Abstract
Helicobacter pylori is the only neutralophile that has been able to colonize the human stomach by using a variety of acid-adaptive mechanisms. One of the adaptive mechanisms is increased buffering due to expression of an acid-activated inner membrane urea channel, UreI, and a neutral pH-optimum intrabacterial urease. To delineate other possible adaptive mechanisms, changes in gene expression in response to acid exposure were examined using genomic microarrays of H. pylori exposed to different levels of external pH (7.4, 6.2, 5.5, and 4.5) for 30 min in the absence and presence of 5 mM urea. Gene expression was correlated with intrabacterial pH measured using 2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein and compared to that observed with exposure to 42°C for 30 min. Microarrays containing the 1,534 open reading frames of H. pylori strain 26695 were hybridized with cDNAs from control (pH 7.4; labeled with Cy3) and acidic (labeled with Cy5) conditions. The intrabacterial pH was 8.1 at pH 7.4, fell to 5.3 at pH 4.5, and rose to 6.2 with urea. About 200 genes were up-regulated and ∼100 genes were down-regulated at pH 4.5 in the absence of urea, and about half that number changed in the presence of urea. These genes included pH-homeostatic, transcriptional regulatory, motility, cell envelope, and pathogenicity genes. The up-regulation of some pH-homeostatic genes was confirmed by real-time PCR. There was little overlap with the genes induced by temperature stress. These results suggest that H. pylori has evolved multifaceted acid-adaptive mechanisms enabling it to colonize the stomach that may be novel targets for eliminating infection.
Helicobacter pylori is a motile neutralophile that is uniquely adapted to colonizing the often highly acidic environment of the human stomach (8, 49). No other neutralophile has adapted to grow in the normal stomach, although many can transit the stomach. Although the exact pH at the surface of the gastric epithelium, the niche occupied by the organism, is not known, experiments with ureI deletion mutants in animal models show that the pH must often be lower than 4.0 (37, 55). For growth under these conditions, specialized mechanisms likely have evolved.
Neutralophilic bacteria, such as Escherichia coli, have an acid tolerance response in log phase and an acid resistance response in stationary phase (9). These responses allow these neutralophiles to survive extremes of pH but do not allow growth. In the case of H. pylori, changes of expression of some genes under acidic conditions could be unique in allowing growth and therefore may be considered acid-adaptive rather than acid tolerance or resistance genes. Aside from their microbiological interest, identification of these genes might allow the design of novel eradication strategies for this type I carcinogen (41, 53).
The initial response of E. coli and other enteric neutralophiles to acute cytoplasmic acidification is activation of outward H+ pumps, in particular the F1F0 ATPase (27). However, survival with prolonged acid exposure requires alteration of gene expression. Several genes involved in acid tolerance or resistance have been described in E. coli. One set of products of such genes that has been studied extensively is the amino acid decarboxylases, particularly arginine and glutamate decarboxylases. These enzymes are membrane embedded, function best at an internal pH (pHin) of ∼5.0 (52), and are important in the buffering of intracellular H+. Additional acid tolerance responses include induction of a protein that modifies the lipid composition of the inner membrane by making cyclopropane fatty acids (12), thereby altering the proton permeability of the inner membrane either directly or by prevention of the insertion of proton-permeable inner membrane proteins. Acid conditioning of E. coli has been observed, since preexposure to acid improves acid survival upon exposure to a more extreme pH, implying gene regulation as an additional mechanism for acid survival (57).
In the case of another enteric organism, Salmonella enterica serovar Typhimurium, although an external pH (pHout) of 5.8 results in simple pH homeostasis, a lower pH induces an acid tolerance response by altering the expression of several regulatory genes, including the σs factor encoded by rpoS, the iron regulatory factor fur, and the two-component signal transduction system phoPQ (21).
Changes in the functions of these regulatory factors may result from stabilization against protease activity or phosphorylation, as well as increased translation. The bacteria can transit the acidity of the gastric lumen, but they cannot grow in the normal stomach, since their internal pH falls below the pH range of most of their enzyme systems, even with the acid tolerance or resistance response (21).
A different acid resistance mechanism has been adopted by Yersinia enterocolitica. This microbe expresses a urease with a sharp pH optimum of 5.0 that is active only when the cytoplasmic pH falls to the value observed when the external pH falls below 3.0. The enhanced urease activity prevents the cytoplasmic pH from falling below 5.0, but at that pH, protein synthesis is halted. Hence, this urease mechanism allows transit through, but not habitation of, the stomach (56). Thus, although other neutralophiles are able to respond to acidity of the medium by expression of acid tolerance or resistance genes, only gastric Helicobacter spp. are likely to express or regulate expression of what might be termed acid-adaptive genes that may be involved in allowing this neutralophile to flourish in the gastric environment.
One such acid-adaptive gene of gastric Helicobacter spp. is ureI, included within the urease gene cluster (20). There are seven genes in this cluster, ureA, -B, -I, -E, -F, -G, and -H. ureA and ureB encode the structural subunits of the neutral-pH-optimum urease and are generally constitutively expressed. The last four genes are involved in the assembly of nickel into the apoenzyme to generate active urease. Homologs of these genes are expressed by other ureolytic organisms (11, 36, 38, 40). However, the third gene in the cluster, ureI, encodes an inner membrane pH-gated urea channel (42, 46, 54). Its function is to increase the access of urea to intrabacterial urease in an acidic milieu. With this increase in urea entry, intrabacterial urease activity is able to buffer the cytoplasm and periplasm at acidic pH values that are usually incompatible with growth (10, 48, 54). The essential roles of UreI and urease in allowing gastric infection establish that the environment of the organism is often acidic. In particular, the demonstration that ureI deletion mutants were able to colonize the gerbil stomach when acid secretion was inhibited but failed to survive when acid secretion was allowed to return showed that the pH of the gastric habitat often must be <4.0, the in vitro limit for survival in the absence of urea (37, 54). Presumably, the ammonia and carbon dioxide generated by intrabacterial urease are able to buffer both the cytoplasm and the periplasm. It has been shown that the presence of urea, and hence intrabacterial urease activity, is essential for protein synthesis at a pH of <5.0 (48). Since the organism inhabits an environment where the pH often falls below 4.0, urease activity at this external pH permits growth that otherwise would not be possible.
The information summarized above indicates that the organism can flourish at acidic pH, although the exact pH limits of its ability to colonize the gastric surface are not known. The pH of the contents of the stomach varies widely, falling to ∼1.0 in the absence of food and perhaps rising to 5.0 during the digestive phase. Gastric juice urea is ∼1 mM and may at times be insufficient to ensure the gastric survival of H. pylori. The migration of the organism from antrum to fundus with inhibition of acid secretion in humans and in gerbil suggests that an optimal pH is sought by the bacteria (28, 29, 30, 37).
Various other genes, including genes that would be capable of adding other pH-homeostatic mechanisms to the cytoplasm or periplasm, have been identified as either improving acid survival or changing expression in acidic media (13, 33, 50). There have been recent reports of systematic studies investigating genes of H. pylori that change under acidic conditions (32, 34). These have either investigated acid shock or measured gene expression as a function of time of exposure.
Examination of the level of gene expression using DNA microarrays over a limited medium external pH range from 7.4 to 4.5 using impermeant buffers in the absence and presence of physiological levels of urea is likely to simulate the majority of the conditions the bacteria encounter in the stomach. Furthermore, correlation of these changes with alterations in cytoplasmic pH allows conclusions as to the site of regulation of gene expression. In the absence of urea, 187 genes appeared to be up-regulated and 113 genes appeared to be down-regulated more than twofold. The presence of urea reduced the number of genes affected by acidic pH by ∼50%. Different genes showed distinct patterns of response to pH and to the presence of urea, indicating different sites or mechanisms of acid-adaptive regulation. The pattern of response to temperature stress was quite distinct from that of acidic pH exposure.
These results indicate H. pylori has evolved an acid-adaptive rather than an acid tolerance or resistance response to an acidic external environment. This acid-adaptive response enables growth in the acidic environment of the stomach, allowing colonization by rather than just survival of the organism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strain 26695 was obtained from the American Type Culture Collection. Primary plate cultures of H. pylori were grown from glycerol stocks on blood agar medium for 2 to 3 days in a microaerobic environment (5% O2, 10% CO2, and 85% N2) at 37°C. In preparation for an experiment, bacteria were scraped from the plates, suspended in 1 mM phosphate HP buffer (138 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 1 mM glutamine), pH 7.0, and transferred to fresh plates for 24 h. For exposure to experimental low-pH conditions, the overnight cultures of H. pylori strain 26695 on Trypticase soy agar plates supplemented with 5% sheep blood were suspended in brain heart infusion (BHI) medium (Difco) to an optical density at 600 nm of 0.20 to 0.25. The pH of BHI was adjusted to 7.4, 6.2, 5.5, or 4.5 using concentrated HCl followed by filtration to remove any precipitate. The H. pylori organisms were then incubated in the presence or absence of 5 mM urea with shaking (120 rpm) under microaerobic conditions at 37°C for 30 min at 10-fold dilution with medium to ensure the presence of urea at the end of the experiment. The presence of urea at the end of each incubation was confirmed by adding jack bean urease to the supernatant after centrifugation of the organisms for RNA preparation and demonstration of a rapid rise in the medium pH.
For temperature stress experiments, the bacteria were exposed to 42°C for 30 min at a medium pH of 7.4 and then processed as for the pH exposure conditions by pelleting them by centrifugation (at 1,500 × g for 10 min at 4°C), followed by isolation of the RNA.
RNA preparation.
Total RNA was isolated from H. pylori strain ATCC 26695 using TRIzol reagent (Invitrogen, Carlsbad, Calif.) combined with RNeasy columns (Qiagen, Valencia, Calif.). The bacterial pellet was resuspended in 500 μl of TRIzol reagent and lysed at room temperature for 5 min before 100 μl of chloroform was added. After the pellet was spun at 12,000 × g for 10 min at 4°C, the supernatant was mixed with 250 μl of ethanol and applied to an RNeasy spin column, and RNA purification was performed following the manufacturer's instructions (beginning with the application to the column). To remove contaminating genomic DNA from purified RNA, samples were treated with RQ1 RNase-free DNase (Promega), followed by proteinase K digestion, phenol-chloroform extraction, and precipitation with ethanol. The RNA concentration was quantified by absorbance at 260 nm, and the quality was evaluated by capillary electrophoresis using a model 2100 Bioanalyzer with an RNA 6000 Nano Assay kit (Agilent Technologies).
Microarray preparation.
PCR products that contained 1,534 predicted open reading frames (ORFs) of H. pylori strain 26695 were purchased from Invitrogen Corporation. The concentration of the PCR product was estimated based on the density of the band on an ethidium bromide-stained agarose gel. After purification of the PCR products using a QIAquick 96-well PCR purification kit (Qiagen), ∼4 μg of PCR product for each ORF was loaded into a fresh 96-well PCR plate. After the plates were completely dried with a speed vacuum, they were resuspended by adding 10 μl of sodium bicarbonate buffer (NaHCO3-Na2CO3 at a concentration of 350 mM at pH 9.0) to each well for printing (University of California—Los Angeles Microarray Core Facility). All steps of glass slide preparation, printing, and processing after printing were performed as described by Eisen and Brown (18). The final array contained 1,534 unique sequences and spots of genomic DNA of H. pylori with known concentrations as external controls.
Fluorescent cDNA labeling.
For each comparative array hybridization, labeled cDNA was synthesized by reverse transcription from experimental RNA (pH 7.4, 6.2, 5.5, or 4.5 with or without 5 mM urea) in the presence of Cy5-dCTP, and from control RNA (pH 7.4) with Cy3-dCTP, using the Superscript II reverse-transcription kit (Invitrogen). H. pylori-specific cDNA-labeling primers (Sigma-Genosys) priming cDNA synthesis of total H. pylori RNA were used to prepare fluorescently labeled probes for array hybridization. For each reverse-transcription reaction, H. pylori cDNA-labeling primers that preferentially prime mRNAs in H. pylori and total H. pylori RNA (40 μg) were mixed in a final volume of 12.5 μl, heated to 90°C for 2 min, and ramped to 42°C for 20 min prior to probe synthesis. Fluorescent-cDNA-probe synthesis was performed at 42°C for 3 h in a 25-μl reaction volume containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dTTP, 0.2 mM dCTP, 0.1 mM either Cy3- or Cy5-conjugated dCTP (Amersham), 40 U of RNAsin (Promega), and 200 U of Superscript II reverse transcriptase (Invitrogen.). Following reverse transcription, the RNA template was degraded by incubation for 40 min at 65°C in 0.27 M NaOH, followed by neutralization with Tris-HCl buffer. The labeled cDNA was purified and concentrated prior to hybridization using Microcon 30 concentrators (Amicon).
Genomic-DNA labeling.
Genomic DNA was labeled by a simple random-priming protocol based on the Gibco-BRL Bioprime DNA-labeling kit. For each labeling reaction, 2 μg of H. pylori genomic DNA and 15 μg of random octamers were mixed in a final volume of 40 μl and heated to 100°C for 5 min. Fluorescent genomic DNA labeling was performed at 37°C for 2 h in a 50-μl reaction volume containing 50 mM Tris-HCl (pH 6.8), 5 mM MgCl2, 10 mM β-mercaptoethanol, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dTTP, 0.2 mM dCTP, 0.1 mM either Cy3- or Cy5-conjugated dCTP (Amersham), and 50 U of Klenow fragment. The labeled cDNA was purified and concentrated prior to hybridization using Microcon 30 concentrators. The labeled DNA was generated from strain 26995 as a template and then hybridized to the microarray.
Microarray hybridization.
H. pylori microarrays were hybridized in a final volume of 20 μl containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.8 mg of salmon sperm DNA/ml, and 0.2% sodium dodecyl sulfate. Prior to hybridization, combined Cy3- and Cy5-labeled probes (representing different pH conditions) were heated to 95°C for 2 min and pipetted directly onto microarray slides. A glass coverslip (22 by 22 mm) was applied, and the arrays were hybridized overnight at 65°C in a humidified hybridization chamber. Following hybridization, the slides were washed for 5 min in 0.2× SSC with 0.1% sodium dodecyl sulfate, followed by two washes of 3 min each in 0.2× SSC and rinsing in 0.05× SSC. To remove residual salts, the slides were spun at 500 rpm for 5 min prior to being scanned.
Array quantification and data processing.
Following hybridization, the array slides were scanned using a GMS 418 scanner (Genetic Microsystems, Inc.), which is available in the University of California—Los Angeles Microarray Core Facility. Separate images were acquired for Cy3 and Cy5. The signal intensity for each spot were determined using ScanAlyze (available at http://www.microarrays.org/software.html) and Phoretix Array (Nonlinear Dynamic) software. Following background subtraction, signal intensities were calculated as the percentage of the total signal from all spots as a means of normalization. These values were used to determine the ratio of experimental to control signals. A threshold of minimum acceptable signal (2 standard deviations above background intensity) was used to eliminate expression ratios that were extremely high or low due to undetectable signal in control or experimental samples. A t test was applied to determine the consistency of ratios across replicate hybridizations. Only those ratios with values showing ≥2-fold change (either increase or decrease) and having a 95% confidence interval as determined by the t test were considered to indicate significantly regulated genes.
Cluster analysis.
Cluster and Treeview (19) were used to select, group, and visualize genes whose expression varied across the samples. We extracted tables (rows of genes and columns of individual microarray hybridizations) of normalized fluorescence ratios from the database. The subsets of genes that were significantly regulated by low pH (as described above) were selected from the 1,534 ORFs on the array. Before the clustering and display, the logarithm of the measured fluorescence ratios for each gene was centered by subtracting the arithmetic mean of all ratios measured for that gene. Then, a hierarchical clustering algorithm was applied to the genes at each experimental condition using the Pearson correlation coefficient as the measure of similarity and average linkage clustering (19).
Real-time PCR.
Primers were designed to 100- to 300-bp regions of the selected up-regulated genes seen in the microarray and were used in standard PCR to ensure that there was only one product. Primer design was aided by the Primer 3 software available at http://www-genome.wi.mit.edu/genomesoftware/other/primer3.html (43). The template for the real-time reaction was the cDNA equivalent to that used in the microarrays, and H. pylori strain 26695 genomic DNA serially diluted 10-fold was used as a standard curve. A reaction mixture containing the master mixture with CYBR green label (Qiagen) and the primers was added to a 96-well plate, along with 1 μl of cDNA or genomic DNA template, for a final reaction volume of 50 μl/well. The negative control contained the reaction mixture but no DNA. Samples were run in an icycler real-time PCR machine (Bio-Rad) for 40 cycles (30 s at 92°C, 30 s at 55°C, and 40 s at 72°C). Data were collected during the extension step and expressed as arbitrary fluorescence units per cycle. A melting curve was run at the end to ensure that there was only one peak and only one product.
Measurement of intrabacterial pH. (i) BCECF loading.
H. pylori strain 26695 was grown overnight on Trypticase soy agar plates supplemented with 5% sheep blood (Becton Dickinson). The bacteria were removed from the plate and resuspended in 5 ml of BHI medium (Difco). The pH-sensitive fluorescent probe, 2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein (BCECF) acetoxymethy ester, was then added to the bacterial suspension to a final concentration of 10 μM. The nonfluorescent ester is converted to the relatively impermeant pH-sensitive probe BCECF by intrabacterial esterase. The bacterial suspension was incubated in a microaerobic (5% O2, 10% CO2, and 85% N2) environment at 37°C for 30 min. To remove external free BCECF, the bacteria were pelleted by gentle centrifugation (3,000 × g; 5 min) and resuspended in 300 μl of Hp medium (140 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 0.5 mM MgSO4, 10 mM glucose, 1 mM glutamine) buffered with either 100 mM 3-(N-morpholino)propane sulfonic acid (HpMOPS), pH 6.5 to 7.7; 100 mM 2-(N-morpholino)ethanesulfonic acid (HpMES), pH 5.5 to 6.5; or 50 mM homopiperazine-N,N′-bis-2(ethanesulfonic acid) (HpHOMOPIPES), pH 4.0 to 5.0. BHI medium is not compatible with BCECF fluorescence measurements. Experiments were performed with and without 5 mM urea. To measure pHin, 20 μl of BCECF-loaded H. pylori organisms were added to 3 ml of HpMOPS, HpMES, or HpHOMOPIPES, depending on the desired pH, and fluorescence was monitored in a dual-excitation (Ex1 = 502 nm; Ex2 = 436 nm) single-emission (Em = 530 nm) fluorimeter.
(ii) Calibration of pHin.
Each experiment was independently calibrated. Once the fluorescence of the internal BCECF was measured, 150 nM protonophore, 3,3′,4′,5-tetrachlorosalicyanilide, was added to equilibrate the pH internally to that of the medium. HCl was then added to obtain minimum fluorescence of the dye, followed by the addition of NaOH to obtain maximum fluorescence of the dye. The internal pH was calculated using the equation pHin = pKa + log[(R − RA)/(RB − R) × FA(λ2)/FB(λ2)], where pKa is the pKa of BCECF (7.0), R = 502 nm/436 nm for each data point, RA is the ratio at minimum fluorescence, RB is the ratio at maximum fluorescence, FA(λ2) is the minimum fluorescence at 436 nm, and FB(λ2) is the maximum fluorescence at 436 nm.
RESULTS
Intrabacterial pH.
Cytoplasmic pH fell as the medium pH was reduced in stepwise fashion from 7.4 to 4.5, as shown in Table 1. The measured cytoplasmic pH was always higher than the external pH throughout this range, even in the absence of urea, showing the presence of internal buffering capacity even in the absence of urea. In the presence of 5 mM urea, additional buffering was seen. Although there are no change of the cytoplasmic pH with the addition of urea at pH 6.2 or 7.4 in the medium, there are significant elevation of the intrabacterial pH in a medium of pH 5.5 and 4.5 with the addition of urea. The increase of cytoplasmic pH to ∼7.0 allows continued protein synthesis at pH 5.5, although at a lower rate than the pH of 8.1 found at neutral medium pH (47). Confocal microscopy showed that at pH 6.2 with the addition of urea there was a significant increase in the fluorescence of BCECF free acid in the periplasm following the addition of 5 mM urea, as previously demonstrated for pH 5.5 in 5 mM phosphate buffer (4) (data not shown).
TABLE 1.
Cytoplasmic pHs at different medium pHs with and without 5 mM urea
| Medium pH | Cytoplasmic pHa
|
|
|---|---|---|
| +Urea | −Urea | |
| 7.4 | 8.0 | 8.1 |
| 6.2 | 7.0 | 7.1 |
| 5.5 | 6.9 | 6.5 |
| 4.5 | 6.5 | 5.3 |
Cytoplasmic pH with (+) or without (−) 5 mM urea.
Validation of the microarray.
Genomic DNA of strain 26695 was labeled with either Cy3 or Cy5 and then hybridized to the array of ORFs from strain 26695. Of the 1,534 sequences present on the slide, all the spots were detected by both the fluorophores, indicating that the microarray slides were well printed and recognized. No significant variability in signal intensities was observed when dye-swap was performed between experiments and reference samples, excluding the experimental artifacts introduced by differential sensitivity of Cy3 and Cy5 to photobleaching.
Gene expression of H. pylori exposed to different acidic medium pHs in the absence and presence of urea.
The microarray method was used to survey the general transcriptional activity of the whole genome in H. pylori maintained at neutral pH (pH 7.4) compared to gradually decreasing external pHs (pH 6.2, 5.5, and 4.5). Throughout the experiment, impermeant buffers were used and the external pH was unchanged. These conditions were kept constant throughout the experiment during an exposure period of 30 min before RNA extraction, and the bacteria were harvested under log-phase growth conditions. This short period of exposure was selected to eliminate a possible transition from log to stationary phase. Identical conditions were used when 5 mM urea was present in order to analyze the effect on gene regulation of intrabacterial urease activity after activation of UreI by acidic medium pH. This might represent the conditions prevailing in the stomach.
The microarrays containing 1,534 ORFs of H. pylori strain 26695 were hybridized with fluorescently labeled cDNAs from the control (pH 7.4) condition (labeled with Cy3; green) and various acidic experimental conditions (labeled with Cy5; red). By comparing cDNA from each experimental condition with a common control (pH 7.4), variation in gene expression across the experimental conditions could be inferred from the observed variation in the normalized Cy5-red/Cy3-green ratio across the hybridization array. Several experiments (at least three) were performed at each pH with and without urea to ensure the reproducibility of the data.
General observations.
There was a progressive increase in the number of genes changing expression as the medium pH was lowered, with a preponderance of up-regulated genes. A twofold up-regulation was selected as an arbitrary cutoff point at pH 4.5. The presence of 5 mM urea during the incubation reduced the number of genes that were activated twofold or more across all pH values. A quantitative tabulation of significant differences in total gene expression under the experimental conditions examined is presented in Table 2.
TABLE 2.
Quantitative summary of low-pH-induced changes in H. pylori gene expressiona
| pH | 5 mM urea | No. (%) of genesb:
|
|
|---|---|---|---|
| Up-regulated | Down-regulated | ||
| 6.2 | Yes | 13 (0.8) | 7 (0.5) |
| 6.2 | No | 28 (1.8) | 32 (2.1) |
| 5.5 | Yes | 36 (2.3) | 22 (1.4) |
| 5.5 | No | 87 (5.7) | 80 (5.2) |
| 4.5 | Yes | 10 (6.6) | 53 (3.5) |
| 4.5 | No | 187 (12.2) | 113 (7.0) |
Almost all of 1,534 ORFs on the array were detectable with intensities above background level in all the experiments.
The percentages in the table are percentages of the 1,534 ORFs on the array.
Exposure to pH 4.5 without urea was the most potent effector of gene regulation in these experiments, resulting in significant (≥2-fold) up-regulation of 187 genes, while pH 4.5 with 5 mM urea resulted in up-regulation of 101 genes. At this pH, 102 genes were down-regulated more than twofold in the absence of urea and 53 were down-regulated more than twofold in the presence of urea. Of the up-regulated genes, 122 had known functions, whereas 65 were of unknown function. For the down-regulated genes, 99 had a known function and 26 are as yet undefined. More genes were up-regulated than were down-regulated by low pH in all the experiments (Table 2). The number of genes changing expression increased with increasing acidity in both the absence and presence of urea.
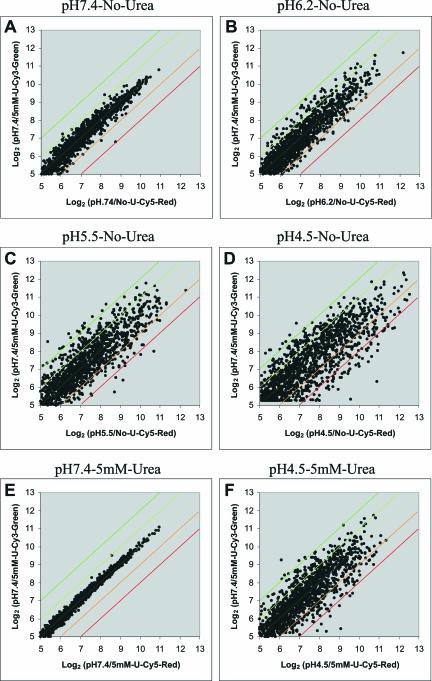
A log2-log2 scatter plot, shown in Fig. 1, illustrates the progressive change in the numbers of genes up-regulated and down-regulated as a function of fixed medium pH in the absence of urea. The total fractions of genes in the microarray containing 1,534 ORFs that showed detectable levels of expression were very similar (∼90%) under the various experimental conditions examined. In contrast, there were striking differences in the general patterns of gene regulation manifested in response to gradually decreasing pH (Fig. 1B to D and F). It can be seen that the correlation of labeling with Cy3 or Cy5 was very high at neutral pH, and there was a progressive number of genes that changed as the external pH was lowered (Fig. 1A to D). Figure 1E and F show the effect of the presence of urea at pH 7.4 and 4.5. There was little change in gene expression when urea was added to H. pylori incubated at pH 7.4, but the number of genes changing expression level decreased dramatically in the presence of urea. Although the presence of urea reduces the number of genes changing expression, there are still a large number of genes whose expressions were altered at acidic pH even in the presence of urea.
FIG. 1.
Scatter analysis of the general patterns of acidic responses of H. pylori genes revealed by transcriptional profiling. Scatter plots (log2-log2) of average normalized intensities representing Cy5-red channel versus Cy3-green channel are shown for experiments at pH 7.4 (n = 3) (A), 6.2 (n = 4) (B), 5.5 (n = 3) (C), and 4.5 (n = 4) (D) without urea (No-U), as well as pH 7.4 (n = 3) (E) and 4.5 (n = 4) (F) with 5 mM urea (U), compared with pH 7.4. Differential expression of a given gene is reflected by deviation from the yellow diagonal line. The orange diagonal defines ≥2-fold up-regulation, the red diagonal defines ≥4-fold up-regulation, light green defines ≥2-fold down-regulation, and dark green defines ≥4-fold down-regulation.
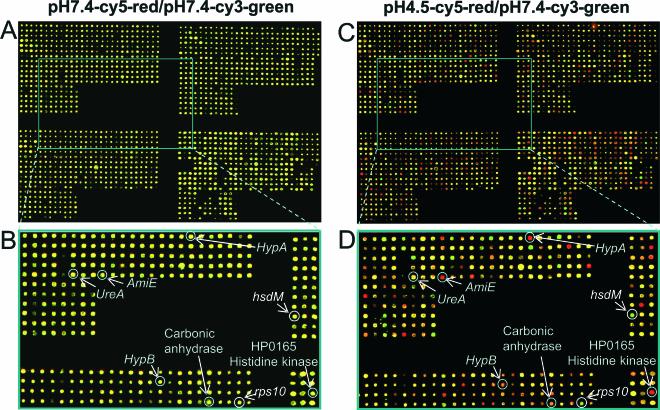
Figure 2 shows two representative microarray images in which hybridization with cDNAs derived from H. pylori maintained at pH 7.4 is compared to the hybridization of the cDNAs generated from H. pylori maintained at pH 4.5 in the presence of 5 mM urea. The constancy of expression of ureA is noteworthy. Expression of ureB is also constant under these conditions. These genes appear to act as housekeeping genes under the conditions employed here. Also shown are some of the genes that can be considered as contributing to pH homeostasis (amiE [GenBank accession no. HP0294]; hypA [HP0869]; hypB [HP0900]; and the carbonic anhydrase gene [HP1186]), as well as the up-regulation of HP0165, a histidine kinase sensor gene. Some down-regulated genes are also shown, such as those encoding the type II restriction enzyme HsdM (HP0092) and a ribosomal protein, Rps10 (HP1320). Several genes change expression even in the presence of 5 mM urea at pH 4.5; this reflects the change of cytoplasmic pH even with urea present in the medium.
FIG. 2.
Representative hybridization with cDNA prepared from H. pylori strain ATCC 26695 incubated at pH 7.4 and 4.5. Microarrays containing 1,534 ORFs from H. pylori strain 26695 were hybridized with fluorescent Cy3-labeled H. pylori strain 26695 cDNA from control conditions (pH 7.4) and Cy5-labeled cDNAs from experimental conditions at pH 7.4 (A and B) and pH 4.5 with 5 mM urea (C and D). A close-up region (boxed in each array image) is also shown (B or D). Genes that are up- or down-regulated by low pH appear in the image as red or green spots, respectively. Genes that are expressed at similar levels in both samples appear as yellow spots. ureA (encoding the urease structural subunit) does not change, whereas amiE (encoding aliphatic amidase), HP1186 (encoding carbonic anhydrase), HypA and HypB (hydrogenase accessory proteins) genes, and HP0165 (encoding histidine kinase) show clear up-regulation, and the type II restriction enzyme gene, hsdM, and a ribosomal protein gene, rps10, are down-regulated.
Acidic medium pH effects. (i) Up-regulated genes.
Tables 3 and 4 summarize the average ratios found for the genes up-regulated more than twofold at pH 4.5. For genes with known or putative functions, Table 3 shows 13 functional categories. Eight up-regulated genes possibly contribute to pH regulation and are categorized as a group of pH-homeostatic genes. This group contains the genes for the urease accessory proteins UreE (HP0070) and UreI (HP0071), periplasmic carbonic anhydrase (HP1186), aliphatic amidase (amiE [HP0294]), l-asparaginase II (ansB [HP0723]), arginase (rocF [HP1399]), and two hydrogenase accessory proteins that are involved in nickel homeostasis, HypA (HP0869) and HypB (HP0900) (39). Other up-regulated genes are classified as follows: 5 genes involved in regulation of transcription, 12 genes involved in motility and chemotaxis, 9 genes involved in pathogenesis, 19 genes implicated in energy metabolism, 3 genes involved in fatty acid and phospholipid metabolism, 3 genes implicated in translation, 11 transporters, 4 protein turnover genes, 8 genes involved in cell envelope structure or biosynthesis, 16 genes of DNA metabolism, 13 genes involved in amino acid and cofactor biosynthesis, and 9 general stress response genes. Table 4 shows 63 genes with unknown functions that are also up-regulated twofold or more at pH 4.5 in the absence of urea. It is clear that a large number of genes are regulated by medium acidification and consequent acidification of the periplasm and cytoplasm.
TABLE 3.
H. pylori genes up-regulated ≥2-fold with decreasing medium pH with and without urea
| Group | Avg ratio (exptl pH-Cy5-red/pH 7.4-cy3-green)a
|
Gene information
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 7.4
|
pH 6.2
|
pH 5.5
|
pH 4.5
|
GenBank accession no. | Description | ||||
| 5 mM U, (n = 3) | 5 mM U (n = 3) | No U (n = 4) | 5 mM U (n = 4) | No U (n = 3) | 5 mM U (n = 4) | No U (n = 4) | |||
| pH homeostasis (8 ORFs) | 0.98 | 1.11 | 1.19 | 1.01 | 0.99 | 1.02 | 2.56 | HP0070 | Urease accessory protein (ureE) |
| 0.95 | 1.09 | 1.01 | 0.98 | 1.16 | 1.49 | 2.35 | HP0071 | Urease accessory protein (ureI) | |
| 0.96 | 0.99 | 1.44 | 0.90 | 1.88 | 2.86 | 7.86 | HP0869 | Hydrogenase expression/formation protein (hypA) | |
| 0.99 | 1.06 | 2.38 | 1.59 | 3.30 | 2.14 | 2.34 | HP0900 | Hydrogenase expression/formation protein (hypB) | |
| 0.96 | 1.12 | 1.49 | 0.97 | 2.01 | 1.85 | 4.76 | HP1186 | Carbonic anhydrase | |
| 0.87 | 1.06 | 1.10 | 1.01 | 4.05 | 5.39 | 17.56 | HP0294 | Aliphatic amidase (amiE) | |
| 0.99 | 0.96 | 1.46 | 1.71 | 2.20 | 1.98 | 2.90 | HP0723 | l-Asparaginase II (ansB) | |
| 0.93 | 1.08 | 1.03 | 1.13 | 1.69 | 1.77 | 6.49 | HP1399 | Arginase (rocF) | |
| Motility and chemotaxis (12 ORFs) | 0.96 | 1.00 | 1.70 | 1.25 | 1.59 | 1.52 | 2.80 | HP0019 | Chemotaxis protein (cheV) |
| 1.14 | 1.57 | 1.50 | 1.46 | 1.56 | 1.52 | 4.13 | HP0082 | Methyl-accepting chemotaxis transducer (tlpC) | |
| 0.95 | 0.98 | 1.42 | 1.21 | 2.07 | 2.01 | 2.45 | HP0115 | Flagellin B (flaB) | |
| 0.92 | 1.00 | 1.43 | 1.07 | 1.97 | 1.58 | 3.23 | HP0325 | Flagellar basal-body L-ring protein (flgH) | |
| 1.00 | 1.08 | 1.19 | 1.05 | 1.17 | 0.99 | 2.24 | HP0393 | Chemotaxis protein (cheV) | |
| 1.10 | 1.27 | 1.79 | 1.86 | 1.92 | 2.06 | 2.03 | HP0601 | Flagellin A (flaA) | |
| 0.99 | 0.91 | 1.27 | 1.04 | 2.09 | 1.97 | 3.37 | HP0751 | Polar flagellin (flaG) | |
| 1.01 | 0.97 | 1.11 | 1.01 | 1.21 | 1.40 | 2.05 | HP0752 | Flagellar-hook-associated protein 2 (fliD) | |
| 0.99 | 0.89 | 1.20 | 1.29 | 1.47 | 1.57 | 2.31 | HP1067 | Chemotaxis protein (cheY) | |
| 1.02 | 1.35 | 3.13 | 4.36 | 4.66 | 6.13 | 14.12 | HP1192 | Secreted protein involved in flagellar motility | |
| 1.05 | 0.98 | 1.02 | 1.07 | 1.15 | 1.01 | 4.81 | HP1557 | Flagellar basal-body protein (fliE) | |
| 1.07 | 0.88 | 0.99 | 0.91 | 1.03 | 1.02 | 2.13 | HP1559 | Flagellar basal-body rod protein (flgB) | |
| Pathogenesis (9 ORFs) | 1.04 | 0.93 | 1.06 | 1.06 | 2.01 | 1.57 | 2.24 | HP0315 | Virulence-associated protein D (vapD) |
| 1.06 | 0.92 | 1.05 | 1.03 | 1.24 | 1.22 | 2.47 | HP0520 | Cag pathogenicity island protein (cag1) | |
| 1.06 | 1.10 | 1.61 | 1.29 | 1.48 | 1.45 | 2.22 | HP0528 | Cag pathogenicity island protein (cag8) | |
| 1.01 | 0.97 | 1.06 | 1.01 | 1.33 | 1.14 | 2.06 | HP0532 | Cag pathogenicity island protein (cag12) | |
| 1.03 | 1.11 | 1.20 | 1.01 | 0.99 | 1.03 | 2.35 | HP0537 | Cag pathogenicity island protein (cag16) | |
| 0.94 | 1.46 | 1.64 | 1.42 | 2.19 | 2.06 | 3.44 | HP0540 | Cag pathogenicity island protein (cag19) | |
| 0.93 | 0.95 | 1.36 | 1.12 | 1.74 | 1.27 | 2.45 | HP0541 | Cag pathogenicity island protein (cag20) | |
| 1.02 | 0.92 | 1.29 | 1.03 | 1.48 | 1.32 | 3.40 | HP0542 | Cag pathogenicity island protein (cag21) | |
| 1.15 | 0.98 | 1.50 | 1.09 | 1.82 | 1.65 | 2.02 | HP0543 | Cag pathogenicity island protein (cag22) | |
| Regulatory functions (5 ORFs) | 1.07 | 2.85 | 1.88 | 2.58 | 2.20 | 2.10 | 2.41 | HP0165 | Signal-transducing protein; histidine kinase |
| 1.01 | 1.36 | 1.23 | 1.01 | 1.06 | 1.40 | 4.21 | HP0166 | Response regulator (ompR-like) | |
| 0.93 | 1.16 | 1.06 | 1.30 | 1.14 | 1.04 | 8.61 | HP0278 | Guanosine pentaphosphate phosphohydrolase (gppA) | |
| 0.95 | 0.98 | 1.04 | 1.10 | 1.28 | 2.24 | 4.97 | HP1287 | Transcriptional regulator (tenA) | |
| 1.01 | 1.53 | 1.63 | 1.45 | 2.09 | 2.89 | 2.65 | HP1572 | Regulatory protein DniR | |
| Cell envelope (8 ORFs) | 0.94 | 0.97 | 1.84 | 1.00 | 2.30 | 2.18 | 2.72 | HP0025 | Outer membrane protein (omp2) |
| 1.00 | 1.39 | 1.37 | 1.60 | 1.21 | 3.51 | 3.61 | HP0079 | Outer membrane protein (omp3) | |
| 1.01 | 1.23 | 1.01 | 1.40 | 1.86 | 2.16 | 3.98 | HP0180 | Apolipoprotein N-acyltransferase (cute) | |
| 0.92 | 1.19 | 2.05 | 1.28 | 2.07 | 2.28 | 5.92 | HP0472 | Outer membrane protein (omp11) | |
| 0.98 | 1.25 | 1.54 | 0.97 | 1.36 | 1.80 | 2.07 | HP0567 | Membrane protein | |
| 0.89 | 0.96 | 1.90 | 1.49 | 2.60 | 1.66 | 4.23 | HP0623 | UDP-N-acetylmuramate-alanine ligase (murC) | |
| 1.07 | 1.03 | 1.08 | 1.06 | 2.66 | 1.11 | 3.97 | HP0896 | Outer membrane protein (omp19) | |
| 0.94 | 0.95 | 1.42 | 1.30 | 1.53 | 2.53 | 3.32 | HP1456 | Membrane-associated lipoprotein (lpp20) | |
| Energy metabolism (19 ORFs) | 0.88 | 0.93 | 1.07 | 0.94 | 2.07 | 2.06 | 2.42 | HP0193 | Fumarate reductase; cytochrome b subunit (frdC) |
| 0.90 | 0.99 | 1.02 | 1.02 | 0.97 | 0.96 | 2.78 | HP0194 | Triosephosphate isomerase (tpi) | |
| 1.10 | 0.92 | 0.91 | 0.92 | 1.21 | 1.22 | 2.65 | HP0277 | Ferredoxin | |
| 0.98 | 1.25 | 1.55 | 2.10 | 2.06 | 2.11 | 3.09 | HP0357 | Short-chain alcohol dehydrogenase | |
| 0.96 | 1.05 | 0.95 | 0.97 | 1.01 | 1.27 | 2.95 | HP0588 | Ferrodoxin-like protein | |
| 0.96 | 1.03 | 1.05 | 1.13 | 1.53 | 1.45 | 2.59 | HP0589 | Ferredoxin oxidoreductase; alpha subunit | |
| 0.97 | 0.96 | 1.06 | 1.07 | 1.71 | 1.52 | 2.63 | HP0590 | Ferredoxin oxidoreductase; beta subunit | |
| 0.98 | 0.97 | 1.17 | 0.97 | 1.90 | 1.27 | 2.58 | HP0591 | Ferredoxin oxidoreductase; gamma subunit | |
| 0.96 | 0.90 | 1.06 | 1.23 | 2.22 | 3.52 | 6.74 | HP0642 | NAD(P)H-flavin oxidoreductase | |
| 0.85 | 1.11 | 1.01 | 1.05 | 1.55 | 1.64 | 4.84 | HP0692 | 3-Oxoadipate CoA-transferase subunit B (yxjE) | |
| 0.93 | 0.95 | 1.24 | 2.10 | 3.50 | 2.75 | 2.49 | HP0824 | Thioredoxin (frxA) | |
| 0.92 | 0.93 | 1.14 | 1.24 | 1.70 | 1.45 | 3.00 | HP1099 | 2-keto-3-deoxy-6-phosphogluconate aldolase (eda)/PICK> | |
| 0.95 | 0.98 | 1.04 | 1.47 | 1.45 | 1.61 | 3.11 | HP1100 | 6-phosphogluconate dehydratase | |
| 1.10 | 0.94 | 1.92 | 1.70 | 2.89 | 3.05 | 9.17 | HP1166 | Glucose-6-phosphate isomerase (pgi) | |
| 0.95 | 0.99 | 1.12 | 1.05 | 1.67 | 1.64 | 3.60 | HP1222 | d-Lactate dehydrogenase (dld) | |
| 1.05 | 0.98 | 1.20 | 1.25 | 1.25 | 1.08 | 2.38 | HP1345 | Phosphoglycerate kinase | |
| 1.08 | 1.36 | 1.61 | 1.12 | 1.83 | 2.23 | 3.34 | HP1386 | d-Ribulose-5-phosphate 3 epimerase (rpe) | |
| 0.99 | 1.01 | 1.39 | 1.07 | 1.28 | 2.87 | 3.07 | HP1461 | Cytochrome c551 peroxidase | |
| 0.99 | 0.87 | 0.95 | 1.51 | 1.34 | 1.52 | 2.56 | HP1508 | Ferrodoxin-like protein | |
| Fatty acid and phospholipid metabolism (3 ORFs) | 1.03 | 1.02 | 1.29 | 0.99 | 1.81 | 2.28 | 7.56 | HP0690 | Acetyl CoA acetyltransferase (thiolase) (fadA) |
| 0.97 | 0.89 | 1.18 | 1.67 | 1.21 | 1.60 | 4.22 | HP0950 | Acetyl-CoA carboxylase beta subunit (accD) | |
| 0.93 | 1.05 | 1.17 | 0.98 | 1.42 | 1.30 | 2.00 | HP1045 | Acetyl-CoA synthetase (acoE) | |
| Translation (3 ORFs) | 1.03 | 1.27 | 1.17 | 1.01 | 1.03 | 1.57 | 8.30 | HP0077 | Peptide chain release factor RF-1 (prfA) |
| 1.02 | 1.58 | 2.19 | 2.32 | 2.12 | 2.47 | 5.19 | HP0182 | Lysyl-tRNA synthetase (lysS) | |
| 1.10 | 1.00 | 1.45 | 1.07 | 1.92 | 1.94 | 2.23 | HP0972 | Glycyl-tRNA synthetase; beta subunit (glyS) | |
| Transporters (11 ORFs) | 1.05 | 0.90 | 0.94 | 0.91 | 0.97 | 1.12 | 3.78 | HP0299 | Dipeptide ABC transporter permease protein (dppB) |
| 0.98 | 1.00 | 0.99 | 0.96 | 1.05 | 1.08 | 3.97 | HP0300 | Dipeptide ABC transporter permease protein (dppC) | |
| 0.86 | 0.96 | 0.93 | 1.01 | 1.41 | 1.00 | 4.04 | HP0301 | Dipeptide ABC transporter ATP-binding protein (dppD) | |
| 0.93 | 0.94 | 0.96 | 1.01 | 1.09 | 1.00 | 2.84 | HP0302 | Dipeptide ABC transporter ATP-binding protein (dppF) | |
| 0.99 | 0.95 | 1.04 | 1.01 | 1.32 | 1.10 | 4.02 | HP0613 | ABC transporter ATP-binding protein | |
| 1.06 | 1.16 | 1.56 | 1.23 | 1.82 | 1.67 | 4.72 | HP0715 | ABC transporter ATP-binding protein | |
| 0.97 | 0.93 | 1.04 | 1.33 | 2.03 | 2.20 | 2.57 | HP0876 | Iron-regulated outer membrane protein (frpB) | |
| 1.20 | 1.18 | 2.01 | 1.28 | 2.34 | 2.58 | 2.51 | HP0889 | Iron(III) dicitrate ABC transporter permease (fecD) | |
| 1.03 | 1.07 | 1.13 | 1.03 | 1.52 | 1.44 | 2.98 | HP0942 | d-Alanine glycine permease (dagA) | |
| 1.07 | 1.26 | 1.55 | 2.39 | 3.36 | 4.16 | 2.64 | HP1172 | Glutamine ABC transporter; glutamine-binding (glnH) | |
| 0.90 | 1.03 | 1.03 | 0.88 | 0.97 | 1.04 | 6.07 | HP1174 | Glucose/galactose transporter (gluP) | |
| Protein turnover (4 ORFs) | 0.91 | 1.01 | 1.01 | 1.61 | 1.44 | 1.25 | 2.79 | HP0382 | Zinc-metalloprotease (YJR117W) |
| 0.87 | 1.01 | 1.16 | 1.10 | 1.01 | 1.06 | 2.43 | HP0570 | Aminopeptidase a/i (pepA) | |
| 0.92 | 1.02 | 1.05 | 1.02 | 1.09 | 1.13 | 2.37 | HP0786 | Preprotein translocase subunit (secA) | |
| 1.02 | 1.17 | 1.15 | 1.26 | 1.25 | 2.33 | 3.08 | HP0793 | Polypeptide deformylase (def) | |
| DNA metabolism (16 ORFs) | 1.15 | 1.32 | 0.97 | 1.05 | 1.07 | 0.98 | 5.32 | HP0259 | Exonuclease VII; large subunit (xseA) |
| 1.05 | 0.94 | 1.07 | 1.06 | 2.24 | 2.63 | 3.79 | HP0478 | Adenine-specific DNA methyltransferase (VSPIM) | |
| 0.90 | 1.01 | 1.33 | 1.01 | 1.93 | 2.96 | 2.88 | HP0593 | Adenine-specific DNA methyltransferase (mod) | |
| 1.03 | 1.25 | 1.62 | 2.16 | 2.15 | 2.17 | 2.45 | HP0717 | DNA polymerase III gamma and tau subunits (dnaX) | |
| 0.98 | 1.40 | 1.58 | 2.90 | 2.10 | 5.14 | 3.03 | HP1022 | Predicted DNA polymeraseI-like 5′-3′ exonuclease | |
| 0.99 | 1.11 | 1.30 | 1.41 | 0.96 | 1.10 | 2.82 | HP1352 | Adenine-specific DNA methyltransferase (HINFIM) | |
| 0.99 | 0.97 | 1.78 | 1.37 | 2.29 | 1.83 | 4.93 | HP1402 | Type I restriction enzyme R protein (hsdR) | |
| 1.01 | 0.99 | 2.07 | 1.79 | 2.01 | 1.95 | 3.23 | HP1403 | Type I restriction enzyme M protein (hsdM) | |
| 1.04 | 0.87 | 1.48 | 1.46 | 2.08 | 2.23 | 10.09 | HP0669 | Remnant of type I restriction-modification peptide | |
| 0.92 | 0.93 | 1.11 | 0.97 | 1.04 | 1.09 | 2.55 | HP1470 | DNA polymerase I (polA) | |
| 1.12 | 1.10 | 1.08 | 1.51 | 1.51 | 2.23 | 6.19 | HP0437 | IS605 transposase (tnpA) | |
| 1.02 | 0.94 | 1.09 | 1.57 | 1.57 | 2.40 | 3.81 | HP0988 | IS605 transposase (tnpA) | |
| 1.07 | 1.12 | 1.07 | 1.40 | 1.25 | 2.11 | 2.51 | HP0998 | IS605 transposase (tnpA) | |
| 1.03 | 1.01 | 1.14 | 1.65 | 1.79 | 1.66 | 4.14 | HP1008 | IS200 insertion sequence from SARA17 | |
| 0.95 | 1.10 | 1.14 | 0.90 | 1.80 | 1.48 | 4.18 | HP1095 | IS605 transposase (tnpB) | |
| 1.14 | 1.35 | 1.20 | 1.68 | 1.73 | 1.96 | 4.07 | HP1096 | Is605 transposase (tnpA) | |
| Biosynthesis (13 ORFs) | 0.97 | 1.19 | 1.67 | 1.06 | 1.26 | 1.11 | 2.38 | HP0183 | Serine hydroxymethyltransferase (glyA) |
| 0.96 | 1.05 | 2.04 | 1.63 | 2.39 | 2.92 | 6.00 | HP0380 | Glutamate dehydrogenase (gdhA) | |
| 0.95 | 0.98 | 1.39 | 0.90 | 1.70 | 1.41 | 2.68 | HP0649 | Aspartate ammonia-lyase (aspA) | |
| 0.96 | 0.81 | 1.45 | 1.01 | 1.90 | 2.02 | 3.64 | HP0695 | Hydantoin utilization protein A (hyuA) | |
| 0.93 | 1.13 | 0.98 | 1.05 | 1.52 | 1.79 | 2.09 | HP1050 | Homoserine kinase (thrB) | |
| 0.96 | 1.05 | 1.17 | 1.01 | 1.20 | 1.13 | 2.57 | HP1468 | Branched-chain amino-acid aminotransferase (ilvE) | |
| 0.88 | 1.43 | 2.52 | 1.96 | 2.08 | 3.35 | 2.28 | HP0306 | Glutamate-1-semialdehyde 2,1-aminomutase (hemL) | |
| 0.93 | 1.59 | 2.28 | 2.35 | 1.91 | 5.93 | 3.21 | HP0376 | Ferrochelatase (hemH) | |
| 1.03 | 0.96 | 1.38 | 1.36 | 1.60 | 2.86 | 2.96 | HP0844 | Thiamine biosynthesis protein (thi) | |
| 1.10 | 0.94 | 1.17 | 1.48 | 2.16 | 2.15 | 3.66 | HP0845 | Thiamin phosphate pyrophosphorylase (thiM) | |
| 1.04 | 1.09 | 1.20 | 1.05 | 1.16 | 1.02 | 2.84 | HP1582 | Pyridoxal phosphate biosynthetic protein J (pdxJ) | |
| 0.87 | 1.01 | 1.23 | 0.95 | 1.05 | 1.02 | 3.22 | HP1583 | Pyridoxal phosphate biosynthetic protein A (pdxA) | |
| 0.94 | 1.43 | 1.90 | 2.47 | 2.14 | 1.67 | 2.32 | HP0854 | GMP reductase (guaC) | |
| General stress response (9 ORFs) | 1.03 | 1.18 | 1.10 | 1.04 | 1.35 | 1.24 | 2.39 | HP0243 | Neutrophil-activating protein (napA) (bacterioferritin) |
| 0.93 | 1.03 | 1.07 | 0.97 | 1.43 | 1.55 | 3.75 | HP0875 | Catalase | |
| 0.99 | 1.04 | 1.07 | 0.93 | 1.52 | 1.67 | 2.32 | HP0020 | Carboxynorspermidine decarboxylase (nspC) | |
| 0.99 | 1.22 | 2.03 | 1.80 | 1.64 | 1.65 | 2.08 | HP0047 | Hydrogenase expression/formation protein (hypE) | |
| 1.10 | 0.99 | 2.10 | 1.62 | 1.91 | 2.38 | 2.88 | HP0899 | Hydrogenase expression/formation protein (hypC) | |
| 1.01 | 1.02 | 0.87 | 1.09 | 1.70 | 1.84 | 2.09 | HP0485 | Catalase-like protein | |
| 0.91 | 0.97 | 1.45 | 1.67 | 1.44 | 2.21 | 3.38 | HP1104 | Cinnamyl-alcohol dehydrogenase ELI3-2 (cad) | |
| 1.04 | 1.01 | 1.10 | 0.93 | 1.26 | 1.09 | 2.79 | HP1193 | Aldo-keto reductase; putative | |
| 0.91 | 1.12 | 1.03 | 1.07 | 1.10 | 1.17 | 2.10 | HP0595 | Predicted DsbB-like protein | |
U, urea.
TABLE 4.
H. pylori genes of unknown function up-regulated ≥2-fold with decreasing medium pH with and without urea
| Avg ratio (exptl pH-Cy5-red/pH 7.4-Cy3-green)
|
GenBank accession no. | ||||||
|---|---|---|---|---|---|---|---|
| pH 7.4
|
pH 6.2
|
pH 5.5
|
pH 4.5
|
||||
| 5 mM urea (n = 3) | 5 mM urea (n = 3) | No urea (n = 4) | 5 mM urea (n = 4) | No urea (n = 3) | 5 mM urea (n = 4) | No urea (n = 4) | |
| 0.92 | 0.78 | 1.00 | 1.08 | 0.85 | 1.12 | 6.47 | HP0309 |
| 0.90 | 1.12 | 0.96 | 1.02 | 1.04 | 1.05 | 3.34 | HP0310 |
| 1.07 | 0.85 | 1.09 | 1.41 | 1.37 | 2.31 | 3.83 | HP0312 |
| 1.00 | 0.98 | 1.44 | 1.73 | 4.14 | 3.39 | 3.75 | HP0318 |
| 0.89 | 0.79 | 1.41 | 0.95 | 1.40 | 2.28 | 4.79 | HP0693 |
| 1.02 | 0.97 | 1.40 | 1.22 | 1.80 | 1.27 | 2.29 | HP0737 |
| 1.02 | 1.20 | 1.03 | 1.08 | 3.89 | 2.47 | 2.68 | HP0891 |
| 1.09 | 0.99 | 1.06 | 1.08 | 2.24 | 1.60 | 5.93 | HP0944 |
| 1.02 | 0.88 | 1.16 | 0.98 | 1.13 | 1.19 | 4.02 | HP1020 |
| 1.05 | 0.99 | 0.96 | 1.13 | 3.51 | 2.02 | 3.06 | HP1225 |
| 1.11 | 0.89 | 0.92 | 0.93 | 1.17 | 1.02 | 4.31 | HP1240 |
| 1.03 | 0.98 | 1.28 | 0.92 | 1.19 | 1.10 | 3.74 | HP0030 |
| 1.03 | 1.72 | 2.42 | 3.76 | 4.16 | 8.69 | 9.12 | HP0078 |
| 1.10 | 0.97 | 1.10 | 1.21 | 3.07 | 1.81 | 6.44 | HP0148 |
| 1.11 | 1.44 | 1.39 | 1.56 | 1.15 | 1.35 | 4.95 | HP0152 |
| 1.00 | 1.56 | 1.41 | 1.44 | 2.10 | 2.14 | 6.26 | HP0167 |
| 0.92 | 1.00 | 2.47 | 1.41 | 2.62 | 1.84 | 4.29 | HP0168 |
| 0.98 | 1.07 | 1.39 | 1.02 | 1.59 | 1.68 | 5.61 | HP0174 |
| 0.99 | 1.90 | 2.48 | 3.48 | 2.52 | 1.85 | 2.14 | HP0181 |
| 1.20 | 1.04 | 1.96 | 1.38 | 1.68 | 1.42 | 2.57 | HP0184 |
| 1.08 | 1.02 | 0.95 | 0.91 | 0.97 | 1.03 | 2.99 | HP0188 |
| 1.01 | 1.04 | 2.07 | 1.02 | 4.84 | 2.44 | 7.60 | HP0204 |
| 1.07 | 0.94 | 0.97 | 1.09 | 1.11 | 0.98 | 2.81 | HP0218 |
| 1.01 | 1.35 | 2.39 | 2.01 | 7.22 | 12.99 | 20.72 | HP0219 |
| 1.14 | 1.01 | 1.19 | 1.06 | 1.77 | 3.19 | 6.69 | HP0242 |
| 1.10 | 0.98 | 1.22 | 1.20 | 1.98 | 2.16 | 3.92 | HP0262 |
| 0.86 | 1.00 | 1.19 | 1.67 | 2.26 | 4.73 | 2.89 | HP0304 |
| 0.98 | 0.97 | 1.78 | 1.40 | 2.02 | 2.07 | 2.36 | HP0305 |
| 0.99 | 0.98 | 0.91 | 1.14 | 1.06 | 1.06 | 2.84 | HP0311 |
| 1.08 | 1.13 | 1.17 | 1.11 | 1.15 | 1.42 | 2.83 | HP0367 |
| 0.97 | 1.43 | 0.98 | 1.17 | 1.09 | 1.05 | 2.70 | HP0398 |
| 0.94 | 1.54 | 1.44 | 1.99 | 1.35 | 1.23 | 4.94 | HP0420 |
| 1.23 | 1.29 | 1.17 | 1.23 | 1.38 | 1.39 | 3.58 | HP0556 |
| 1.19 | 1.40 | 2.51 | 1.98 | 8.19 | 3.69 | 10.00 | HP0614 |
| 1.08 | 0.96 | 1.25 | 1.02 | 2.62 | 1.76 | 4.24 | HP0636 |
| 0.96 | 0.96 | 0.93 | 1.06 | 1.01 | 1.59 | 3.01 | HP0681 |
| 0.87 | 0.99 | 1.15 | 1.15 | 1.96 | 2.05 | 3.96 | HP0697 |
| 1.07 | 1.37 | 1.02 | 1.42 | 2.21 | 2.41 | 3.25 | HP0704 |
| 0.91 | 1.04 | 1.18 | 1.18 | 1.00 | 1.26 | 2.81 | HP0711 |
| 0.94 | 0.95 | 1.03 | 1.00 | 1.03 | 1.17 | 2.31 | HP0780 |
| 1.09 | 1.10 | 1.32 | 1.08 | 1.57 | 2.67 | 2.70 | HP0868 |
| 1.03 | 1.03 | 1.45 | 1.00 | 1.91 | 2.03 | 3.29 | HP0874 |
| 1.12 | 1.08 | 1.19 | 1.75 | 2.20 | 1.08 | 4.58 | HP0895 |
| 0.94 | 0.91 | 1.38 | 1.07 | 2.05 | 2.88 | 2.62 | HP0937 |
| 1.10 | 0.99 | 1.19 | 2.14 | 2.20 | 3.62 | 9.42 | HP0948 |
| 0.95 | 1.03 | 1.18 | 1.10 | 2.13 | 1.97 | 2.03 | HP0953 |
| 1.03 | 0.96 | 1.36 | 1.30 | 1.28 | 1.29 | 2.72 | HP0990 |
| 1.07 | 0.96 | 0.98 | 1.62 | 2.34 | 2.16 | 4.25 | HP1187 |
| 1.05 | 1.07 | 1.13 | 1.44 | 2.11 | 2.04 | 5.17 | HP1188 |
| 1.11 | 1.21 | 1.27 | 1.01 | 2.05 | 1.14 | 4.57 | HP1207 |
| 1.08 | 1.00 | 1.08 | 1.70 | 1.32 | 1.28 | 4.47 | HP1236 |
| 0.99 | 1.03 | 1.11 | 1.05 | 1.05 | 1.10 | 2.32 | HP1288 |
| 0.91 | 1.04 | 0.98 | 1.02 | 1.08 | 1.10 | 5.04 | HP1289 |
| 1.11 | 0.98 | 1.28 | 0.99 | 3.58 | 2.58 | 15.17 | HP1326 |
| 1.04 | 0.88 | 1.30 | 1.04 | 2.23 | 2.11 | 3.48 | HP1327 |
| 0.99 | 0.89 | 1.36 | 1.02 | 2.21 | 2.53 | 25.11 | HP1391 |
| 1.04 | 0.96 | 2.03 | 1.37 | 2.04 | 2.10 | 2.90 | HP1397 |
| 0.99 | 0.84 | 1.11 | 1.05 | 1.31 | 1.37 | 2.50 | HP1412 |
| 1.01 | 1.29 | 1.47 | 1.17 | 1.79 | 2.36 | 3.06 | HP1440 |
| 0.98 | 1.08 | 1.18 | 1.20 | 1.61 | 2.55 | 2.61 | HP1457 |
| 0.92 | 1.02 | 1.49 | 1.12 | 1.72 | 1.59 | 4.94 | HP1499 |
| 1.09 | 1.60 | 1.10 | 1.75 | 2.25 | 2.02 | 2.85 | HP1520 |
| 1.10 | 3.86 | 1.42 | 5.09 | 1.54 | 2.18 | 2.31 | HP0433 |
An intriguing pH-dependent pattern can be seen in that some genes increased expression even at pHout 6.2, where the cytoplasmic pH has not fallen below 7.0, with 10 genes with known functions and 3 of unknown function increasing more than twofold. However, most genes are up-regulated only when the cytoplasmic pH falls below 7.0, i.e., at a medium pH of 5.5 or 4.5. The presence of urea throughout the incubation prevents up-regulation of most, but not all, of these genes: 44 known-function genes and 30 unknown-function genes remained increased more that twofold at pH 4.5, even in the presence of urea. Notably, among the pH-homeostatic genes, those for hydrogenase expression, amidase, asparaginase, arginase, and periplasmic carbonic anhydrase remained up-regulated in the presence of urea at pH 4.5, as did the histidine kinase gene. Almost all the general stress response genes lost the >twofold up-regulation when urea was present at a medium pH of 4.5.
(ii) Down-regulated genes.
Tables 5 and 6 show the genes that were down-regulated ≥2-fold when the external pH was 4.5 in the absence of urea and the genes that were down-regulated ≥2-fold in the presence of urea. Of the genes with known function, 10 were related to cell envelope synthesis or function; 8 were involved in cellular processes, including motility functions; 2 were related to protein or RNA degradation and response regulation; 4 were involved in energy metabolism; 25 were involved in translation; 6 were involved in transport; 6 were involved in DNA metabolism; and 12 were involved in biosynthesis of different small molecules, for a total of 73 genes with known functions. The presence of urea reduced this number to 15 genes in which down-regulation was still twofold or more at pH 4.5; 23 genes of unknown function were also down-regulated in the absence of urea at pH 4.5, with only 4 down-regulated in the presence of urea at pH 4.5.
TABLE 5.
H. pylori genes down-regulated ≥2-fold with decreasing medium pH with and without urea
| Group | Avg ratio (exptl pH-Cy5-red/pH 7.4-Cy3-green)a
|
Gene information
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 7.4
|
pH 6.2
|
pH 5.5
|
pH 4.5
|
GenBank accession no. | Description | ||||
| 5 mM U (n = 3) | 5 mM U (n = 3) | No U (n = 4) | 5 mM U (n = 4) | No U (n = 3) | 5 mM U (n = 4) | No U (n = 4) | |||
| Cell envelope (10 ORFs) | 1.00 | 0.97 | 1.01 | 1.00 | 0.89 | 1.05 | 0.38 | HP0159 | Lipopolysaccharide 1,2-glucosyltransferase (rfaJ) |
| 1.00 | 0.90 | 0.42 | 0.43 | 0.30 | 0.24 | 0.13 | HP0229 | Outer membrane protein (omp6) | |
| 1.00 | 1.06 | 0.88 | 1.01 | 0.55 | 0.92 | 0.36 | HP0379 | Lucosyltransferase | |
| 0.89 | 0.95 | 1.04 | 0.87 | 0.92 | 1.00 | 0.36 | HP0549 | Glutamate racemase (glr) | |
| 1.01 | 0.96 | 1.08 | 1.11 | 1.10 | 1.01 | 0.47 | HP0648 | UDP-N-acetylglucosamineenolpyruvyl transferase (murZ) | |
| 0.95 | 0.99 | 0.71 | 0.99 | 0.56 | 0.82 | 0.36 | HP0651 | Fucosyltransferase | |
| 1.14 | 1.08 | 0.68 | 0.72 | 0.42 | 0.57 | 0.42 | HP0796 | Outer membrane protein (omp18) | |
| 0.84 | 1.08 | 0.73 | 0.63 | 0.47 | 0.43 | 0.27 | HP0957 | 3-deoxy-d-manno-octulosonic-acid transferase (kdtA) | |
| 0.94 | 0.35 | 0.26 | 0.31 | 0.28 | 0.13 | 0.29 | HP1177 | Outer membrane protein (omp27) | |
| 0.89 | 0.93 | 0.92 | 1.09 | 0.53 | 0.71 | 0.39 | HP0228 | Predicted sulfate permease | |
| Cellular processes (8 ORFs) | 1.03 | 0.96 | 0.66 | 0.88 | 0.31 | 0.58 | 0.47 | HP0331 | Cell division inhibitor (minD) |
| 0.94 | 1.05 | 1.03 | 1.14 | 1.09 | 0.96 | 0.49 | HP0351 | Flagellar basal-body M-ring protein (fliF) | |
| 1.00 | 0.98 | 1.03 | 1.07 | 1.11 | 0.91 | 0.37 | HP0517 | GTP-binding protein (era) | |
| 0.91 | 0.88 | 0.48 | 0.71 | 0.35 | 0.45 | 0.43 | HP0753 | Flagellar protein (fliS) | |
| 1.07 | 1.03 | 0.98 | 0.98 | 1.14 | 0.79 | 0.35 | HP0840 | FlaA1 protein | |
| 1.17 | 1.03 | 0.99 | 0.97 | 0.90 | 0.97 | 0.44 | HP0930 | Stationary-phase survival protein (surE) | |
| 0.91 | 0.71 | 1.01 | 0.71 | 0.89 | 0.80 | 0.47 | HP1035 | Flagellar biosynthesis protein (flhF) | |
| 1.03 | 0.53 | 1.04 | 0.75 | 0.99 | 0.97 | 0.35 | HP0390 | Adhesin-thiol peroxidase (tagD) | |
| Transcription and regulatory functions (2 ORFs) | 0.93 | 1.12 | 1.06 | 0.56 | 0.43 | 0.40 | 0.26 | HP1021 | Response regulator |
| 0.94 | 0.93 | 0.71 | 0.75 | 0.56 | 0.88 | 0.31 | HP1448 | Ribonuclease P protein component (rnpA) | |
| Energy metabolism (4 ORFs) | 0.92 | 0.72 | 0.93 | 0.89 | 0.93 | 0.83 | 0.36 | HP0056 | Delta-1-pyrroline-5-carboxylate dehydrogenase |
| 0.99 | 1.01 | 0.69 | 0.79 | 0.75 | 0.60 | 0.45 | HP0828 | ATP synthase F0; subunit a (atpB) | |
| 0.99 | 0.85 | 0.70 | 0.64 | 0.61 | 0.64 | 0.44 | HP1134 | ATP synthase F1; subunit alpha (atpA) | |
| 0.86 | 1.04 | 1.00 | 1.03 | 0.71 | 0.71 | 0.40 | HP1272 | NADH-ubiquinone oxidoreductase subunit (NQO13) | |
| Translation (25 ORFs) | 1.08 | 1.07 | 0.75 | 0.99 | 0.93 | 0.95 | 0.39 | HP0083 | Ribosomal protein S9 (rps9) |
| 0.99 | 0.91 | 1.02 | 0.97 | 0.85 | 0.91 | 0.35 | HP0084 | Ribosomal protein L13 (rpl13) | |
| 0.89 | 0.88 | 0.72 | 0.96 | 0.54 | 0.89 | 0.47 | HP0200 | Ribosomal protein L32 (rpl32) | |
| 1.07 | 1.68 | 1.68 | 0.88 | 0.94 | 0.94 | 0.31 | HP0296 | Ribosomal protein L21 (rpl21) | |
| 1.12 | 1.05 | 1.08 | 1.00 | 0.68 | 0.78 | 0.39 | HP0827 | Single-stranded-DNA binding protein 12RNP2 precursor | |
| 1.17 | 0.97 | 1.03 | 0.89 | 1.10 | 1.00 | 0.40 | HP1047 | Ribosome-binding factor A (rbfA) | |
| 1.00 | 0.95 | 0.95 | 0.82 | 0.82 | 0.70 | 0.30 | HP1148 | tRNA (guanine-N1)-methyltransferase (trmD) | |
| 1.03 | 0.99 | 0.44 | 1.00 | 0.39 | 0.45 | 0.45 | HP1199 | Ribosomal protein L7/L12 (rpl7/l12) | |
| 1.13 | 0.99 | 0.57 | 0.88 | 0.42 | 0.56 | 0.35 | HP1200 | Ribosomal protein L10 (rpl10) | |
| 1.00 | 0.96 | 0.39 | 0.66 | 0.52 | 0.60 | 0.43 | HP1202 | Ribosomal protein L11 (rpl11) | |
| 0.99 | 0.88 | 0.81 | 0.68 | 0.74 | 0.54 | 0.42 | HP1205 | Translation elongation factor EF-Tu (tufB) | |
| 1.23 | 1.00 | 0.89 | 1.00 | 0.51 | 0.55 | 0.40 | HP1296 | Ribosomal protein S13 (rps13) | |
| 1.06 | 1.01 | 0.84 | 0.87 | 0.43 | 0.67 | 0.36 | HP1303 | Ribosomal protein L18 (rpl18) | |
| 1.07 | 1.05 | 0.52 | 0.60 | 0.47 | 0.50 | 0.29 | HP1305 | Ribosomal protein S8 (rps8) | |
| 0.90 | 1.22 | 0.64 | 0.84 | 0.54 | 0.95 | 0.42 | HP1307 | Ribosomal protein L5 (rpl5) | |
| 1.00 | 1.06 | 1.01 | 0.91 | 0.44 | 0.65 | 0.37 | HP1309 | Ribosomal protein L14 (rpl14) | |
| 0.92 | 1.18 | 0.82 | 0.84 | 0.98 | 0.84 | 0.40 | HP1311 | Ribosomal protein L29 (rpl29) | |
| 1.17 | 0.96 | 0.59 | 0.74 | 0.45 | 0.62 | 0.32 | HP1316 | Ribosomal protein L2 (rpL2) | |
| 1.02 | 0.59 | 0.62 | 0.65 | 0.46 | 0.37 | 0.26 | HP1317 | Ribosomal protein L23 (rpl23) | |
| 0.99 | 0.95 | 1.06 | 1.05 | 0.45 | 1.11 | 0.42 | HP1318 | Ribosomal protein L4 (rpl4) | |
| 0.97 | 0.97 | 0.98 | 0.97 | 0.48 | 0.60 | 0.39 | HP1319 | Ribosomal protein L3 (rpl3) | |
| 1.07 | 0.79 | 1.07 | 0.84 | 0.45 | 0.41 | 0.30 | HP1320 | Ribosomal protein S10 (rps10) | |
| 0.99 | 1.13 | 1.11 | 0.96 | 0.97 | 0.95 | 0.18 | HP1497 | Peptidyl-tRNA hydrolase (pth) | |
| 0.93 | 0.97 | 0.84 | 1.00 | 1.09 | 1.01 | 0.36 | HP0553 | Predicted 23s rRNA methyltransferase | |
| 0.92 | 1.07 | 0.76 | 0.89 | 0.55 | 0.77 | 0.34 | HP1149 | Predicted 16s rRNA processing protein | |
| Transporter (6 ORFs) | 0.86 | 0.63 | 0.66 | 0.64 | 0.41 | 0.48 | 0.42 | HP0497 | Sodium-and chloride-dependent transporter |
| 0.87 | 1.02 | 0.97 | 0.84 | 0.71 | 0.88 | 0.34 | HP0498 | Sodium- and chloride-dependent transporter | |
| 0.99 | 1.11 | 0.71 | 0.75 | 0.67 | 0.78 | 0.49 | HP1181 | Multidrug efflux transporter | |
| 1.07 | 0.77 | 0.58 | 0.51 | 0.50 | 0.75 | 0.42 | HP1445 | Biopolymer transport protein (exbB) | |
| 0.94 | 0.99 | 1.25 | 1.01 | 0.99 | 1.02 | 0.49 | HP1464 | Predicted ABC transport system substrate binding protein | |
| 0.90 | 0.76 | 0.64 | 0.66 | 0.40 | 0.35 | 0.45 | HP1512 | Iron-regulated outer membrane protein (frpB)/PICK> | |
| DNA metabolism (6 ORFs) | 1.06 | 0.89 | 0.45 | 0.41 | 0.25 | 0.26 | 0.27 | HP0091 | Type II restriction enzyme R protein (hsdR) |
| 1.09 | 0.87 | 0.44 | 0.47 | 0.47 | 0.32 | 0.29 | HP0092 | Type II restriction enzyme M protein (hsdM) | |
| 1.12 | 0.92 | 0.86 | 0.97 | 0.62 | 0.80 | 0.40 | HP0481 | Adenine-specific DNA methyltransferase (MFOKI) | |
| 0.86 | 0.98 | 0.34 | 0.58 | 0.40 | 0.42 | 0.44 | HP1208 | Ulcer adenine-specific DNA methyltransferase | |
| 0.88 | 0.68 | 0.33 | 0.34 | 0.24 | 0.35 | 0.27 | HP1209 | Ulcer-associated gene restriction endonuclease (lceA) | |
| 0.99 | 1.00 | 0.82 | 0.94 | 0.62 | 0.89 | 0.45 | HP1421 | Conjugative transfer regulon protein (trbB) | |
| Biosynthesis (2 ORFs) | 1.22 | 0.99 | 1.13 | 1.02 | 1.21 | 0.88 | 0.38 | HP0157 | Shikimic acid kinase I (aroK) |
| 1.02 | 0.85 | 1.03 | 0.79 | 1.10 | 0.96 | 0.33 | HP0512 | Glutamine synthetase (glnA) | |
| 0.98 | 1.00 | 0.99 | 1.01 | 1.08 | 0.95 | 0.37 | HP0736 | Phosphoserine aminotransferase (serC) | |
| 0.88 | 0.93 | 0.92 | 0.68 | 0.72 | 0.87 | 0.34 | HP0598 | 8-Amino-7-oxononanoate synthase (bioF) | |
| 0.89 | 1.13 | 0.82 | 1.02 | 0.47 | 0.67 | 0.38 | HP0604 | Uroporphyrinogen decarboxylase (hemE) | |
| 1.13 | 1.14 | 1.17 | 0.97 | 0.64 | 0.92 | 0.43 | HP0799 | Molybdopterin biosynthesis protein (mog) | |
| 0.97 | 1.12 | 1.24 | 1.05 | 0.85 | 0.98 | 0.47 | HP1360 | 4-Hydroxybenzoate octaprenyltransferase (ubiA) | |
| 0.98 | 0.99 | 1.01 | 1.01 | 1.02 | 1.06 | 0.35 | HP1545 | Folylpolyglutamate synthase (folC) | |
| 0.95 | 1.02 | 0.92 | 0.84 | 0.99 | 0.87 | 0.32 | HP0255 | Adenylosuccinate synthase (purA) | |
| 0.97 | 0.87 | 1.04 | 0.73 | 0.96 | 0.60 | 0.35 | HP0919 | Carbamoyl-phosphate synthase (pyrAb) | |
| 0.97 | 0.95 | 1.25 | 1.35 | 0.98 | 0.93 | 0.47 | HP1011 | Dihydroorotate dehydrogenase (pyrD) | |
| 0.82 | 0.94 | 0.39 | 0.67 | 0.21 | 0.26 | 0.21 | HP0956 | Predicted pseudouridine synthase C | |
U, urea.
TABLE 6.
H. pylori genes of unknown function down-regulated ≥2-fold with decreasing medium pH with and without urea
| Avg ratio (exptl pH-Cy5-red/pH 7.4-Cy3-green)a
|
GenBank accession no. | ||||||
|---|---|---|---|---|---|---|---|
| pH 7.4
|
pH 6.2
|
pH 5.5
|
pH 4.5
|
||||
| 5 mM urea (n = 3) | 5 mM urea (n = 3) | No urea (n = 4) | 5 mM urea (n = 4) | No urea (n = 3) | 5 mM urea (n = 4) | No urea (n = 4) | |
| 1.03 | 1.00 | 0.97 | 1.09 | 1.07 | 0.99 | 0.38 | HP0022 |
| 1.04 | 1.08 | 1.20 | 1.14 | 1.05 | 0.98 | 0.39 | HP0162 |
| 1.01 | 1.08 | 1.13 | 0.98 | 1.07 | 0.73 | 0.48 | HP0234 |
| 0.91 | 0.94 | 1.04 | 0.92 | 1.27 | 0.96 | 0.29 | HP0552 |
| 0.96 | 0.86 | 0.99 | 0.78 | 0.65 | 0.86 | 0.31 | HP0707 |
| 1.03 | 0.94 | 0.98 | 0.97 | 0.76 | 0.78 | 0.35 | HP0926 |
| 0.86 | 0.96 | 1.05 | 0.91 | 0.68 | 0.79 | 0.35 | HP0946 |
| 1.01 | 0.99 | 1.10 | 0.93 | 0.95 | 1.00 | 0.35 | HP0161 |
| 0.91 | 0.97 | 0.85 | 0.80 | 0.51 | 0.62 | 0.42 | HP0270 |
| 1.07 | 0.92 | 1.11 | 0.94 | 0.67 | 0.89 | 0.48 | HP0369 |
| 0.92 | 1.10 | 1.05 | 0.91 | 0.97 | 0.89 | 0.41 | HP0582 |
| 0.94 | 0.56 | 0.31 | 0.31 | 0.35 | 0.36 | 0.20 | HP0603 |
| 1.01 | 0.96 | 0.91 | 0.69 | 0.43 | 0.73 | 0.37 | HP0838 |
| 0.92 | 0.95 | 0.84 | 0.85 | 0.65 | 0.52 | 0.44 | HP0863 |
| 0.99 | 0.75 | 0.71 | 0.78 | 0.69 | 0.41 | 0.45 | HP0864 |
| 1.17 | 1.11 | 0.99 | 0.91 | 0.94 | 0.96 | 0.47 | HP1029 |
| 0.98 | 0.98 | 1.06 | 0.93 | 0.79 | 0.88 | 0.33 | HP1106 |
| 0.97 | 1.03 | 1.21 | 0.94 | 0.93 | 1.05 | 0.38 | HP1124 |
| 1.00 | 0.94 | 0.29 | 0.63 | 0.12 | 0.32 | 0.42 | HP1217 |
| 1.09 | 0.68 | 0.43 | 0.43 | 0.37 | 0.45 | 0.45 | HP1334 |
| 1.08 | 0.91 | 0.65 | 0.59 | 0.28 | 0.64 | 0.48 | HP1377 |
| 1.02 | 0.73 | 0.88 | 0.79 | 0.41 | 0.52 | 0.31 | HP1502 |
| 0.99 | 1.00 | 1.10 | 0.92 | 0.88 | 0.97 | 0.41 | HP1546 |
Clusters of genes expressed in response to gradually decreasing medium pH.
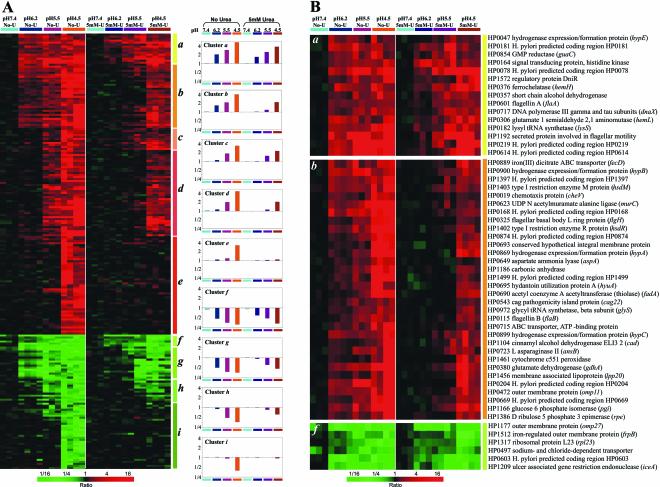
The tables mentioned above show the average ratios derived from three or four different experiments for each condition. To identify patterns of low-pH-regulated gene expression, we performed an average-linking hierarchical cluster analysis (19) on those genes that were regulated more than twofold (either up or down) by a pHout of 4.5. The object of the clustering was to group genes whose expression levels among samples were similarly regulated under each of the experimental conditions. The data presented in Fig. 3A are all the genes that change expression as a function of varying medium pH in the absence or presence of urea, as shown by color coding, and the average level of up- or down-regulation (right-hand column). The figure shows that there is reasonable consistency across different experiments at any given pH, whether in the presence or absence of urea.
FIG. 3.
Cluster analysis of gene expression patterns at different medium pHs with and without urea. (A) Hierarchical clustering was applied to expression data from a set of 278 genes measured across eight experimental pH conditions. The 278 genes were those (of 1,534 total) with transcript levels that varied by at least twofold relative to the control condition (pH 7.4) under conditions of pH 4.5 without urea. Data from 28 hybridizations were used: 3 for pH 7.4, 4 for pH 6.2, 3 for pH 5.5, and 4 for pH 4.5 without urea, as well as 3 for pH 7.4, 3 for pH 6.2, 4 for pH 5.5, and 4 for pH 4.5 with 5 mM urea. The color scale used to represent the expression ratios is shown on the bottom. The colored bars with labels (a to i) refer to the identified clusters of genes. A profile bar graph for each cluster in which the mean log ratio of that cluster in each experimental condition is plotted is shown on the right. (B) Enlargements of regions of the cluster diagram in panel A. The genes in up-regulated cluster a (top) and down-regulated cluster f (bottom), beginning at pH 6.2 and independent of the presence of urea, are shown. The genes up-regulated at pH 6.2 where the presence of urea results in up-regulation only when the medium pH is 4.5 are shown in cluster b (middle image). Gene names are shown corresponding to their positions in the cluster diagram.
The genes that respond to medium acidity fall into two general categories, those that show regulation in the presence or absence of urea (clusters a to d and f to h) and those that show regulation at pH 4.5 only in the absence of urea (clusters e and i) (Fig. 3A and B). These can be further subdivided into those that respond to pH 6.2 (early acidity response) and those that respond only to greater acidification. This classification possibly reflects the effect of the degree of change of pH, periplasmic and cytoplasmic, due to urease activity.
For example, cluster a contains 15 genes that are up-regulated and cluster f contains 6 genes that are down-regulated at pH 6.2 in the absence or the presence of urea (Fig. 3A and B). These can be thought of as early acid-adaptive genes. The addition of urea at pH 6.2 did not affect the cytoplasmic pH based on BCECF measurements. Nevertheless, there are also genes that are up-regulated at pH 6.2 and that lose their up-regulation in the presence of urea (cluster b). This correlates with an elevation of periplasmic pH by the addition of urea at pH 6.2, as visualized in confocal microscopy performed previously (4). The genes that change at pH 6.2 in the presence of urea thus respond to changes in periplasmic, not cytoplasmic, pH.
There are also a large number of genes that are up-regulated at lower medium pH (≤5.5) and maintain up-regulation in the presence of urea (cluster c), and there are also genes that lose up-regulation in the presence of urea (cluster d), with similar groups visible in the down-regulated genes. Perhaps the genes in cluster c respond to the cytoplasmic pH of 6.5 to 6.9 that is found in the presence of urea at pH 4.5 and 5.5, whereas the genes in cluster d respond only to a cytoplasmic pH of <6.0, such as the pH of 5.3 that is found in the absence of urea.
The specific genes contained within these clusters are shown in Fig. 3B. Clusters a and f contain the genes that change regulation at pH 6.2 in the absence or presence of urea. Some of the up-regulated genes in this group are hypE (HP0047), the histidine kinase gene (HP0165), flaA (HP0601), and the methyl-accepting chemotaxis transducer gene, tlpC (HP0082). Down-regulated genes of this type include the Na+- and Cl−-dependent transporter gene (HP0497) and the pathogenicity gene, iceA (HP1209). Cluster b contains genes that, while up-regulated at pH 6.2 in the absence of urea, fail to up-regulate with the addition of urea until pHout 4.5 is reached. Urea still decreases the level of expression in those genes, as shown in the bar graph in Fig. 3. The ORFs coding for the urease accessory proteins UreE (0070) and UreI (HP0071), carbonic anhydrase (HP1186), and a number of gene products involved in energy metabolism and motility and chemotaxis and ORFs rocF (HP1399), hypA (HP0869), cag1 (HP0520), cag8 (HP0528), cag12 (HP0532), cag16 (HP0537), and cag19 to -22 (HP0540-HP0543), as well as the ompR-like response regulator (HP0166), are found in this cluster.
Temperature stress.
The effect of pH on gene regulation of this organism that has adapted for life in the gastric environment may be relatively selective. When a comparison is made with the effect of exposure to 42°C on the genes present in the microarray, very few genes that are up-regulated overlap with those genes that are up- or down-regulated by acid exposure, as shown in Table 7. Of the genes discussed below, flaA (HP0601) is up-regulated by temperature stress, as is another gene involved in motility, HP1192, indicating that these are likely general stress response genes, like the outer membrane protein gene omp11 (HP0472).
TABLE 7.
42°C-regulated H. pylori genes that overlap with pH 4.5-regulated genes
| Regulation | Ratio (42°C-Cy5-red/ 37°C-Cy3-green) | GenBank accession no. | Description |
|---|---|---|---|
| Up-regulated | 2.2 | HP0472 | Outer membrane protein (omp11) |
| 3.1 | HP1192 | Secreted protein involved in motility | |
| 2 | HP0601 | Flagellin A (flaA) | |
| 2 | HP1096 | IS605 transposase (tnpA) | |
| 2.3 | HP0793 | Polypeptide deformylase (def) | |
| 2.3 | HP1326 | H. pylori predicted coding region HP1326 | |
| 2.2 | HP0556 | H. pylori predicted coding region HP0556 | |
| 2.1 | HP0078 | H. pylori predicted coding region HP0078 | |
| 2 | HP1288 | H. pylori predicted coding region HP1288 | |
| 2 | HP0872 | Alkylphosphonate uptake protein (phnA) | |
| Down-regulated | 0.4 | HP0092 | Type II restriction enzyme (hsdM) |
| 0.5 | HP1209 | Ulcer-associated gene endonuclease (iceA) | |
| 0.4 | HP0956 | Conserved hypothetical protein | |
| 0.32 | HP0296 | Ribosomal protein L21 (rpl21) | |
| 0.44 | HP1318 | Ribosomal protein L4 (rpl4) | |
| 0.49 | HP1305 | Ribosomal protein S8 (rps8) |
Real-time PCR.
Three genes from the pH homeostasis gene cluster that were up-regulated by acid exposure were selected for confirmation by real-time PCR amplification, and the results are shown in Fig. 4. This method confirmed the up-regulation of the genes for amidase (HP0294), arginase (HP1399), and the periplasmic carbonic anhydrase (HP1186). Additionally, the accessory genes of the urease gene cluster were also analyzed by real-time PCR amplification, even though the up-regulation of some of them was not detected on the microarray, perhaps due to their relatively low levels of expression. Up-regulation of ureE (HP0070) and ureI (HP0071) was confirmed. The real-time PCR analysis of ureF (HP0069), ureG (HP0068), and ureH (HP0067), which were not detected as changed on the microarray, showed an increase in RNA synthesis under acidic conditions (data not shown). Temperature stress did not result in an enhanced signal in amidase (HP0294) on the microarray, and real-time amplification confirmed that there was no change in the expression level of this gene under this stress condition.
FIG. 4.
Real-time PCR. (A to C) The cDNAs encoding periplasmic carbonic anhydrase (A), aliphatic amidase (B), and arginase (C) from typical experiments of the microarray analysis at pH 7.4 and 4.5 in the absence of urea were subjected to PCR amplification. The lines represent amplifications from cDNAs generated from the RNAs isolated at pH 7.4 and 4.5. (D) Similar amplification for the amidase cDNA resulting from temperature stress (42°C) compared to the cDNA generated from the RNA isolated from bacteria maintained at 37°C.
DISCUSSION
Colonization of the human stomach is a large evolutionary challenge for a neutralophile such as H. pylori. Acid resistance or acid tolerance responses that are expressed by other neutral-pH-dwelling bacteria are perhaps sufficient for gastric transit, but not for growth in the gastric environment. The urease of H. pylori is essential for colonization in animal models (17), but many other organisms express urease, although perhaps at a lower level. The acid-activated urea channel, UreI, is essential for infection in animal models (37, 51) and is a unique adaptation to life in the stomach for gastric Helicobacter spp. (22).
The pH of the gastric mucus or the gastric surface is a subject of controversy. Some suggest that the pH is close to neutral, but microelectrode and fluorimetric methods have generally shown a loss of pH regulation in the gastric mucus when the pH falls below 2.0 (14, 45). This pH, representing a concentration of 10 mM HCl, is not able to be buffered by the maximal capacity of the stomach to secrete bicarbonate, which is 10% of acid output. In the absence of food in the human stomach, gastric juice is unbuffered and there is continuing acid secretion, maintaining a fairly constant highly acidic pH. During the digestive phase, there is buffering by food and its digestion products, with a transient rise in pH. The median intragastric pH is 1.4 due to the long periods of high acidity between meals. The finding that ureI-deficient organisms were able to infect only if acid secretion was inhibited and then were eradicated when acid secretion was restored shows that a pH of <4.0 is present in their environment for a period sufficient for eradication (37). Over a 24-h period, H. pylori will be exposed to levels of acid at which ordinary neutralophiles do not divide or grow. The changes in internal or periplasmic pH of the organism thus must be compatible with colonization of its niche in the stomach, and experimental and physiological experiments show that this niche is highly acidic much of the time.
Changes in gene expression can depend on changes in periplasmic pH, cytoplasmic pH, or both. For efficient gastric colonization, changes in either compartment must be limited. The pH of the cytoplasm may be the most important factor determining whether the organism is able to survive in its niche in the stomach. Based on the measurements made here and previously (48), exposure to a medium pH of 6.2 results in a cytoplasmic pH of 7.0, and at this pH, the organism is able to synthesize protein without the addition of urea, although at a lower rate than at neutral pH. At a pH of 4.5, cytoplasmic pH drops to 5.3 in the absence of urea and protein synthesis, and hence, growth stops. However, at a medium pH of 4.5, the addition of urea elevates the cytoplasmic pH to 6.2 and protein synthesis is restored. This elevation of cytoplasmic pH may occur at an even lower gastric pH. It is also necessary for the bacteria to maintain their proton motive force, the electrochemical hydrogen ion gradient across the inner membrane, to allow growth at the acidic pH of the environment. This can be achieved by buffering the periplasm to above medium pH, as has been previously shown by the restoration of inner membrane potential to approximately −101 mV with urea addition at a medium pH as low as 3.0 (48). Gene expression after acid exposure can therefore be controlled by the level to which the cytoplasmic pH falls and also by the periplasmic pH, perhaps due to activation of one or more two-component systems.
Genes other than those in the urease gene cluster have already been identified as contributing to acid survival by a variety of methods. This implies that, although urease activity and its regulation by UreI are necessary for infection, that alone may not be sufficient. Other genes may be activated to improve the maintenance of infection, and some may even play a role in periplasmic or cytoplasmic buffering in addition to urease activity.
Up-regulated genes.
The changes in the increase of gene expression varied among different genes as a function of pH. Some were up-regulated at a medium pH of 6.2; others did not change expression until a pH of 4.5 was applied. Genes that lose up-regulation at a medium pH of 6.2 with the addition of urea are responding to a change in periplasmic pH, since the cytoplasmic pH does not change (Table 1). It was observed that BCECF free-acid fluorescence at pH 6.2 increased under the conditions used here with the addition of urea, showing an elevation of periplasmic pH. Genes that lose regulation with the addition of urea at more acidic pH presumably change expression only due to changes in the cytoplasmic pH. Thus, up-regulation of genes detected at an external pH of 4.5 that is absent with the addition of urea is due to a shift in the cytoplasmic pH from 5.3 to 6.5 that is found with the addition of urea.
As can be seen from the tables and the cluster analysis, a large number of genes change expression when exposed to acid. In some cases, it is possible to deduce or suggest the functions of up-regulated genes; in other cases, the rationale for changes of expression is at present obscure. Hence, the effect of acid exposure with and without urea will be discussed for only a few categories of genes illustrated in Tables 3 and 5 and displayed in Fig. 3B.
(i) pH-homeostatic genes.
The genes that theoretically could contribute to alkalization of the cytoplasm are categorized as pH homeostatic. Aliphatic amidase and l-asparaginase are both capable of generating NH3, and arginase generates urea. Each has been described as improving acid survival in vitro (33, 50). The hydrogenase expression genes hypA and hypB are among the genes that are up-regulated at pH 6.2, but their regulation is suppressed by the addition of urea until a pH of 5.5 or 4.5 is applied. These genes appear to be involved in the accumulation of Ni2+, the metal cofactor essential for activation of the urease apoenzyme. The urease accessory genes encoding UreI and UreE are up-regulated at pH 4.5. UreI is the acid-activated urea channel, and UreE is a urease assembly protein. It appears that the promoter between ureB and ureI is activated as a function of acidity, facilitating urea entry and assembly of urease without requiring de novo synthesis of the enzyme itself. This would enable a more rapid response in urease activity in spite of constant production of UreA and UreB. Apparently, longer acid exposure also up-regulates ureA and ureB (34). The periplasmic carbonic anhydrase gene HP1186 is progressively up-regulated as a function of the decrease of medium pH and may contribute to the regulation of the pH of the periplasm, since its deletion decreases survival at pH 4.0 even in the presence of urea (Y.-C. Lee, T. Tohru, H. Y. Shan, B. Grandjean, A. T. Charney, I. G. Perez-Perez, and M. J. Blaser, abstract from the Digestive Disease Week and 103rd Annual Meeting of the American Gastroenterological Association 2002, Gastroenterology 122:A423, 2002). This enzyme could function to accelerate the conversion of CO2 to bicarbonate in the periplasm, thus generating NH4HCO3 as the periplasmic buffer. It is likely that these pH-homeostatic genes contribute to regulation of cytoplasmic and/or periplasmic pH, in addition to increasing urease activity. Such mechanisms could compensate for variations in the urea concentration in the habitat of the organism.
(ii) Motility genes.
The up-regulation of 12 genes involved in motility or chemotaxis may be regarded as contributing to increased motility of the organism in acid, which perhaps acts as both an acid-adaptive response and an acidic stress response. flaA (HP0601) and HP1192, the gene for a secreted protein involved in flagellar motility, are among those genes up-regulated at pH 6.2 both in the absence and presence of urea. Although flaA has been described as pH regulated (24, 32), both flaA and HP1192 are also up-regulated by temperature stress and hence are not selective acid-adaptive genes. flaB and cheV are among the genes increased at pH 6.2 but suppressed by the presence of urea (Fig. 3B). cheY is also up-regulated, but not >2-fold until pH 4.5. The change of distribution of H. pylori with inhibition of acid secretion may be due in part to the up-regulation of motility genes (30, 32, 37).
(iii) Pathogenesis genes.
Eight genes in the pathogenicity island are up-regulated, as well as vapD, the gene for a virulence-associated protein. Many of the genes in the pathogenicity island are often considered responsible for type IV secretion, and perhaps an acidic environment prepares the organism for association with the plasma membranes of its gastric cell hosts. The cagA gene was not found to be up-regulated under our conditions.
(iv) Regulatory genes.
Various regulatory genes are up-regulated by the presence of mild acidity. Some of these may signal changes in the periplasmic pH. The gene for the inner membrane histidine kinase, HP0165, is one of those up-regulated at pH 6.2, independently of the presence of urea (Fig. 3B). This is part of a two-component signaling system in H. pylori. Although E. coli has 29 and 32 ORFs encoding histidine kinase and response regulators, respectively (35), only four and six two-component sensor and response regulator genes are found in the genome of H. pylori. Deletion of the sensor protein gene, HP0165, that phosphorylated HP0166, ompR-like, produced no change in the protein pattern at neutral pH, whereas deletion of HP0166 is lethal (5, 15). The paucity of signaling systems in H. pylori suggests that the ones that remain are multifunctional and interact with many genes. Hence, HP0165 activation may be responsible for many of the activated genes seen in this profiling of the genes adapted to mild acidity. The protein product of this gene has two transmembrane segments and a periplasmic domain replete with histidines, which suggests the possibility that a pH of 6.2 might determine protonation of one or more of these, resulting in autophosphorylation. The partner of this kinase, the ompR-like element, is also up-regulated, although not until pH 4.5.
Other regulatory genes are also increased, such as tenA and dniR, and up-regulation persists in the presence of urea at pH 4.5. gppA, a stringency response element encoding a guanosine pentaphosphate hydrolase, is up-regulated at pH 4.5, but only in the absence of urea. The high level of up-regulation of this gene is seen because of the low level of expression of the stringency response component at neutral pH. A stringency response may also be part of the acid adaptation of H. pylori.
(v) Outer envelope genes.
Of the eight outer envelope genes that show up-regulation, six are in fact outer membrane proteins and two, cutE and murC, are involved in outer membrane biosynthesis. Perhaps membrane composition determines the permeability of the outer membrane. Since some outer membrane components are down-regulated, it appears that outer membrane synthesis is selectively affected by exposure of the organism to acidity.
Many of the genes described above retain significant up-regulation in the presence of urea at pH 4.5. These include the various pH-homeostatic genes, with the exception of ureI and ureE; the motility genes flaA, flaB, and HP1192; cag19 and the regulatory genes HP0165 (histidine kinase) and tenA; and the gene encoding DniR protein and all the outer membrane genes with the exception of omp19. It seems likely that changes of expression of these genes must occur during gastric colonization.
Previously, genes encoding the heat shock protein Hsp70 and cell wall composition, as well as cagA (23, 25, 31), have been observed to be up-regulated in acid, and random mutagenesis showed that at least 10 genes were essential for growth at pH 4.8 (6). Gamma-glutamyl transpeptidase has been shown to be essential for colonization (13). The regulatory protein ferric uptake regulator gene (fur) is involved in acid resistance, since its deletion impaired growth in acid (7). The genes for pH-homeostatic enzymes, such as arginase (33) and amidase (50), found to be up-regulated in the gene array, increase acid survival in vitro. Since the first of these produces intracellular urea and the second produces intracellular NH3, they possibly have a role in increasing buffering by the organism in an acidic environment. It has also been shown that there is increased assembly of active urease from apoenzyme during incubation at pH 5.5, which also increases survival after exposure to pH 2.5 (47). This is consistent with increased activity of the assembly complex, which may be due in part to up-regulation of the genes involved in urease assembly (ureE, -F, -G, and -H), since these genes, in contrast to ureA and ureB, are not constitutively expressed at moderate acidity and have much lower expression levels. Since assembly is rate limiting for total urease activity, regulation of the promoter (ureIp) in front of ureI would be beneficial to the organism in terms of increased synthesis of ureI, -E, -F, -G, and -H, resulting in increased and rapid assembly of the urease apoenzyme and enhanced acid-activated urea entry. It has been suggested that acid inhibits degradation of the mRNA translated after ureB rather than up-regulation of gene expression (1), but both up-regulation and stabilization may occur.
The contributions of the above-mentioned genes to acid resistance were discovered individually. Differential display of H. pylori grown for eight passages at pH 5.5 and 7.4 showed elevation of eight genes, including ureB, atoE, and flaA (16). Microarray methods, used to detect differences between strains of H. pylori (44), have been applied to studies of the effects of pH. Such a study has been performed using radioactive hybridization to a microarray with cDNA prepared from bacteria incubated at pH 4.0 or 7.0 in buffered medium without urea. About eight genes were identified as up-regulated and three were identified as down-regulated under these conditions, using PCR amplification of the hybridization spots. Among these was cagA, as previously found by direct measurement (25), as well as the genes encoding an enzyme involved in lipopoloysaccharide biosynthesis (HP1052) and a component of the membrane secretory apparatus (secF) (2). A comparison of gene expression after incubation for 48 h at pH 7.2 and 5.5 showed 53 genes highly expressed and 445 stably expressed under both conditions, although 952 genes were undetected by this biotin-labeling method. Eighty genes were reported as >5-fold up-regulated. These include the genes for subunits of the F1F0 ATPase, an Na+-dependent transporter, amidase, a flagellin B homolog, flagellar biosynthesis protein, thioredoxin reductase, methyl-accepting chemotaxis protein, and arginase, as well as omp11 (3). The duration of these experiments could have resulted in transition from log- to stationary-phase growth, preventing conclusions as to the roles of the genes in acid adaptation.
A previous study using microarrays identified other genes up-regulated twofold or more following exposure for various times to pH 5.0, but these arrays were performed after exposure to permeant buffer and only in the absence of urea. Several consistencies with our results were observed (34). For example, trhB (HP1050; encoding homoserine kinase), gdhA (HP0380; encoding glutamate dehydrogenase), hemL (HP0306; encoding glutamate semialdehyde amino mutase), and glnH (HP 1172; encoding glutamine ABC transporter) as genes of amino acid metabolism; the gene for a DNA polymerase-like transcript (HP1022); the genes ureI (HP0071) and ureF (HP0069) and the gene for amidase (HP0294) as pH-homeostatic genes; flgB (HP1559) and flaB (HP0155) as motility genes; the gene for a transcriptional regulator (HP1021); HP0175, an ABC transporter gene; frpB (HP0876; encoding an iron-regulated outer membrane protein); and various hypothetical genes (HP0367, HP0118, HP1327, HP1076, HP1022, HP1440, and HP1457) were also among these found to be increased. Longer exposure to pH 5.0 also resulted in the up-regulation of other urease cluster genes, such as ureA, -B, -G, and -H (HP0073, HP0072, HP0068, and HP0067). Hence, motility genes; amino acid-metabolic and ammonia-generating genes, such as those in the urease gene cluster; and the gene for amidase have been shown to be up-regulated at acidic pH in both types of experiments.
Of the genes also found to be up-regulated with exposure to pH 4.0 in permeant buffer, only flaA and HP0636 overlapped with the genes identified here (32). The method used in these experiments, exposure to permeant buffer at the extreme of survival in the absence of urea, would result in rapid cytoplasmic acidification and thus inhibition of transcription. Presumably, the genes that show up-regulation are those whose transcription or breakdown survives the acidic cytoplasm.
There was some degree of overlap between the genes essential for survival in the gerbil stomach and those with exposure to pH 4.0. These were discovered by signature tag mutagenesis. flaA (HP0601), fliD (HP0725), fliF (HP0351), and fliS (HP0753), which are motility genes; cheV (HP0393), a chemotaxis protein gene; trbB (HP1421), a virB11 homolog; dppF (HP0302), a dipeptide transporter gene; and ureI (HP0071), the urea channel, were all found to be essential, as was surE (HP0930), encoding a stationary-phase survival protein. However, the genes ureE, ureF, and ureG were not identified as essential by this method, although they are clearly required for the generation of active urease. Not all acid-activated genes will be essential, and conversely, not all of the essential genes revealed by this method will be acid activated. The exposure to the gerbil's stomach acidity may result in both log- and stationary-phase transformation, accounting for the detection of surE as essential for colonization (26). In terms of acid adaptation under conditions of log-phase growth in vitro as described here, surE was found to be down-regulated (Table 5).
Down-regulated genes.
Fewer genes are down-regulated than up-regulated, and again, only a few will be discussed here. The response to acidity is therefore different from a response in which cell division is inhibited as the organism is transiting from log to stationary phase or from a stringency response where cellular processes are restricted.
(i) Motility genes.
Four flagellum-associated genes are down-regulated >2-fold at pH 4.5, and of these, none are up-regulated by this factor in the presence of urea. How these genes interface with the motility and chemotaxis genes that are up-regulated is unknown, but clearly the organism is adapting its motility system to deal with medium acidity.
(ii) Outer envelope genes.
There are 10 outer envelope genes that are down-regulated. Four are actual membrane proteins, and the rest are implicated in cell wall biogenesis. Again, how this down-regulated response integrates with the up-regulated response to change the properties of the outer membrane for a more effective acid-adaptive response remains to be determined. Perhaps there are pH-adaptive changes in the permeability of the outer membrane. Two of the genes, omp27 and frpB, are down-regulated at pH 6.2 and below independently of the presence of urea.
(iii) Miscellaneous down-regulated genes.
Among the genes observed to be down-regulated were two encoding components of the F1F0 ATP synthase, atpB and atpA. This is somewhat surprising, since this system is the first to respond to acidity with outward proton pumping in other neutralophiles (9). HP1021, previously identified as essential and as a response regulator, was down-regulated more than twofold in the absence or presence of urea, as was the gene for a newly described pathogenesis factor, iceA. The Na+ and Cl− transporter was also down-regulated with this pattern.
Of the genes down-regulated following exposure to pH 5.0 in permeant buffer and in the absence of urea, omp27 (HP1177), ptuA (HP0056), and frpB2 (HP1512; encoding another iron-regulated outer membrane protein) were also down-regulated when exposed to impermeant buffer at acidic pH for 30 min. However, several of the genes down-regulated by exposure to pH 5.0 (34) were seen to be up-regulated under the assay conditions described here. For example, the gene for carbonic anhydrase (HP1186), ansB (HP0721; encoding asparaginase), and acoE (HP1045; encoding acetyl-coenzyme A [CoA] synthetase), as well as genes of unknown function (HP0310, HP0387, HP0485, and HP1193), were up-regulated in our experiments. The cytoplasmic pH is significantly lower in permeant buffers that in impermeant buffers, and perhaps this accounts for some of the differences observed.
The different methods used to identify acid-regulated genes in H. pylori have yielded some similar and some dissimilar results. This may be due to the exact conditions under which the analyses were performed. For example, long-duration experiments may result in transition from log to stationary phase. Exposure to acidity in the presence of permeant buffers and in the absence of urea may result in larger changes in cytoplasmic pH than when impermeant buffers are used, and certainly, when urea is present the pH changes are blunted. The design of the present studies permits analysis of progressive adaptation of gene expression with increasing acidity and also determines the contribution of urea to acid adaptation under these conditions. The conditions used here may be a better representation of the state of the gastric environment of the organism adhering to the gastric surface.
The transcriptional response of H. pylori to mild acidity is pleiotropic, involving many functional classes of genes, and most of the genes showing a response to acidification of the medium are distinct from the usual stress response genes. The difference between these responses and a stress response, such as that to 42°C, as well as the specialty of the niche occupied by this organism, suggests that the responses described are part of a generalized acid-adaptive response enabling colonization of the acidic environment of the mammalian stomach. Evidently, not all the genes up-regulated are dedicated only to pH adaptation, but those that remain in the presence of urea presumably do adapt the organism to its natural habitat. A systematic investigation of these genes as they relate to acid survival in vitro and in vivo with gastric infection should elucidate target genes specific for H. pylori other than ureI or urease activity.
Acknowledgments
This work was supported in part by USVA and NIH grants DK46917, 53462, 41301, and 17294.
Editor: J. N. Weiser
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 3.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athmann, C., N. Zeng, T. Kang, E. A. Marcus, D. R. Scott, M. Rektorschek, A. Buhmann, K. Melchers, and G. Sachs. 2000. Local pH elevation mediated by the intrabacterial urease of Helicobacter pylori cocultured with gastric cells. J. Clin. Investig. 106:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma, J. J., A. L. M. Lie, I. C. Nootenboom, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J., B. Waidner, A. H. Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser, M. J. 1996. The bacteria behind ulcers. Sci. Am. 274:104-107. [DOI] [PubMed] [Google Scholar]
- 9.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bury-Mone, S., S. Skouloubris, A. Labigne, and H. De Reuse. 2001. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol. Microbiol. 42:1021-1034. [DOI] [PubMed] [Google Scholar]
- 11.Cesareo, S. D., and S. R. Langton. 1992. Kinetic properties of Helicobacter pylori urease compared with jack bean urease. FEMS Microbiol. Lett. 78:15-21. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y. Y., and J. E. Cronan, Jr. 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33:249-259. [DOI] [PubMed] [Google Scholar]
- 13.Chevalier, C., J. M. Thiberge, R. L. Ferrero, and A. Labigne. 1999. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 31:1359-1372. [DOI] [PubMed] [Google Scholar]
- 14.Coskun, T., S. Chu, and M. H. Montrose. 2001. Intragastric pH regulates conversion from net acid to net alkaline secretion by the rat stomach. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G870-G877. [DOI] [PubMed]
- 15.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, Q., D. Hyde, C. Herra, C. Kean, P. Murphy, C. A. O'Morain, and M. Buckley. 2001. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol. Lett. 196:245-249. [DOI] [PubMed] [Google Scholar]
- 17.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 19.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrero, R. L., and A. Labigne. 1993. Cloning, expression and sequencing of Helicobacter felis urease genes. Mol. Microbiol. 9:323-333. [DOI] [PubMed] [Google Scholar]
- 21.Foster, J. W., and M. P. Spector. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49:145-174. [DOI] [PubMed] [Google Scholar]
- 22.Gueneau, P., and S. Loiseaux-De Goer. 2002. Helicobacter: molecular phylogeny and the origin of gastric colonization in the genus. Infect. Genet. Evol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 23.Huesca, M., A. Goodwin, A. Bhagwansingh, P. Hoffman, and C. A. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 25.Karita, M., and M. J. Blaser. 1998. Acid-tolerance response in Helicobacter pylori and differences between cagA+ and cagA− strains. J. Infect. Dis. 178:213-219. [DOI] [PubMed] [Google Scholar]
- 26.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroll, R. G., and I. R. Booth. 1983. The relationship between intracellular pH, the pH gradient and potassium transport in Escherichia coli. Biochem. J. 216:709-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin, C. J., R. G. P. Watson, J. M. Sloan, M. Stevenson, J. E. Ardill, and D. Buchanan. 2000. Distribution of atrophy in Helicobacter pylori-infected subjects taking proton pump inhibitors. Scand. J. Gastroenterol. 35:578-582. [DOI] [PubMed] [Google Scholar]
- 29.Lee, A., M. F. Dixon, S. J. Danon, E. Kuipers, F. Megraud, H. Larsson, and B. Mellgard. 1995. Local acid production and Helicobacter pylori: a unifying hypothesis of gastroduodenal disease. Eur. J. Gastroenterol. Hepatol. 7:461-465. [PubMed] [Google Scholar]
- 30.Logan, R. P., M. M. Walker, J. J. Misiewicz, P. A. Gummett, Q. N. Karim, and J. H. Baron. 1995. Changes in the intragastric distribution of Helicobacter pylori during treatment with omeprazole. Gut 36:12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 32.McGowan, C. C., A. S. Necheva, M. H. Forsyth, T. L. Cover, and M. J. Blaser. 2003. Promoter analysis of Helicobacter pylori genes with enhanced expression at low pH. Mol. Microbiol. 48:1225-1239. [DOI] [PubMed] [Google Scholar]
- 33.Mendz, G. L., E. M. Holmes, and R. L. Ferrero. 1998. In situ characterization of Helicobacter pylori arginase. Biochim. Biophys. Acta 1388:465-477. [DOI] [PubMed] [Google Scholar]
- 34.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 36.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mollenhauer-Rektorschek, M., G. Hanauer, G. Sachs, and K. Melchers. 2002. Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res. Microbiol. 153:659-666. [DOI] [PubMed] [Google Scholar]
- 38.Moncrief, M. B., and R. P. Hausinger. 1996. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J. Bacteriol. 178:5417-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 40.Park, I. S., and R. P. Hausinger. 1995. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 177:1947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsonnet, J. 1993. Helicobacter pylori and gastric cancer. Gastroenterol. Clin. N. Am. 22:89-104. [PubMed] [Google Scholar]
- 42.Rektorschek, M., A. Buhmann, D. Weeks, D. Schwan, K. W. Bensch, S. Eskandari, D. Scott, G. Sachs, and K. Melchers. 2000. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol. Microbiol. 36:141-152. [DOI] [PubMed] [Google Scholar]
- 43.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 44.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schade, C., G. Flemstrom, and L. Holm. 1994. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology 107:180-188. [DOI] [PubMed] [Google Scholar]
- 46.Scott, D. R., E. A. Marcus, D. L. Weeks, A. Lee, K. Melchers, and G. Sachs. 2000. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect. Immun. 68:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 48.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 49.Sipponen, P., and H. Hyvarinen. 1993. Role of Helicobacter pylori in the pathogenesis of gastritis, peptic ulcer and gastric cancer. Scand. J. Gastroenterol. Suppl. 196:3-6. [DOI] [PubMed] [Google Scholar]
- 50.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 51.Skouloubris, S., J. M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stim, K. P., and G. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 54.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 55.Weeks, D. L., and G. Sachs. 2001. Sites of pH regulation of the urea channel of Helicobacter pylori. Mol. Microbiol. 40:1249-1259. [DOI] [PubMed] [Google Scholar]
- 56.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zilberstein, D., V. Agmon, S. Schuldiner, and E. Padan. 1984. Escherichia coli intracellular pH, membrane potential, and cell growth. J. Bacteriol. 158:246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]