-17 mutant hermaphrodites, resulting in severely posteriorly displaced SMs. Removal of the gonad in egl-15 and egl-17 mutants allows the SMs to migrate further anteriorly, implicating the gonad in the posterior displacement of the SMs in these mutants. The dramatic effects of these mutations on the interactions between the gonad and the SMs implicates egl-15 and egl-17 in the gonad-dependent attractive mechanism.
egl-15 was cloned and shown to encode a C. elegans fibroblast growth factor (FGF) receptor (FGFR), implicating FGFR signaling in the guidance of SM migration (4). In this paper we report the cloning and molecular characterization of egl-17.
MATERIALS AND METHODS
SM Migration Mutants.
The systematic screen for mutants with severely displaced SMs in a clr-1(e1745ts); sem-5(n1779) sensitized background has been described elsewhere (5). The mutations obtained from this screen fall into two classes. The first class of mutations confer an SM migration-defective phenotype on their own. Complementation tests with egl-15 and egl-17 revealed these to be new alleles of either of these two genes. The second class of mutations are dependent on the sem-5(n1779) mutant background to reveal their effects on SM migration. The analysis of these mutations is described elsewhere (5). egl-17(n1377) was isolated in a smaller similar screen in a wild-type background, and the mutations n1450, n1451, n1452, and n1479 were isolated in a noncomplementation screen in trans to egl-17(e1313) (M.J.S. and H. R. Horvitz, unpublished observations). All egl-17 mutants are viable and display similar distributions of posteriorly displaced SMs (data not shown). Genetic analyses are, thus, consistent with these mutations being null alleles of egl-17 on the basis of three arguments: (i) the frequency at which egl-17 alleles are isolated (see Results); (ii) the uniformity of the severity of the phenotype observed in all egl-17 mutants; and (iii) the types of alleles isolated in a noncomplementation screen that had the potential to identify null alleles of egl-17.
egl-17 Rescue by Germ-Line Transformation.
For cosmid rescue and narrowing of the cosmid T28F1 to the 9.1-kb rescuing fragment, DNA fragments (20 μg/ml) were injected into egl-17(n1377) hermaphrodites as described (4, 6), using the plasmid pRF4 (100 μg/ml) as the cotransformation marker. Stable transgenic lines were scored as rescued if at least 40% of array-bearing Roller animals were non-Egl. egl-17(n1377) is 83% penetrant for the Egl phenotype. In germ-line transformation experiments where SM positions were scored, DNA fragments (50 μg/ml) were injected into dpy-20(e1282ts); egl-17(n1377) hermaphrodites using the plasmid pMH86 (50 μg/ml), which carries the dpy-20(+) gene (7) as the cotransformation marker. Rescued lines were maintained at 25°C and the SMs were scored as described previously (3) in the array-bearing (non-Dpy) animals.
Insertion Mutations in the egl-17 Rescuing Fragment.
Four-base-pair insertion mutations were introduced by restriction endonuclease digestion followed by extension with the Klenow fragment of DNA polymerase and religation. The SalI and HindIII insertions were confirmed by dideoxynucleotide sequencing.
Reverse Transcription (RT)-PCR and Rapid Amplification of cDNA Ends (RACE).
The SalI–HindIII portion of the egl-17 cDNA was obtained as a RT-PCR product by using mixed-stage, poly(A)+ RNA from wild-type (N2) hermaphrodites (4) and the oligonucleotide primers 5′-CCTGATGTTGTCGACAAACTTTCGG-3′ and 5′-CGGACAAGCTTGATCCAGTCAAAAC-3′. cDNA fragments corresponding to the 5′ and 3′ ends of the egl-17 transcript were isolated by using the Marathon RACE Kit (CLONTECH) and oligonucleotide primers complementary to those described above. Genomic organization of egl-17 was determined by comparison of cDNA sequences to genomic sequences (data not shown).
Sequence Analysis and Alignments.
The signal sequence and potential cleavage site were predicted as described (8, 9). The residues constituting the internal homologous region of FGFs (10) were used in alignments. The program megalign (DNAstar, Madison, WI) was used to align EGL-17 to the other FGF family members. The program gap (GCG) was used for individual comparisons, yielding the following similarities and identities, respectively, to EGL-17: murine FGF-1 (11), 40%/15%; murine FGF-2 (11), 37%/16%; murine FGF-5 (11), 41%/20%; rat FGF-9 (12), 46%/18%; and murine FGF-8a (13), 45%/19%.
Analysis of egl-17 Mutations.
Genomic DNA from each of the egl-17 strains (14) was used in Southern analysis and for amplifying the egl-17 coding regions by PCR. For Southern analysis, DNA from wild type and from egl-17 mutants (ay6, n1377, ay59) was digested with HindIII and probed with the 32P-labeled 9.1-kb insert from NH#75. Allele-specific deletions were further mapped using Southern analysis, and the end points were determined by sequencing after PCR amplification. Mutations were identified by sequencing the entire egl-17 coding region from PCR pools derived from the genomic DNA and confirmed by sequencing independently derived PCR pools.
RESULTS
Screen to Identify Genes Involved in SM Migration.
To identify genes involved in the migrations of the hermaphrodite SMs, we performed a systematic screen of animals representing 35,000 ethyl methanesulfonate-mutagenized haploid genomes for mutant hermaphrodites with severely misplaced SMs (see Materials and Methods). Five mutations were isolated that confer dramatically posteriorly displaced SMs in a wild-type background. Complementation tests showed that these mutations are new alleles of either of two genes previously known to affect SM migration (2). Four mutations (ay6, ay8, ay16, and ay59) are alleles of egl-17 and arose at a frequency of 1.1 × 10−4. This frequency is consistent with the rate of obtaining complete loss-of-function alleles in this type of mutagenesis (15). One mutation (ay1) is a hypomorphic, class I allele of the gene egl-15 that encodes a C. elegans FGFR (4). The most frequently obtained mutations in egl-15 result in apparent elimination of egl-15 function (4). These putative null mutations result in a larval-arrest phenotype that precludes their isolation in our screen for SM migration defects. This may account for the lower frequency of egl-15 mutations isolated in our screen.
egl-15 and egl-17 are the only two genes identifed in this and in other similar screens (M.J.S. and H. R. Horvitz, unpublished observations) which can be individually mutated to confer dramatically posteriorly displaced SMs. Since only egl-17 mutations arise at knock-out frequency, egl-17 is likely to be the only gene in which null mutations confer this kind of phenotype.
Cosmid Rescue of egl-17.
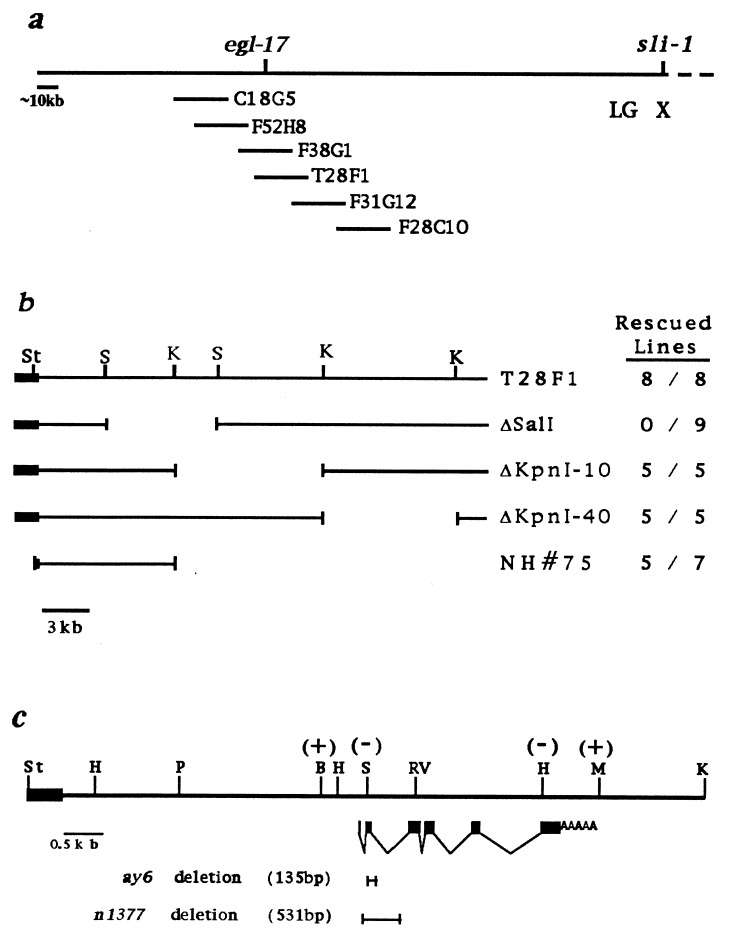
egl-17 maps to the left end of the X chromosome, a region which is covered by a set of overlapping yeast artificial chromosomes (YACs) and cosmids (16). We cloned egl-17 by identifying two cosmids from this region, F38G1 and T28F1, that rescued the Egl phenotype when introduced by germ-line transformation into egl-17(n1377) animals (Fig. 1a). This transformation rescue assay was used to narrow the rescuing DNA to a 9.1-kb fragment capable of rescuing both the Egl and the SM migration defects (Figs. 1b and 2).
Figure 1.
Molecular mapping of egl-17. (a) Cosmids from the left end of the X chromosome contig are shown. The Egl defect of egl-17 mutants is rescued by cosmids F38G1 and T28F1, but not by the adjacent cosmids shown. (b) Restriction map of cosmid T28F1 and subclones used to determine the smallest region containing egl-17-rescuing activity. Rescued lines, fraction of stable lines rescued out of the total number of lines scored. The solid bar represents cosmid vector sequences. (c) Genomic structure of the egl-17 transcript within the 9.1-kb rescuing fragment. The 7.0-kb PstI–KpnI fragment also shows egl-17 rescuing activity albeit at somewhat reduced efficiency. Vector sequences are depicted as in b. Solid boxes represent exons; poly(A) tail is indicated. The effects of 4-bp insertion mutations are indicated within parenthesis: +, rescuing activity retained; −, rescuing activity abolished. Extent of allele-specific deletions are depicted by bracket bars. B, BglII; RV, EcoRV; H, HindIII; K, KpnI; M, MluI; P, PstI; S, SalI; St, StuI.
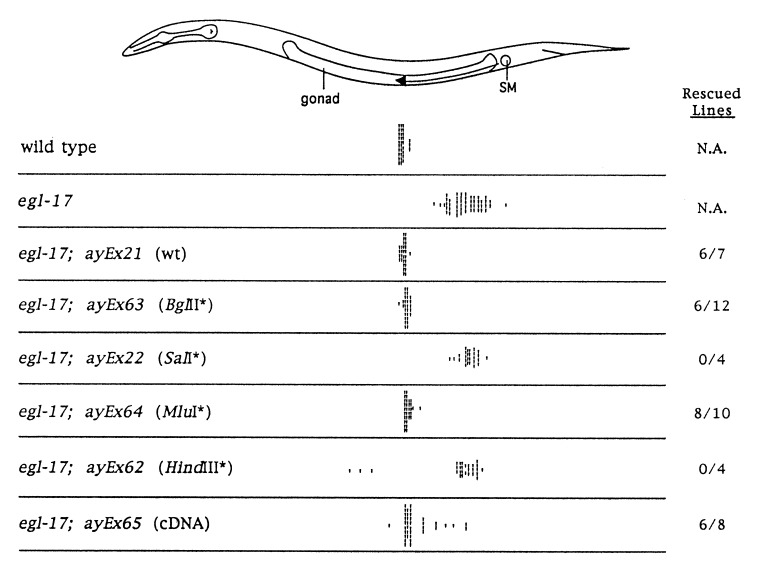
Figure 2.
SM positions in wild-type, egl-17, and transgenic animals. Hash marks represent the final positions of individual SMs. Data are shown for individual transgenic lines; similar SM distributions were observed for additional lines obtained with the same transformation mixture. Rescued lines, fraction of stable lines rescued for the Egl phenotype of egl-17(n1377). N.A., not applicable. The nature of the egl-17 portion of the extragenic array is shown in parentheses. All are modifications of the 9.1-kb genomic rescuing fragment of NH#75 shown in Fig. 1b. wt, wild type; ∗, 4-bp insertions at the respective sites; cDNA, SalI–HindIII portion of the cDNA inserted into the corresponding sites of the genomic rescuing fragment. The cDNA construct does not remove the first intron from the 5′ untranslated region of egl-17. SM distributions from wild-type (3) and egl-17 (2) hermaphrodites are shown for comparison. The split SM distribution seen in egl-17; ayEx62 animals has been observed previously in egl-17 animals with partially ablated gonads (2).
Identification of the egl-17 Transcript.
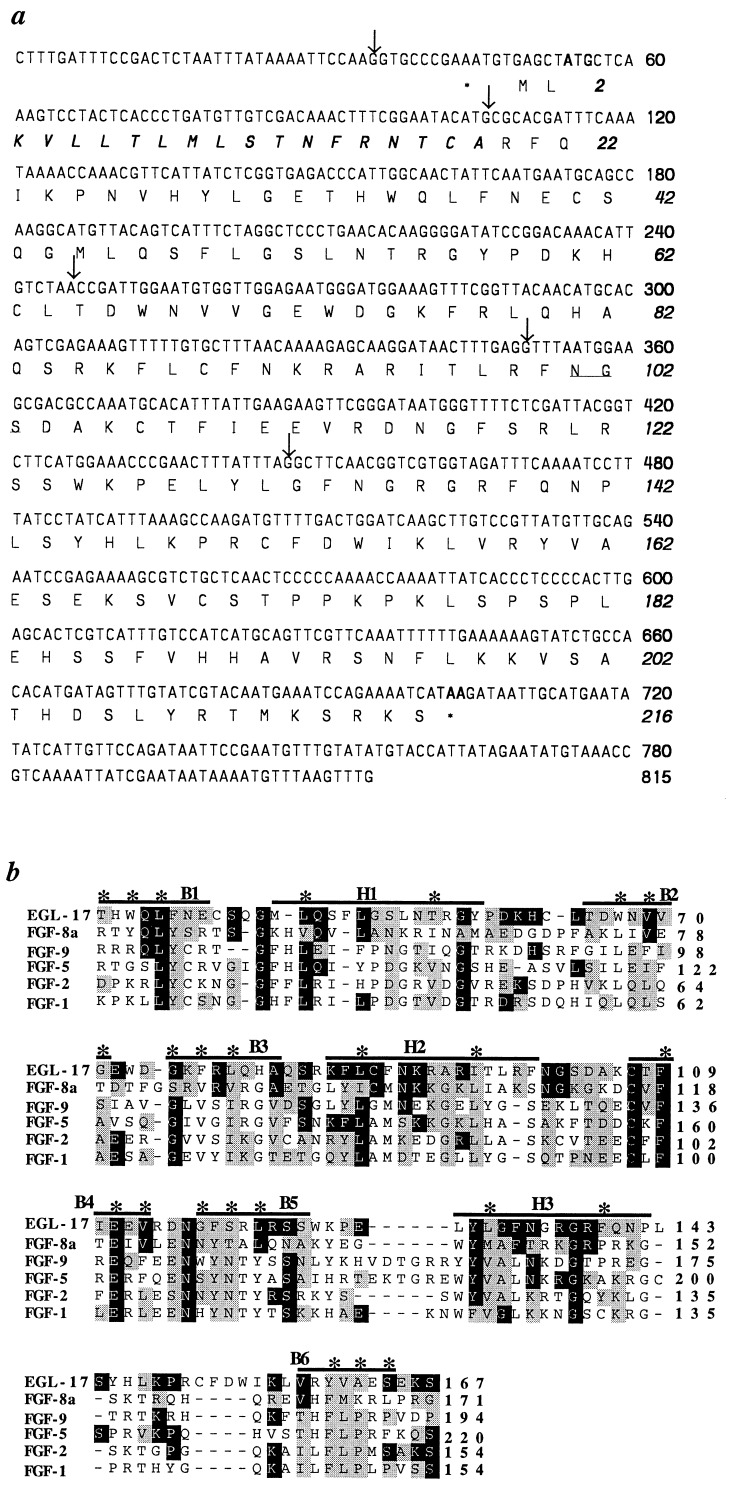
egl-17 coding sequences were identified by an RT-PCR-based strategy. The egl-17 mRNA appears to be a rare transcript, since we were unable to detect it by Northern analysis or identify a corresponding cDNA in a number of cDNA libraries. In the absence of a cDNA, we used the transformation rescue assay to identify potential coding regions in the egl-17 genomic rescuing fragment. Four-base-pair insertions were introduced at various restriction sites in the genomic rescuing fragment. Insertion in the egl-17 open reading frame would cause a frameshift mutation that might abolish rescuing activity. Two sites were found where insertions abrogated rescue, and oligonucleotide primers corresponding to these sites were used to obtain a partial cDNA by RT-PCR (Figs. 1c and 2). cDNA fragments corresponding to the 5′ and 3′ ends of the egl-17 transcript were obtained by RACE. The assembled 815-bp cDNA contains a single long open reading frame with the potential to encode a protein of 216 amino acids (Fig. 3a). This cDNA, when introduced in the context of upstream and downstream genomic sequences, is capable of rescuing both the Egl and the SM defects of egl-17 mutants at high frequency in the transformation rescue assay, confirming that it encodes a functional egl-17 gene product (Fig. 2).
Figure 3.
(a) Nucleotide sequence of the egl-17 cDNA and its predicted amino acid sequence. Nucleotides and amino acids are numbered at the right in roman and italic, respectively. Asterisks mark in-frame stop codons just upstream of the putative start methionine and at the end of egl-17. The putative signal sequence is in boldface italics, the potential glycosylation sequence is underlined, and splice site positions are marked with arrows. (b) Alignment of EGL-17 with other members of the FGF family. The internal homologous region common to all FGFs is shown. Amino acid positions are numbered at the right. Horizontal lines mark the 12 β-sheets that form the six barrel strands (B) and three hairpin regions (H) that make up the β-trefoil fold (17). The asterisks mark the residues inside the barrels and between the hairpins, based on crystallographic studies of FGFs 1 and 2 (17, 18). Residues identical or similar to those in EGL-17 are shaded in dark grey or light grey, respectively.
The EGL-17 protein shows significant amino acid sequence homology to members of the FGF family and is one of the first functional FGF family members known in invertebrates. The conservation between the vertebrate FGFs and EGL-17 is highest within the internal homologous region shared by all FGFs (10), 37–46% similar and 15–20% identical (Fig. 3b). The internal homologous region contains key residues that are required to form the distinctive tertiary structure common to FGFs, the Kunitz inhibitors, and interleukins 1β and 1α (17, 18). The EGL-17 protein preserves the nature of many of these residues, suggesting EGL-17 also may form this unique tertiary structure.
Two other observations provide additional evidence indicating that EGL-17 is an FGF family member: (i) the strikingly similar phenotypes conferred by egl-17 mutations and certain mutations affecting the egl-15-encoded FGFR (2, 4); and (ii) retention of the intron/exon boundaries conserved in all FGF genes (10) and in the FGF-homologous factor-2 gene (19).
Analysis of egl-17 Mutations.
The molecular characterization of egl-17 mutants provides compelling evidence that the SM migration defect seen in egl-17 mutant animals represents the null phenotype for egl-17. Analysis of the molecular lesions associated with egl-17 in these alleles shows that all have the potential to abolish egl-17 function (Fig. 4): two alleles are deletions that remove part or all of exon 2, which encodes the initiating methionine; six alleles are nonsense mutations; one allele alters an absolutely conserved residue (20) of the splice donor site at the end of exon 5; and one allele is a missense mutation that alters the codon encoding the initiating methionine. These molecular data are supported by the genetic analyses that are consistent with all extant egl-17 mutations being null alleles (see Materials and Methods). It is interesting that no mutations were found that would alter only a single EGL-17 residue without affecting translation of the egl-17 transcript. This result suggests that only drastic disruptions of EGL-17 structure can confer SM migration defects and that most EGL-17 residues are not absolutely required for its function. This finding may account for the ability of several different FGF family members to be roughly functionally equivalent despite a large amount of amino acid diversity (21).
Figure 4.
Mutational analysis of egl-17. (a) Point mutation alterations. The alteration corresponding to n1452 changes the essential guanosine residue at the splice donor junction of exon 5. (b) Southern blotting analysis of deletion mutations ay6 and n1377.
DISCUSSION
We have cloned and characterized egl-17 and found it to be an invertebrate member of the FGF family. We propose that egl-17 encodes the ligand for the EGL-15 FGFR in its role in SM migration. Consistent with their functioning in the same pathway, SM distributions in double mutants bearing SM-defective mutations in egl-15 and egl-17 are no more affected than in egl-15 single mutants (M.J.S., unpublished observations).
The identification of EGL-15 as an FGFR implicated FGFR signaling in the proper guidance of the SMs (4). The finding that egl-17 encodes an FGF, coupled with egl-15 and egl-17 being the only two genes easily mutated to result in severely posteriorly displaced SMs, further underscores the importance of FGFR signaling in this cell migration event. Downstream components of FGFR signaling were not identified in this screen, perhaps due to functional redundancy of downstream components or pathways, or the potential lethality or sterility of such mutations.
FGFR pathways have been implicated in cell motility processes in both mammalian and invertebrate systems. FGFs are chemotactic for many types of cells (22), and cell lines transfected with FGFs show higher rates of motility. This increased motility can be blocked either by antibodies to the FGFs or by suramin, which interferes with the abilities of heparin-binding growth factors to bind to their receptors (23, 24). In Drosophila, an FGFR pathway has been shown to be important in tracheal cell migrations (25) and the migrations of the border cells during oogenesis (26). The involvement of an FGFR signaling pathway in the migrations of the hermaphrodite SMs in C. elegans supports FGFR signaling as a common mechanism for controlling some cell migration events.
Further study of egl-15 and egl-17 may provide insight into why mutations in these genes can result in SMs that are repelled by the cells that they are normally attracted to. The EGL-17 signal is required for the gonad-dependent attraction of the SMs. However, since the gonad still affects (by repulsion) the distribution of SMs in the complete absence of EGL-17, an additional signal besides EGL-17 must be able to affect SM migration guidance. We have proposed two models that might describe the normal roles for EGL-17 and this additional signal. In the first model, EGL-17 is the attractive signal that guides the SMs to their final positions and the additional signal is a latent gonad-dependent repelling signal. Mutations in egl-15 and egl-17 compromise the attraction, revealing the underlying repulsion. In the second model, the additional signal acts instructively to guide the migrations of the SMs, and EGL-17 acts permissively to allow the SMs to interpret this signal as an attraction. In the absence of EGL-17-mediated signaling, the SMs misinterpret the attractive signal as a repellent. The identification of EGL-17 as a putative ligand for EGL-15 should allow experiments to distinguish between these two models. If EGL-17 acts as an attractant, ectopic expression of EGL-17 should be able to guide the SMs to the ectopically expressed positions. By contrast, if EGL-17 acts permissively, ectopic expression of EGL-17 could only restore the guidance of the SMs to their normal positions.
Besides affecting SM migration, mutations in egl-15 can result in larval arrest, scrawny body morphology, and the ability to suppress the effects of mutations in clr-1 (4). Since elimination of egl-17 function results only in SM migration defects, EGL-17 is likely to be an SM migration-specific ligand, and additional ligands presumably play a role in the other functions of EGL-15. A similar situation occurs in Drosophila, in which different subsets of cellular responses requiring the epidermal growth factor receptor homologue appear to be activated by two different ligands (27).
Many growth factors, including FGF family members, can initiate a large array of different cellular responses. Signaling specificity is likely to be conferred by a combination of localized expression, differing ligand–receptor association affinities, and variations in the subsets of signaling components that are present in different cellular environments. The analysis of FGF signaling pathways in C. elegans may provide a more detailed explanation of how FGF family members can initiate the specific responses required in particular developmental contexts.
Acknowledgments
We thank N. Theodosiou for analysis of deletion alleles, J. Shyu for restriction mapping of cosmid T28F1, and members of the Stern laboratory for discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Lucille P. Markey Charitable Trust. R.D.B. is a Howard Hughes Medical Institute Predoctoral Fellow.
ABBREVIATIONS
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- SMs
sex myoblasts, Egl, egg-laying-defective
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Sulston J E, Horvitz H R. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 2.Stern M J, Horvitz H R. Development. 1991;113:797–803. doi: 10.1242/dev.113.3.797. [DOI] [PubMed] [Google Scholar]
- 3.Thomas J H, Stern M J, Horvitz H R. Cell. 1990;62:1041–1052. doi: 10.1016/0092-8674(90)90382-o. [DOI] [PubMed] [Google Scholar]
- 4.DeVore D L, Horvitz H R, Stern M J. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen E B, Branda C S, Stern M J. Dev Biol. 1997;182:88–100. doi: 10.1006/dbio.1996.8473. [DOI] [PubMed] [Google Scholar]
- 6.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark D V, Suleman D S, Beckenback K A, Gilchrist E J, Baillie D L. Mol Gen Genet. 1995;247:367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 9.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas K A. In: Neurotrophic Factors. Loughlin S E, Fallon J H, editors. San Diego: Academic; 1993. pp. 285–312. [Google Scholar]
- 11.Hébert J M, Basilico C, Goldfarb M, Haub O, Martin G R. Dev Biol. 1990;138:454–463. doi: 10.1016/0012-1606(90)90211-z. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Mol Cell Biol. 1993;13:4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur C A, Shankar D B, Shackleford G M. J Virol. 1995;69:2501–2507. doi: 10.1128/jvi.69.4.2501-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 15.Herman R K. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 17–45. [Google Scholar]
- 16.Yoon C H, Lee J, Jongeward G D, Sternberg P W. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 17.Murzin A G, Lesk A M, Chothia C. J Mol Biol. 1992;223:531–543. doi: 10.1016/0022-2836(92)90668-a. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Komiya H, Chirino A, Faham S, Fox G M, Arakawa T, Hsu B T, Rees D C. Science. 1991;251:90–93. doi: 10.1126/science.1702556. [DOI] [PubMed] [Google Scholar]
- 19.Smallwood P M, Munoz-Sanjuan I, Tong P, Macke J P, Hendry S H C, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. Proc Natl Acad Sci USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause M. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism, Methods in Cell Biology. Epstein H F, Shakes D C, editors. Vol. 48. San Diego: Academic; 1995. pp. 483–512. [Google Scholar]
- 21.Johnson D E, Williams L T. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 22.Mason I J. Cell. 1994;78:547–552. doi: 10.1016/0092-8674(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 23.Bikfalvi A. Eur J Cancer. 1995;31A:1101–1104. doi: 10.1016/0959-8049(95)00169-j. [DOI] [PubMed] [Google Scholar]
- 24.Taylor W R, Greenberg A H, Turley E A, Wright J A. Exp Cell Res. 1993;204:295–301. doi: 10.1006/excr.1993.1036. [DOI] [PubMed] [Google Scholar]
- 25.Klämbt C, Glazer L, Shilo B-Z. Genes Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- 26.Murphy A M, Lee T, Andrews C M, Shilo B-Z, Montell D J. Development. 1995;121:2255–2263. doi: 10.1242/dev.121.8.2255. [DOI] [PubMed] [Google Scholar]
- 27.Neuman-Silberberg F, Schüpbach T. Cell. 1993;75:165–174. [PubMed] [Google Scholar]