Abstract
In order to generate marked insertions in the myxobacterium Sorangium cellulosum, a transposon based on the eukaryotic mariner transposon was developed. The transposition frequency was increased with the use of a mutated tnp gene. The transposon randomly inserts into the chromosome, as demonstrated by targeted mutagenesis of the epoK gene.
Myxobacteria are soil-dwelling gram-negative bacteria. They survive by secreting a variety of hydrolytic enzymes that break down the organic matter as well as other living microorganisms in their environment. They are most noted for their ability to form fruiting-body structures when they are starved for nutrients (2). These fruiting bodies house thousands of dormant myxospores that are resistant to a variety of environmental stresses. Within the last decade, they have gained prominence as producers of secondary metabolites, some of which are currently being exploited as potential drug candidates (12). The most prominent of these metabolites are the epothilones (1).
Analysis of myxobacteria reveals that the genus Sorangium is a rich source of unique bioactive secondary metabolites (12-14). However, Sorangium strains are some of the most difficult myxobacteria with which to work. They have the longest doubling time of myxobacteria, up to 16 h, and very few genetic tools are available. Conjugation into S. cellulosum has been developed to introduce DNA into the cell, but the recombination frequency in this host is very low and, therefore, integrating DNA with regions of homology of less than 1,000 bp can be extremely difficult (6). Thus, making knockout mutations by insertion of a vector containing a small region of homology is problematic.
The ability to make mutations in Sorangium would be extremely useful for identifying the gene clusters responsible for the synthesis of secondary metabolites; a single strain of Sorangium can produce several different known secondary metabolites, such as So ce12, which makes four known compounds (13), and in addition may harbor gene clusters that synthesize compounds that have not been identified. Many of the secondary metabolites isolated from myxobacteria are complex polyketides synthesized by type I polyketide synthases (PKS), which are large multimodular proteins (for reviews, see references 5, 8, and 18). Analysis of one strain of S. cellulosum, SMP44, revealed that PKS sequences represent approximately 3.2% of the genome or over 380 kb of DNA, assuming a genome size of approximately 12 Mb (11, 17). With many PKS gene clusters requiring 40 to 50 kb of DNA to encode the necessary proteins for synthesis of the corresponding compounds, it can be predicted that six to eight PKS gene clusters are present in SMP44. Thus, a transposon would provide a valuable tool for generating mutations in S. cellulosum to determine which PKS gene cluster is responsible for synthesizing which molecule, facilitating the sequencing of a desired gene cluster.
A transposon based on the eukaryotic mariner family of transposons has been used for eubacteria, archaebacteria, and eukaryotic cells (3, 16, 21, 22). This mariner-based transposon has been shown to function in Myxococcus xanthus (20). It has a higher frequency of transposition and inserts more randomly into the chromosome than Tn5 (P. L. Hartzell, D. J. Lampe, and P. Youderian, unpublished data). Analysis of the site of insertion for Tn5 reveals a preference for the sequence A-GNTYWRANC-T, whereas the mariner transposon requires only the dinucleotide TA (4, 15). Because of the advantages of the mariner transposon, a version was developed for use in S. cellulosum.
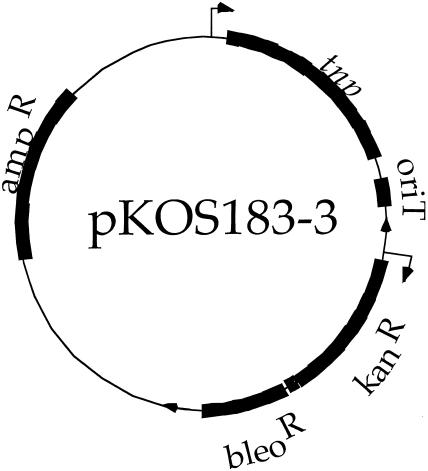
A conjugative plasmid harboring both a mariner tnp gene and the mariner inverted repeats flanking the antibiotic resistance genes for kanamycin and bleomycin was constructed. This plasmid, pKOS183-3, is shown in Fig. 1. It contains the tnp gene under the LacI repressible T7A1 promoter and is outside of the inverted repeats (10). This promoter has been used successfully to drive expression of the Tn5 transposase in M. xanthus (B. Julien, R. Calendar, and D. Kaiser; unpublished data).
FIG. 1.
Map of plasmid pKOS183-3. Raised arrowheads represent transcriptional start sites. Arrows on the circle designate the transposon inverted repeats.
By use of a procedure previously described by Jaoua et al. (6) with S. cellulosum strain So ce90, the number of mutants generated ranged from 16,000 to 80,000 per conjugation. Since approximately 109 S. cellulosum cells were used for the conjugation, this translates into a transposition frequency of 10−4 to 10−5 per cell. The frequency of transposition did not change if the S. cellulosum cells were heat shocked at 50°C for 10 min or if the Escherichia coli strain harbored mutations in dam and dcm, genes required for methylating DNA. Either heat shock or the use of the methylation-free E. coli strain improves the efficiency of homologous recombination in S. cellulosum, but they appear not to be necessary for transposition (6, 11).
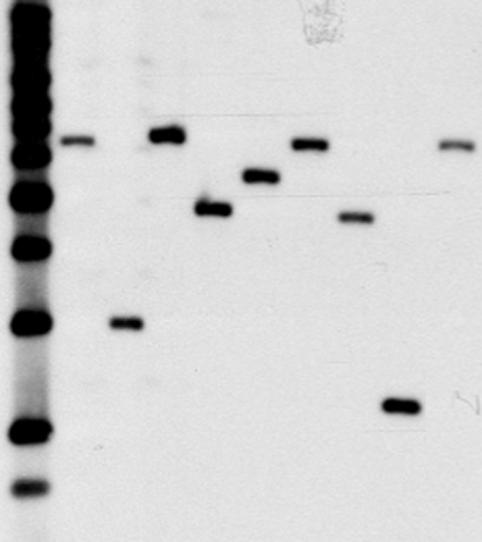
To demonstrate that the phleomycin-resistant colonies contain random insertions of transposon in the chromosome, DNA from nine isolates was analyzed by Southern blotting. Figure 2 shows the autoradiogram of chromosomal DNA cleaved with BamHI, at a site not found within the transposon, and probed with the kanamycin and bleomycin resistance genes. The figure shows a different banding pattern for each isolate, indicating apparent random insertion into the chromosome. The parent strain does not contain a sequence that hybridizes to this probe, and no antibiotic-resistant colonies were obtained in the absence of the transposase gene (data not shown).
FIG. 2.
Southern blot of transposon insertion strains. Lane 1, 1-kb ladder (the smallest band is 1.6 kb); lanes 2 to 10, nine independent transposon insertion strains.
Several point mutations have been isolated in the mariner tnp gene, which produces a transposase protein that results in a higher transposition frequency. The mutant with the greatest increase in transposition frequency in E. coli, approximately 50-fold, has two amino acid substitutions: one is the glutamine-to-arginine change at position 131, and the other is the glutamic acid-to-lysine change at position 137 (9). An attempt to construct this mutant failed due to difficulties with cloning the altered gene into the expression vector. However, the single amino acid change at position 137 results in a 20-fold increase in transposition in E. coli. This single amino acid substitution in the Tnp protein increases the transposition frequency fourfold in S. cellulosum relative to that of the wild type in three independent experiments, with a variability of less than 12% between experiments.
To demonstrate that the mariner transposon constructed had the potential to insert into a gene of interest, the 1,260-bp epoK gene was chosen for targeting. This gene is a cytochrome P450 gene that adds an epoxide to epothilones C and D to make epothilones A and B, respectively (7, 19). Insertions in epoK would provide an S. cellulosum strain that produces epothilones C and D. Using the transposon, approximately 12,000 insertion mutant strains were generated with So ce90, and pools of 1,000 mutants were grown in liquid medium. DNA was isolated from each of the pools, and PCR analysis using primers annealing to the inverted repeat of the transposon and the sequence upstream of epoK was performed. Five of the pools gave a PCR product. Sequencing of the PCR products showed that the transposon had inserted into 5 out of 21 TA sequences within the epoK gene, at nucleotides 277, 342, 377, 781, and 1016.
In summary, it has been demonstrated that a derivative of the mariner transposon is able to transpose in a strain of S. cellulosum at a frequency greater than 10−4 per cell. Although all of the experiments done in the present study were performed with strain So ce90, we have performed the same experiments successfully with S. cellulosum strain So ce12, although the frequency of transposition was reduced. This may be due to the reduction in the transfer efficiency of the transposon into this strain. S. cellulosum cells have a tendency to aggregate, which significantly reduces the conjugation efficiency. Thus, it is necessary to grow Sorangium strains in a medium in which they are dispersed or to isolate a mutant strain that no longer aggregates. The engineered mariner transposon described here provides a valuable method of generating mutations in S. cellulosum that would be difficult to generate and select for by other methods.
REFERENCES
- 1.Altmann, K. H. 2001. Microtubule-stabilizing agents: a growing class of important anticancer drugs. Curr. Opin. Chem. Biol. 5:424-431. [DOI] [PubMed] [Google Scholar]
- 2.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golden, N. J., A. Camilli, and D. W. Acheson. 2000. Random transposon mutagenesis of Campylobacter jejuni. Infect. Immun. 68:5450-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goryshin, I. Y., J. A. Miller, Y. V. Kil, V. A. Lanzov, and W. S. Reznikoff. 1998. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95:10716-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopwood, D. A., and D. H. Sherman. 1990. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24:37-66. [DOI] [PubMed] [Google Scholar]
- 6.Jaoua, S., S. Neff, and T. Schupp. 1992. Transfer of mobilizable plasmids to Sorangium cellulosum and evidence for their integration into the chromosome. Plasmid 28:157-165. [DOI] [PubMed] [Google Scholar]
- 7.Julien, B., S. Shah, R. Ziermann, R. Goldman, L. Katz, and C. Khosla. 2000. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum. Gene 249:153-160. [DOI] [PubMed] [Google Scholar]
- 8.Khosla, C., R. S. Gokhale, J. R. Jacobsen, and D. E. Cane. 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68:219-253. [DOI] [PubMed] [Google Scholar]
- 9.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanzer, M., and H. Bujard. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. USA 85:8973-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pradella, S., A. Hans, C. Sproer, H. Reichenbach, K. Gerth, and S. Beyer. 2002. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch. Microbiol. 178:484-492. [DOI] [PubMed] [Google Scholar]
- 12.Reichenbach, H. 2001. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 27:149-156. [DOI] [PubMed] [Google Scholar]
- 13.Reichenbach, H., and G. Höfle. 1999. Myxobacteria as producers of secondary metabolites, p. 149-179. In S. Grabley and R. Thiericke (ed.), Drug discovery from nature. Springer Verlag, Berlin, Germany.
- 14.Reichenbach, H., and G. Höfle. 1993. Production of bioactive secondary metabolites, p. 347-397. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 15.Robertson, H. M., and D. J. Lampe. 1995. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 12:850-862. [DOI] [PubMed] [Google Scholar]
- 16.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santi, D. V., M. A. Siani, B. Julien, D. Kupfer, and B. Roe. 2000. An approach for obtaining perfect hybridization probes for unknown polyketide synthase genes: a search for the epothilone gene cluster. Gene 247:97-102. [DOI] [PubMed] [Google Scholar]
- 18.Shen, B. 2003. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7:285-295. [DOI] [PubMed] [Google Scholar]
- 19.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 20.Youderian, P., N. Burke, D. J. White, and P. L. Hartzell. 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49:555-570. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, J. K., M. A. Pritchett, D. J. Lampe, H. M. Robertson, and W. W. Metcalf. 2000. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. USA 97:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L., U. Sankar, D. J. Lampe, H. M. Robertson, and F. L. Graham. 1998. The Himar1 mariner transposase cloned in a recombinant adenovirus vector is functional in mammalian cells. Nucleic Acids Res. 26:3687-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]