Abstract
To investigate the molecular basis of antigenic mimicry by peptides, we studied a panel of closely related mAbs directed against the cell-wall polysaccharide of group A Streptococcus. These antibodies have restricted V-gene usage, indicating a shared mechanism of binding to a single epitope. Epitope mapping studies using synthetic fragments of the cell-wall polysaccharide supported this conclusion. All of the mAbs isolated crossreactive peptides from a panel of phage-displayed libraries, and competition studies indicated that many of the peptides bind at or near the carbohydrate binding site. Surprisingly, the peptides isolated by each mAb fell into distinct consensus-sequence groups that discriminated between the mAbs, and in general, the peptides bound only to the mAbs used for their isolation. Similar results were obtained with polyclonal antibodies directed against synthetic oligosaccharide fragments of the streptococcal cell-wall polysaccharide. Thus, the peptides appear to be specific for their isolating antibodies and are not recognized by the same mechanism as their carbohydrate counterparts.
Keywords: carbohydrate mimicry, synthetic oligosaccharide mapping, phage-displayed peptide libraries, group A Streptococcus
Carbohydrates (CHOs) have proven to be valuable tools in demonstrating immunologic mimicry. Anti-idiotypic antibodies (Abs) directed against the V domains of anti-CHO Abs can, in some instances, elicit CHO-binding Ab responses when used themselves as immunogens (e.g., refs. 1–4). This has been attributed to chemical similarity (known as the “internal image”) between an anti-idiotypic Ab and the corresponding CHO antigen (1). Likewise, crossreactive peptides have been identified for several anti-CHO mAbs (4–7). In one case, the peptide was shown to elicit Abs having the same idiotype as the cognate, anti-CHO mAb (5), and in another to elicit a CHO-binding response (4).
The work described here addresses the molecular basis of crossreactivity between CHO and protein antigens with Abs. Our goal was to determine if the crossreactive peptides recognized by anti-CHO Abs would bind by the same mechanism as the corresponding epitope on the CHO target; if so, the basis of crossreactivity would be structural mimicry. We assembled a panel of five closely related mAbs against the cell-wall polysaccharide (CWPS) of group A Streptococcus (GAS) and showed by oligosaccharide mapping studies that they indeed bind a similar, if not identical, epitope. Each of four anti-GAS CWPS mAbs and three polyclonal Abs (PCAbs) against synthetic oligosaccharide fragments of the GAS CWPS isolated peptides bearing unique, chemically distinct consensus sequences. Moreover, representative peptides from each consensus group were functionally specific, because they usually bound only to their isolating Ab. Thus, these Abs were more restricted in their peptide reactivity than in their CHO recognition. We conclude that the predominating basis of peptide recognition by anti-CHO Abs differs between Abs, with true CHO mimics being relatively rare. We propose that the antigenic mimicry observed for CHO-crossreactive peptides is determined mainly by the binding sites of anti-CHO Abs (including small differences between them), rather than by chemical similarity to the corresponding CHO epitope.
MATERIALS AND METHODS
The peptide NH2ADGADRPVPYGACGOrn(biotin)–NH2 (DRPVPY-peptide) was synthesized and HPLC-purified by the Alberta Peptide Institute (Edmonton, Alberta). The anti-GAS CWPS mAbs, SA-3 (8), Strep 9, (a gift from J. B. Pitner, Becton Dickinson Research Center, Durham, NC), HGAC 39, HGAC 47, and HGAC 101 (9), were raised against a heat-killed, pepsin-treated GAS (dGAS) vaccine (10). SA-3 is an IgM, and the others are IgG3; all use κ light chains. The dGAS used in this work was provided by J. B. Pitner and D. R. Bundle (University of Alberta, Edmonton). The production and characterization of the PCAbs were described previously (8). mAbs SE155.4 and SYA/J6 (provided by D. R. Bundle) were raised against Salmonella serogroup B and Shigella flexneri Y, respectively. The amino acid sequences are published for mAbs SE155.4 (11), HGAC 39, HGAC 47, and HGAC 101 (9); those of Strep 9 (J. B. Pitner, W. F. Beyer, S. L. Harris, C. Nycz, T. Venetta, M. J. Mitchell, and B.M.P.), SA-3 (D. C. Watson, M. Yaguchi, B. Sinnott, D. R. Bundle, and N. M. Young), and SYA/J6 (D. C. Watson, D. Bilous, S.-J. Deng, M. A. J. Gidney, D. R. Bundle, and N. M. Young) are unpublished. The syntheses of the GAS oligosaccharides and glycoconjugates have been published (8, 12, 13). The lipopolysaccharides and oligosaccharides of Salmonella serogroup B and Shigella flexneri Y were gifts from D. R. Bundle.
Eleven different peptide libraries, displayed as fusions to coat protein VIII of the phage vector f88.4, and their screening with mAbs (HGAC 39, HGAC 47, HGAC 101, Strep 9, SE155.4, and SYA/J6) have been described, as well as the isolation and analysis of phage clones (14). Briefly, 1011 to 1012 virions from each library were affinity-selected on biotinylated mAbs that had been immobilized in avidin- or streptavidin-coated microwells (14). Enrichment for Ab-binding phage was assessed by titering after each round of panning. Enriched, amplified phage pools were tested for binding by ELISA after the third and fourth rounds of screening. Ten individual clones were isolated from the two or three pools of enriched phage displaying the highest enrichment and/or ELISA signal. The clones were analyzed by ELISA and their displayed peptide sequences were determined. Hexamer (15) and 15-mer peptide libraries (16), displayed as fusions to coat protein III of the phage vector fUSE5, were screened by SA-3 and the PCAbs as described (14); the 15-mer library (16) was provided by H. Saya (University of Kumamoto, School of Medicine, Japan). Phage pools and clones from these latter screens were analyzed by ELISA (15) and DNA sequencing (17).
ELISAs.
All washes were performed with Tris-buffered saline (TBS) and 0.1% Tween 20. Except where noted, wells were blocked with 200 μl of blotto (5% milk powder in TBS) for 2 h at 4°C, IgG Abs were used at 100 nM in 35 μl of blotto, and the IgM (SA-3) was used at 20 nM in 35 μl of blotto; incubation times were 4 h at 4°C. Biotinylated mAbs were detected with avidin·horseradish-peroxidase complexes (14), and nonbiotinylated mAbs and PCAbs were detected with secondary Abs conjugated to horseradish peroxidase (Pierce). Absorbances are reported as (A405 − A490) × 1000 (14).
Phage ELISAs were performed as described previously (14), using sample phage or controls without peptide inserts [fd-tet (15) or f88.4 (14)]. Briefly, microwells were coated overnight at 4°C with 35 μl of TBS containing 1 μg of anti-phage Ab. After blocking and washing three times, 1010 virions in 35 μl of TBS were added to the wells and incubated for 2 h at 4°C (14). Biotinylated Abs were added after three washes. For the peptide ELISA, microwells were coated overnight with 1 μg of streptavidin in 35 μl of TBS and blocked. The wells then were washed and incubated for 30 min at room temperature in 40 μl of 4 nM DRPVPY-peptide diluted in TBS, or in TBS only. Remaining streptavidin sites were blocked by the addition of 1.5 mM biotin in blotto and incubation for 1 h at 37°C, then biotinylated mAb was added after three washes. For competition ELISAs, the immobilized antigens were either phage, a synthetic glycoconjugate, or the native CHO antigen (dGAS or a lipopolysaccharide) that had been immobilized by adsorption, or the DRPVPY-peptide immobilized in streptavidin-coated microwells. Competition was established by the addition of equal volumes of an Ab and an inhibitor to antigen-coated or control microwells.
When synthetic oligosaccharides were used as inhibitors, microwells were coated overnight at 4°C with 35 μl of 0.1 M bicarbonate buffer, pH 9.0, containing 4 × 106 dGAS (for mAbs HGAC 39 and Strep 9) or 4 × 107 dGAS (for mAbs SA-3, HGAC 47, and HGAC 101), then blocked with BSA. After washing the blocked wells three times, increasing concentrations of the oligosaccharide inhibitor were added to wells along with a constant concentration of mAb. MAb SA-3 and biotinylated HGAC mAbs were used at 10 nM; Strep 9 (not biotinylated) was used at 20 nM. Bound mAb was detected after overnight incubation at room temperature, followed by six washes.
Microcalorimetry.
A solution of SA-3 was titrated with either the DRPVPY-peptide or a hexasaccharide, Hexa 2 (structure 9 in Fig. 1), using a Microcal Omega titration microcalorimeter (18) and methods described previously (19). The concentration of SA-3 was such that the product of the concentration of binding sites and the binding constant was in the range of 1 to 1000. The ligand concentration in the syringe was such that the final ligand concentration was at least 10 × Kd. For the SA-3–Hexa 2 interaction, a cell with a volume of 1.3678 ml was loaded with 68.4 μM SA-3 dissolved in 10 mM Na2PO4/127 mM NaCl, pH 7.0, then 1.25 mM Hexa 2 in PBS was added in 16 15-μl aliquots at 37°C. For the SA-3–DRPVPY-peptide interaction, 20.5 μM SA-3 was injected with 10-μl aliquots of 0.168 mM DRPVPY-peptide. All injections were 5 s with 3-min intervals between injections; the data were analyzed as described (19).
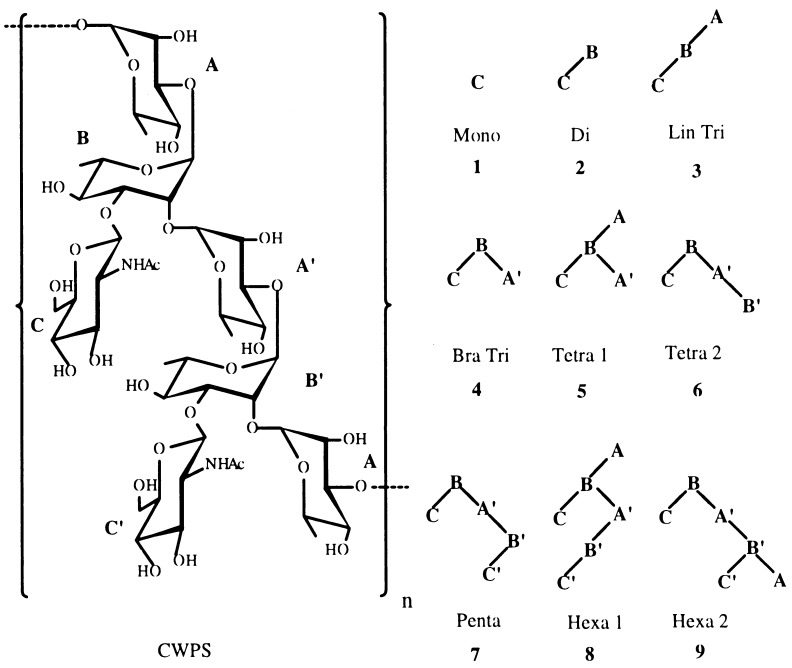
Figure 1.
The CWPS of GAS and synthetic oligosaccharides corresponding to portions (structures 1–9) of the CWPS. The CWPS chemical structure is: AB′A′B -α-l-Rhap-(1→2)-α-l-Rhap-(1→3)-α-l-Rhap-(1→2)-α-l-Rhap-(1→3) 33 ↑↑ 11 β-d-GlcpNAcβ-d-GlcpNAc C′C
RESULTS
The Five Anti-dGAS mAbs All Have Similar CHO Fine Specificities, Sequences, and Restricted V-gene Usage.
The minimal epitope on the CWPS of GAS recognized by the anti-dGAS mAbs, SA-3, HGAC 39, HGAC 47, HGAC 101, and Strep 9, was deduced by competition ELISAs using a panel of synthetic oligosaccharides, which comprise portions of the CWPS (Fig. 1). The data in Table 1 show that all of the oligosaccharides inhibited mAb binding to dGAS. The mAbs could be separated into two groups: SA-3, Strep 9, and HGAC 39, which bound the oligosaccharides relatively tightly, and the weaker-binding mAbs, HGAC 47 and HGAC 101. All of the mAbs were best inhibited by structures containing a branch point (CBA′), which has been shown to be a conformationally restricted feature of GAS oligosaccharides (20). The branched trisaccharide (Bra Tri, 4), which bears this minimal epitope, was a significantly better inhibitor for all of the mAbs than the linear trisaccharide (Lin Tri, 3). An extended CHO surface also appears to be important for mAb recognition, because all the mAbs were best inhibited by the pentasaccharide (Penta, 7).
Table 1.
Affinity constants of synthetic oligosaccharides and one synthetic peptide for the anti-dGAS mAbs
| mAb | Affinity constant for hapten, 104 M−1
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mono | Di | Lin Tri | Bra Tri | Tetra 1 | Tetra 2 | Penta | Hexa 1 | Hexa 2 | CWPS* | DRPVPY-peptide | |
| SA-3 | 0.1 | 2.4 | 0.6 | 5.6 | 7.0 | 3.8 | 3.6 | 4.0 | 3.3 (5.71 ± 0.49)† | 3.6 | (160 ± 0.43)† |
| Strep 9 | 0.2 | 0.3 | <0.1 | 0.7 | 0.6 | 2.0 | 2.7 | 1.6 | 3.7 (3.09 ± 0.67)† | 10.4 | ND |
| HGAC 39 | ND | 0.4 | 0.1 | 0.9 | ND | 2.1 | 2.3 | ND | 1.7 | ND | ND |
| HGAC 47 | ND | 0.1 | <0.1 | 0.1 | ND | 0.1 | 1.3 | ND | 0.1 | ND | ND |
| HGAC 101 | ND | 0.2 | <0.1 | 0.2 | ND | <0.1 | 0.3 | ND | <0.1 | ND | ND |
Affinity constants are the inverse of the IC50 values (11) determined by competition ELISA against immobilized dGAS. The error in the values determined by ELISA is approximately 10%. ND, not determined.
The CWPS isolated and purified from GAS (12).
Data in parentheses were determined by titration microcalorimetry.
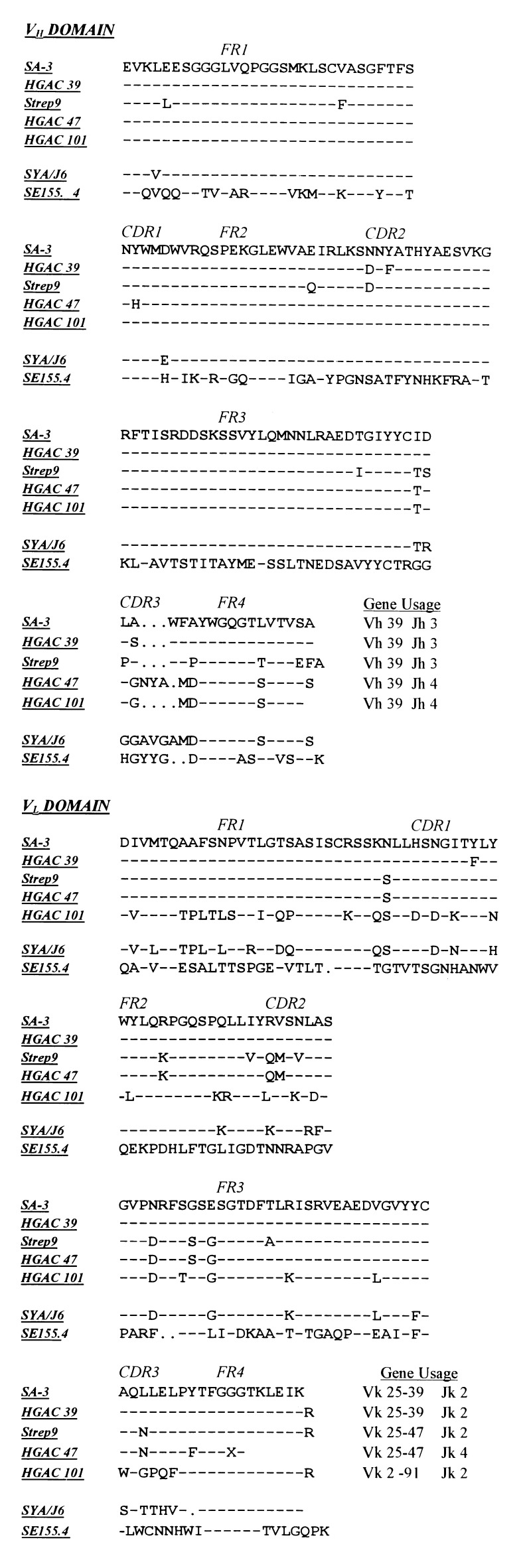
As for other anti-CHO responses (21, 22), the VH-gene usage for the anti-dGAS mAbs (Fig. 2) is highly restricted; identical germ-line VH genes are used for all five mAbs. The tighter-binding mAbs, SA-3, Strep 9, and HGAC 39, all use the same VH and JH germ-line genes, and they also have the same number of residues at the D–J junctions, as compared with the weaker-binding mAbs HGAC 47 and HGAC 101. Together, the oligosaccharide mapping studies and the VH and JH gene usage suggest that the anti-dGAS mAbs recognize similar, if not identical, epitopes.
Figure 2.
The amino acid sequences and gene usage of the anti-dGAS mAbs, SE155.4 and SYA/J6.
SA-3 Isolates a CHO-Crossreactive Peptide That Is mAb Specific.
The data in Table 2 show that the phage clones isolated from a 6-mer library by SA-3 bear peptides sharing the consensus sequence RPX2Y. In competition ELISA, the synthetic DRPVPY-peptide inhibited SA-3 from binding any of the CHO antigens, including dGAS, suggesting that the DRPVPY-peptide binds at or near the CHO-binding site (data not shown). The thermodynamics of SA-3 binding to the DRPVPY-peptide and to Hexa 2 (9) were investigated further by titration microcalorimetry (data not shown). As shown in Table 1, the Ka of SA-3 for the DRPVPY-peptide was 30 times greater than for Hexa 2 (9), an oligosaccharide whose inhibitory potency was similar to that of purified CWPS. Using 1 M peptide or oligosaccharide as the reference state, the binding energies for the DRPVPY-peptide and Hexa 2 (9) were −8.46 ± 0.35 kcal/mol and −6.49 ± 0.05 kcal/mol, respectively. The differences in affinity were due to the calculated entropies of binding, because these haptens had similar enthalpies of binding [−6.30 ± 0.33 kcal/mol for the DRPVPY-peptide and −5.62 ± 0.38 kcal/mol for Hexa 2 (9)]. Identical calculated stoichiometries of mAb binding for the DRPVPY-peptide (0.376 ± 0.015) and Hexa 2 (9) (0.398 ± 0.023) indicate that the peptide binds in the CHO-binding site. The data in Table 3 demonstrate that peptide binding is specific to SA-3.
Table 2.
The phage-displayed peptide sequences isolated by the anti-dGAS mAbs, SE155.4 and SYAJ/6
| mAb | Library* | Peptide sequence† | ΔA × 1000 |
|---|---|---|---|
| SA-3 | X6 | DRPVPY | 765 |
| KSPTPY | 641 | ||
| ARPLWY | 583 | ||
| VRPQVP | 524 | ||
| fd-tet‡ | — | 169 | |
| Strep 9 | XCX8CX | MCPPLYSPSACA | 957 |
| ECNFLYPGFTCA | 202 | ||
| X8CX8 | YPYCGHALCPGLYADAS | 1020 | |
| VILPYDNNCALCLNLYP | 644 | ||
| VIDAPTPNCAWPNGRRG | 256 | ||
| MPPAGTGTCFLYALSCS | 153 | ||
| ADLSPTPYCQPSTMHTN | 144 | ||
| NEYINQDHCLLYAMLCP | 38 | ||
| X15CX | EIAPQGPSKCLLYAYCQ | 13 | |
| f88‡ | — | 14 | |
| HGAC 39 | X15 | ADAAPSPTPYLPRLS | 643 |
| ATYRPVPAEFARKHL | 425 | ||
| TITATDSPTPWPFER | 244 | ||
| XCX8CX | MCRPSPYNPPCT | 112 | |
| f88‡ | — | 72 | |
| HGAC 47 | XCX8CX | MCRPGIPTHHCA | 174 |
| HCSPGQRPGTCQ | 165 | ||
| DCGNMLHAEVCR | 149 | ||
| DCRPGVPLLSCP | 115 | ||
| f88‡ | — | 87 | |
| HGAC 101 | XCX6CX | SCISAACFCI | 141 |
| X6 | KQLMAP | 138 | |
| f88‡ | — | 29 | |
| SE155.4 | X6 | NYPMDH | 141 |
| MYPMSH | 104 | ||
| YPMGHL | 27 | ||
| IYPMPA | 13 | ||
| QQYPMG | 12 | ||
| QSTYPM | 10 | ||
| X15 | EPYPMSEANYVRPMP | 267 | |
| YPMPASSDNAQWLLK | 15 | ||
| DGTNAYPMNEDISVS | 15 | ||
| HSTRNYSYLGSPYPM | 13 | ||
| NYPMSGARIEPLLHA | 13 | ||
| YAATEPRYMIPYPMP | 13 | ||
| YPMGETCQRIRSCVW | 11 | ||
| XCX8CX | VCPAPYPAGTCA | 11 | |
| f88‡ | — | 14 | |
| SYAJ/6 | X4CX4CX4 | YTTQCGYGGCMNFE | 938 |
| MGVICMNMECDRNM | 799 | ||
| LHEYCNMETCPYNH | 585 | ||
| QYPQCHNMDCKSIT | 472 | ||
| PTHVCYNMECQGGD | 275 | ||
| TPTNCYNMTCQNQP | 187 | ||
| X6 | MDWNMH | 993 | |
| f88‡ | — | 66 |
The listed peptides displayed on cpIII are preceded by the N-terminal sequence ADGA. In the sequence of the peptide library, X represents “randomized” amino acids and C stands for fixed cysteines.
Sequences in boldface were chosen for further study; see Table 3. Consensus sequences are underlined.
Wild-type phage vector without a random peptide insert.
Table 3.
Crossreactivity of clones isolated by an anti-CHO mAb with other anti-CHO mAbs
| Immobilized antigen | ΔA × 1000 with mAb
|
||||||
|---|---|---|---|---|---|---|---|
| SA-3 | Strep 9 | HGAC 39 | HGAC 47 | HGAC 101 | SE155.4 | SYAJ/6 | |
| DRPVPY-peptide | 300 | 28 | 21 | 20 | 24 | ND | ND |
| Streptavidin* | 53 | 22 | 16 | 15 | 17 | ND | ND |
| MCPPLYSPSACA | 92 | 446 | 40 | 44 | 25 | 10 | 33 |
| YPYCGHALCPGLYADAS | 7 | 674 | 42 | 45 | 22 | 8 | 29 |
| ADAAPSPTPYLPRLS | 47 | 14 | 726 | 62 | 31 | 12 | 35 |
| MCRPGIPTHHCA | 6 | 13 | 44 | 148 | 29 | 11 | 33 |
| DCGNMRQAEVCR | 7 | 12 | 41 | 160 | 26 | 10 | 32 |
| SCISAACFCI | 6 | 12 | 80 | 120 | 51 | 12 | 36 |
| KQLMAP | 6 | 12 | 95 | 121 | 43 | 11 | 34 |
| NYPMDH | 6 | 12 | 58 | 78 | 37 | 112 | 36 |
| EPYPMSEANYVRPMP | 6 | 11 | 56 | 74 | 32 | 145 | 35 |
| MDWNMH | 6 | 16 | 57 | 69 | 34 | 13 | 689 |
| MGVICMNMECDRNM | 6 | 12 | 47 | 54 | 28 | 10 | 283 |
| YTTQCGYGGCMNFE | 8 | 14 | 55 | 55 | 32 | 12 | 483 |
| f88† | 7 | 11 | 48 | 55 | 26 | 10 | 34 |
Numbers in boldface indicate that a phage clone bearing the listed sequence was isolated by the corresponding mAb (e.g., SA-3 isolated the clone bearing the DRPVPY sequence). Underlined numbers indicate significant crossreactivity with a mAb not used to select the clone. ND, not determined.
Streptavidin-coated wells used as a negative control for the DRPVPY-peptide-coated wells.
Wild-type phage vector without a random peptide insert.
All of the Closely Related Anti-dGAS mAbs Isolate Peptides Having Restricted Specificity.
The remaining four anti-dGAS mAbs were used to screen 11 peptide libraries (14). The random peptide sequences in each library varied in length, and in the number and position of fixed cysteine residues within the randomized region (14). The data in Table 2 illustrate that each mAb isolated binding clones, and that Strep 9, HGAC 39, and SA-3 isolated tighter-binding clones than did HGAC 47 or HGAC 101, paralleling their reactivities with the oligosaccharides. Moreover, each of the anti-dGAS mAbs, other than HGAC 101, isolated unique consensus groups (underlined in Table 2); CX1–2LY and PTPXC for Strep 9, PXPX1–2P for HGAC 39, and CXPG or RPG for HGAC 47. Thus, the mAbs that bound the GAS CWPS epitope tightly isolated tight-binding peptides forming consensus sequences; whereas, the weak-binding mAbs bound peptides relatively weakly, and either isolated a weak consensus sequence or none at all. Two other anti-dGAS mAbs, SA-2 and SA-4 (8), also isolated binding peptides having unique consensus sequences (data not shown). Further examination of the sequences in Table 2 reveals that two common motifs, ADX1–3SPTPY and RPX1–2P, were identified by several of the mAbs. The functional relevance of these motifs is unclear, because peptides bearing them do not crossreact with each other.
The observations of distinct consensus groups and shared sequence motifs between groups led us to question the specificity of the peptides for each mAb. To test this, we determined the reactivity of each mAb with the best-binding phage pools isolated by each mAb (data not shown). In some cases, more than one mAb bound detectably to a given phage pool; however, none of the pools bound all of the mAbs, or even the three mAbs that bound the GAS CWPS tightly. To confirm these results, each mAb was tested for binding by ELISA to the best-binding clones from each pool; their peptide sequences are shown in bold in Table 2. The reactivity patterns of the clones paralleled those of the phage pools from which the clones were derived. The data in Table 3 show that each clone was bound best by the mAb that isolated it, with the exception of the clones isolated by HGAC 101. The best examples of peptides reacting with more than one mAb were for clones isolated by Strep 9 and HGAC 39; these clones also were bound by SA-3 (Table 3). Because the reactivity of any clone was limited to no more than two antibodies, we conclude that the reactivity patterns of the peptides are more restricted than those observed with the branched oligosaccharides.
We also tested a tight-binding synthetic oligosaccharide for its ability to compete with immobilized phage for binding to mAb (data not shown). A relatively high concentration of Penta (7, 1.1 mM) inhibited the binding of Strep 9 and HGAC 39 to their respective clones, indicating that peptide binding to these mAbs occurs at or near the CHO-binding site, whereas it did not inhibit the weaker binding of HGAC 47 or HGAC 101, suggesting that peptide binding occurs at a separate site. Because these latter mAbs appear to have a relatively lower affinity for CHOs than for peptides, it is also possible that inhibition may occur with a higher concentration of Penta (7).
PCAbs Directed Against Synthetic Oligosaccharide Fragments of the GAS CWPS Also Isolate Crossreactive Peptides.
Our study of peptide reactivity with closely related anti-CHO Abs was extended to PCAbs directed against BSA conjugates of the Lin Tri (3), Bra Tri (4), and Penta (7), respectively (8). The specificities of the PCAbs were similar to those of the anti-dGAS mAbs, in that the anti-Lin Tri and anti-Bra Tri PCAbs bound only their cognate trisaccharide and Penta (7), whereas anti-Penta PCAb bound all three haptens (8); moreover, dGAS was bound best by the anti-Penta PCAb (data not shown). Peptide library screening yielded several unique consensus groups for each PCAb (Table 4), reflecting the different specificities within each. The specificity of the phage pools, obtained after three rounds of panning, for their isolating PCAb was tested by ELISA. Each PCAb was specific for the phage pool it isolated (Table 5), even though each of the PCAbs reacted with at least two of the three glycoconjugates used for their production (8). Therefore, as with the anti-dGAS mAbs, each PCAb was more specific for a given set of peptides than for the oligosaccharide antigens.
Table 4.
The peptide sequences isolated by the PCAbs
| PCAb | Peptide sequences* |
|---|---|
| Anti-Lin Tri (3) | CVFHQDa YLFTQDb CVFHQDa |
| YLFTQDb GYMFTQ SKCNQP | |
| KCSIRQ | |
| LLACSY | |
| LCQTCA | |
| Anti-Bra Tri (4) | SIKWLEVSFWDW |
| LIKWLEFIFYPW | |
| YWKYES | |
| KFGDLF | |
| AVWGPAGPAFRPRWSc | |
| DWRFSFRPWGLDLSS | |
| AVWGPAGPAFRPRWSc | |
| QMWFPAGPAWSSSCL | |
| RDHLVFWTTSGPIFG | |
| RDWHGAPYEVAVRSR | |
| Anti-Penta (7) | KCCVSVYGYLYI |
| LCCEGSYSNLYL | |
| CCSRFLVNYSFY | |
| CCPTPCYRNLLF | |
| AVCCPCPSGSLPFFL | |
| LVFYDDdLFFAWY | |
| DLLWDHTRCLFFRGLSHCDVD | |
| FEFDYNeCDRQPPPVRCFRLVD | |
| VPVWLATFRWEFYPF | |
| WLLCVLVSDGFEFCAF | |
| FEFDYNe | |
| LVFYDDd | |
| WYWCYCIPLQLDDGC |
Random peptide sequence displayed on cpIII are preceded by the N-terminal sequence ADGA. Each superscript (a–e) marks a sequence that appears twice because it aligns into two different consensus groups.
Table 5.
Crossreactivity of pooled phage with PCAbs
| Immobilized phage | ΔA × 1000 with PCAb
|
||
|---|---|---|---|
| Anti-Lin Tri | Anti-Bra Tri | Anti-Penta | |
| X6 | 461 | 101 | 36 |
| X6 | 52 | 222 | 25 |
| X6 | 61 | 117 | 262 |
| X15 | 286 | 111 | 21 |
| X15 | 67 | 299 | 21 |
| X15 | 86 | 130 | 90 |
| fd-tet* | 80 | 158 | 37 |
X6 and C15 indicate that phage pools were derived from the 6-mer and 15-mer libraries, respectively. Numbers in boldface indicate that the listed phage pool was selected by the corresponding PCAb.
Wild-type phage clone without a random peptide insert.
Two mAbs Against Non-GAS CHOs Also Isolate Crossreactive Peptides.
We extended our investigation to two well characterized mAbs, SE155.4 and SYA/J6, which are specific for two different O-antigens. These “out-group” mAbs do not bind the GAS CWPS, even though the VH-gene usage of SYA/J6 is similar to that of the anti-dGAS mAbs (Fig. 2). A variety of physical methods, including epitope mapping (23–25) and x-ray crystallography (26–28), have been used to determine the features of the O-antigen of Salmonella serogroup B recognized by SE155.4 and the O-antigen of Shigella flexneri Y recognized by SYA/J6. As shown in Table 2, the peptide sequences isolated by SE155.4 share the very strong consensus sequence YPM, indicating strong selection by SE155.4 despite low ELISA signals, whereas SYA/J6 isolated tight-binding peptides that form the unique consensus sequence CXNM(E/D). Two or three of the best-binding clones were tested for reactivity with the other mAbs (Table 3). Each clone was specific for its isolating mAb; neither SE155.4 nor SYA/J6 bound clones isolated by the other mAbs in the panel. In competition ELISA, the SYA/J6-specific trisaccharide methyl 2-acetamido-2-deoxy-3-O-[(3-O-α-l-rhamnopyranosyl)-α-l-rhamnopyranosyl]-β-d-glucopyranoside at 1.8 mM (23) inhibited the binding of SYA/J6 to phage; however, no detectable inhibition was observed for SE155.4 with 2.0 mM methyl 3-O-(3,6-dideoxy-α-d-xylohexopyranosyl)-2-O-α-d-galactopyranosyl-α-d-mannopyrnoside (26), a trisaccharide specific for this mAb (data not shown). These results suggest that peptide binding occurs at or near the CHO-binding site of SYA/J6, but not of SE155.4. The requirement for disulfide bridges within the peptides shown in Table 3 containing two cysteines was determined in the presence and absence of 5 mM DTT (data not shown). In every case, except SYA/J6, disruption of the disulfide bridging significantly decreased binding to the phage-borne peptides, but not to immobilized CHO antigens.
DISCUSSION
Our results with a panel of closely related anti-dGAS mAbs suggest that the mechanism of peptide binding differs from that of CHO binding. Most or all of the anti-dGAS mAbs probably bind CHOs by a similar mechanism, because they recognized the minimal, branched-trisaccharide epitope and have similar VH-gene usage. In contrast, each mAb recognized a restricted, mostly nonoverlapping, subset of peptides. If peptide binding was solely due to mimicry of the GAS CWPS, the peptides also should have bound all of the anti-dGAS mAbs. Yet, crossreactive peptides forming chemically distinct consensus sequences were isolated for every anti-CHO mAb; moreover, the predominant reactivity of each peptide studied (besides those isolated by HGAC 101) was associated with the mAb that isolated it. The work with related PCAbs further supports this trend.
With regard to the definition of the CHO epitope recognized by each anti-dGAS MAb, our oligosaccharide mapping studies indicate that all of the anti-dGAS mAbs recognize an epitope presented by a minimal, branched-trisaccharide unit. The mAbs may recognize different subsites within the branched-trisaccharide epitope and/or different conformations of the epitope, which would cause the crucial interactions between mAb and CHO epitope to vary. Our recent study of the binding of the Bra Tri (4) to Strep 9 shows that this mAb selects a local minimum conformation that differs significantly from the global minimum conformation of the free trisaccharide (20). Thus, the other anti-dGAS mAbs in the panel may recognize the branched-trisaccharide epitope in alternative conformations and/or at other key subsites.
The restricted reactivity of the peptides with the anti-dGAS mAbs shows that the peptides discriminate between the mAbs far better than the branched-trisaccharide epitope. This discrimination occurs even if some of the shared residues present in different consensus sequences are acting as structural mimics of the CHO epitope; other residues must be responsible for discrimination. We conclude that the peptides bind the mAbs by different mechanisms than the branched-trisaccharide epitope. Alternatively, peptide binding may indeed be due to structural CHO mimicry; however, this would require the unlikely restriction that each mAb recognize the branched-trisaccharide epitope by a different mechanism (e.g., at different subsites).
Our work with nine anti-CHO mAbs and three PCAbs, when taken together with previous work (4–7), demonstrates that crossreactive peptides can be found for most, if not all, anti-CHO Abs. This may be explained, in part, by the Ab structures that bind them. In several instances, peptide- and CHO-reactive mAbs have been shown to possess groove-like binding sites (28–31). Work by Vargas-Madrazo et al. (32) suggests that the majority of anti-CHO Abs use a limited subset of V genes, indicating that this class of Abs uses a restricted structural repertoire. In contrast to the antibodies, at most, only two of the many lectins screened have been shown to bind peptides (33–35); this probably will not be the case for the CHO-specific enzymes (36, 37).
The relationship between antigenic mimicry and immunologic mimicry by peptides is unclear. Westerink et al. (4) demonstrated that a peptide designed from an anti-idiotypic Ab could elicit CHO-reactive Abs, whereas a peptide isolated by Valadon et al. (5) failed to elicit CHO-reactive Abs; yet, it elicited the correct idiotype (2H1). Previous work with anti-idiotypic Abs indicates that competition with the target CHO-antigen and high-affinity binding often are not sufficient for immunologic mimicry, and at times, may not be necessary (3). We suggest that the CHO-crossreactivity of Abs produced against CHO-mimic peptides (and anti-idiotypic mAbs) is determined by factors other than their structural similarity to the CHO antigen. These factors, being specific to a subset of CHO-binding Abs, will only elicit a specific subset of Abs from within an anti-CHO response.
Acknowledgments
This work was funded by grants from the U.S. Army (Grant DAA L03–92-G-0178), the President’s Office at Simon Fraser University, Terrapin Technologies, Inc., the Heart and Stroke Foundation of British Columbia and the Yukon, the Natural Sciences and Engineering Research Council of Canada, the National Institutes of Health (Grant GM48653), the Camile and Henry Dreyfus Foundation, and the Alfred P. Sloan Foundation. We gratefully acknowledge studentships from the Medical Research Council of Canada, the British Columbia Science Council, I.D. Biomedical Corporation, and the Natural Sciences and Engineering Research Council, and a scholarship from the British Columbia Health Research Foundation.
ABBREVIATIONS
- Ab
antibody
- PCAbs
polyclonal antibodies
- CHO
carbohydrate
- CWPS
cell-wall polysaccharide
- GAS
group A Streptococcus
- dGAS
heat-killed, pepsin-treated GAS
- TBS
Tris-buffered saline
References
- 1.Köhler H, Kaveri S, Kieber-Emmons T, Morrow W J W, Müller S, Raychaudhuri S. Methods Enzymol. 1989;178:3–35. doi: 10.1016/0076-6879(89)78003-4. [DOI] [PubMed] [Google Scholar]
- 2.Stein K E, Söderström T. J Exp Med. 1984;160:1001–1011. doi: 10.1084/jem.160.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monafo W J, Greenspan N S, Cebra-Thomas J A, Davie J M. J Immunol. 1987;139:2702–2707. [PubMed] [Google Scholar]
- 4.Westerink M A J, Giardina P C, Apicella M A, Kieber-Emmons T. Proc Natl Acad Sci USA. 1995;92:4021–4025. doi: 10.1073/pnas.92.9.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadon P, Nussbaum G, Boyd L F, Margulies D H, Scharff M D. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 6.Hoess R, Brinkmann U, Handel R, Pastan I. Gene. 1993;128:43–49. doi: 10.1016/0378-1119(93)90151-r. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi E, Folgori A, Wallace A, Nicotra M, Acali S, Phalipon A, Barbato G, Bazzo R, Cortese R, Felici F, Pessi A. J Mol Biol. 1995;247:154–160. doi: 10.1006/jmbi.1994.0129. [DOI] [PubMed] [Google Scholar]
- 8.Reimer K B, Gidney M A J, Bundle D R, Pinto B M. Carbohydr Res. 1992;232:131–142. doi: 10.1016/s0008-6215(00)91000-0. [DOI] [PubMed] [Google Scholar]
- 9.Nahm M H, Clevinger B L, Davie J M. J Immunol. 1982;129:1513–1518. [PubMed] [Google Scholar]
- 10.Krause R M. Adv Immunol. 1970;12:1–56. doi: 10.1016/s0065-2776(08)60167-4. [DOI] [PubMed] [Google Scholar]
- 11.Bundle D R, Eichler E, Gidney M A J, Meldal M, Ragauskas A, Sigurskjold B W, Sinnott B, Watson D C, Yaguchi M, Young N M. Biochemistry. 1994;33:5172–5182. doi: 10.1021/bi00183a022. [DOI] [PubMed] [Google Scholar]
- 12.Pinto B M. In: Carbohydrate Antigens, ACS Symp. series 519. Garegg P J, Lindberg A A, editors. Washington, DC: American Chemical Society; 1993. pp. 111–131. [Google Scholar]
- 13.Auzanneau F-I, Pinto B M. Bioorg Med Chem Lett. 1996;4:2003–2010. doi: 10.1016/s0968-0896(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 14.Bonnycastle L L C, Mehroke J S, Rashed M, Gong X, Scott J K. J Mol Biol. 1996;258:747–762. doi: 10.1006/jmbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- 15.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 16.Nishi T, Tsurui H, Saya H. Exp Med. 1993;11:1759–1764. [Google Scholar]
- 17.Haas S J, Smith G P. Biotechniques. 1993;15:422–431. [PubMed] [Google Scholar]
- 18.Wiseman T, Welliston S, Brandts J F, Lin L-N. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 19.Chervenak M C, Toone E J. Biochemistry. 1995;34:5685–5695. doi: 10.1021/bi00016a045. [DOI] [PubMed] [Google Scholar]
- 20.Weimar T, Harris S L, Pitner J B, Bock K, Pinto B M. Biochemistry. 1995;34:13672–13681. doi: 10.1021/bi00041a049. [DOI] [PubMed] [Google Scholar]
- 21.Insel R A, Adderson E E, Carroll W L. Int Rev Immunol. 1992;9:25–43. doi: 10.3109/08830189209061781. [DOI] [PubMed] [Google Scholar]
- 22.Kabat E A. J Immunol. 1988;141:S25–S36. [PubMed] [Google Scholar]
- 23.Bundle D R, Altman E, Auzanneau F-I, Baumann H, Eichler E, Sigurskjold B W. In: Complex Carbohydrates in Drug Research, Alfred Benzoin Symposium 36. Bock K, Clausen H, editors. Copenhagen: Munksgaard; 1994. pp. 168–181. [Google Scholar]
- 24.Carlin N I A, Gidney M A J, Lindberg A A, Bundle D R. J Immunol. 1986;137:2361–2366. [PubMed] [Google Scholar]
- 25.Bundle D R, Eichler E, Gidney M A J, Meldal M, Ragauskas A, Sigurskjold B W, Sinnott B, Watson D C, Yaguchi M, Young M N. Biochemistry. 1994;33:5172–5182. doi: 10.1021/bi00183a022. [DOI] [PubMed] [Google Scholar]
- 26.Bundle D R, Baumann H, Brisson J R, Gagné S M, Zdanov A, Cygler M. Biochemistry. 1994;33:5183–5192. doi: 10.1021/bi00183a023. [DOI] [PubMed] [Google Scholar]
- 27.Cygler M, Rose D R, Bundle D R. Science. 1991;253:442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- 28.Vyas M N, Vyas N K, Meikle P J, Sinnott B, Pinto B M, Bundle D R, Quiocho F A. J Mol Biol. 1993;231:133–136. doi: 10.1006/jmbi.1993.1262. [DOI] [PubMed] [Google Scholar]
- 29.Evans S V, Sigurskjold B W, Jennings H J, Brisson J R, To R, Tse W C, Altman E, Frosch M, Weisgerber C, Kratzin H D, Baesen M, Bitter-Suermann D, Rose D R, Young N M, Bundle D R. Biochemistry. 1995;34:6737–6744. doi: 10.1021/bi00020a019. [DOI] [PubMed] [Google Scholar]
- 30.Wilson I A, Ghiara J F, Stanfield R L. Res Immunol. 1994;145:73–78. doi: 10.1016/s0923-2494(94)80049-9. [DOI] [PubMed] [Google Scholar]
- 31.Padlan E A, Kabat E A. Proc Natl Acad Sci USA. 1988;85:6885–6889. doi: 10.1073/pnas.85.18.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vargas-Madrazo E, Lara-Ochoa F, Almagro J C. J Mol Biol. 1995;254:497–504. doi: 10.1006/jmbi.1995.0633. [DOI] [PubMed] [Google Scholar]
- 33.Oldenburg K R, Loganathan D, Goldstein I J, Schultz P G, Gallop M A. Proc Natl Acad Sci USA. 1992;89:5393–5397. doi: 10.1073/pnas.89.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott J K, Loganathan D, Easley R B, Gong X, Goldstein I J. Proc Natl Acad Sci USA. 1992;89:5398–5402. doi: 10.1073/pnas.89.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens C L, Cwirla S E, Lee R Y-W, Whitehorn E, Chen E Y-F, Bakker A, Wagstrom C, Gopalan P, Smith S W, Tate E, Koller K J, Schatz P J, Dower W J, Barrett R W. J Biol Chem. 1995;270:21129–21136. doi: 10.1074/jbc.270.36.21129. [DOI] [PubMed] [Google Scholar]
- 36.Etzkorn F A, Guo T, Lipton M A, Goldberg S D, Bartlett P A. J Am Chem Soc. 1994;116:10412–10425. [Google Scholar]
- 37.Eichler J, Lucka A W, Pinilla C, Houghten R A. Mol Diversity. 1995;1:233–240. doi: 10.1007/BF01715527. [DOI] [PubMed] [Google Scholar]