Abstract
Apparent alterations in DNA methylation have been observed in many cancers, but whether such alterations represent a persistent alteration in the normal methylation process is not known. In this study, we report a striking difference in the expression of exogenously introduced retroviral genes in various colorectal cancer cell lines. Extinguished expression was associated with DNA methylation and could be reversed by treatment with the demethylating agent 5-azacytidine. A striking correlation between genetic instability and methylation capacity suggested that methylation abnormalities may play a role in chromosome segregation processes in cancer cells.
DNA methylation is one of the most intriguing chemical phenomena affecting the genome. It is essential in prokaryotes, dispensable in lower eukaryotes such as Saccharomyces cerevisiae, yet present and presumably important in mammals (1, 2). As gene expression is abnormal in cancer and DNA methylation plays a role in controlling gene expression (3), it was supposed that aberrant DNA methylation might be responsible for expression differences (4). Accordingly, many cancers were shown to have a global hypomethylation of DNA compared with normal tissues (5–7). Treatment of cells or animals with 5-azacytidine (5-aza-C), a demethylating agent that irreversibly inactivates the methyltransferase (8–10), was shown to be oncogenic in vitro and in vivo (11). Conversely, other studies have shown that hypermethylation of specific sequences is found in some tumors and can be associated with the inactivation of tumor suppressor gene expression (12–14). Mice genetically deficient in methyltransferase were found to be resistant to colorectal tumorigenesis initiated by mutation of the APC tumor suppressor gene, and treatment of these mice with 5-aza-C enhanced the resistance (15).
One of the impediments to the interpretation of any of these somewhat conflicting studies is the absence of knowledge about DNA methylation in the precursor cells of the studied cancers. Colorectal tumors, for example, arise from stem cells that form only a minor component of the total epithelial population. Comparison of DNA methylation patterns in colorectal tumors to those of total colorectal epithelium may therefore yield misleading results, as there is no guarantee that the methylation pattern in the stem cells is identical to that of their much more numerous progeny. Thus, statements about “hypo” or “hyper” methylation must be viewed cautiously. Moreover, the mechanisms responsible for the putative altered methylation in tumors, and the time at which such aberrant methylation occurs, are obscure. In particular, whether the putative alteration in DNA methylation represents a historical event that occurred during tumorigenesis rather than a persistent defect in the process of methylation is unknown.
In the present study, we have made some unexpected observations which shed light on these issues. While studying retrovirus-mediated gene expression, we noted a qualitative difference among colorectal cancer cell lines which we later tied to DNA methylation. Because all colorectal cancer cell lines are presumably derived from the same stem cell precursor, the methylation differences observed were likely to be related to the tumorigenic process rather than to differences in cellular origin. Moreover, because de novo methylation was assessed, we could conclude that the differences represented alterations that persisted throughout the lifetime of the tumor cell. Furthermore, a striking correlation between genetic instability and methylation capacity suggested that methylation abnormalities might play a role in chromosomal segregation processes in cancer.
MATERIALS AND METHODS
Cell Culture.
Cells were cultured in McCoy’s 5A medium (modified) supplemented with 10% fetal bovine serum (HyClone), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Monolayer cultures were grown at 37°C in a 5% CO2 atmosphere. The cell lines Caco2, Colo205, DLD1, HCT116, HT29, LoVo, SW480, SW48, and SW837 were obtained from the American Type Culture Collection (Rockville, MD), and RKO was generously provided by M. Brattain (Baylor College of Medicine, Houston, TX). The status of these lines with respect to mismatch repair capacity has been analyzed previously (16–21).
Vector Construction and Retrovirus Production.
The hygromycin-resistance gene (HygR) was PCR amplified from the pCEP4 plasmid (CLONTECH) using the primers 5′-CGA TCA AGC TTC CGT GTT TCA GTT AGC-3′ and 5′-CAT GCG ATA TCC ACC ATG AAA AAG CCT GAA CTC AC-3′ (DNAgency, Malvern, PA) at an annealing temperature of 63°C. The PCR fragment was gel purified using a Spin-X centrifugal filter (Costar), digested with HindIII and EcoRV, and inserted into the HindIII and StuI sites of the plasmid pGal/Neo (a derivative of G1BgSVNa kindly provided by Genetic Therapy Inc., Bethesda, MD) to yield plasmid pGal/Hyg. Retroviral vector DNA was prepared by cesium chloride banding and transfected into the ecotropic retrovirus packaging cell line BOSC 23 (22). The virus supernatant was harvested 48 hr after transfection and used to infect the amphotropic packaging line PA317 (23). Colonies were selected for 21 days with hygromycin C (Calbiochem). Stable producer clones were cultivated, and virus supernatants were titered (106 colony-forming units/ml) and used for infection.
Transfection and Virus Infection.
Plasmid DNA was transfected using Lipofectin (GIBCO/BRL), and colonies were selected for 23 days with the neomycin analog G418 (GIBCO/BRL). For virus infection, subconfluent cells were treated with 8 μg/ml Polybrene and infected with the retrovirus Gal/Neo (derived from the retroviral construct G1BgSvNa, Genetic Therapy, Bethesda, MD) or the retrovirus Gal/Hyg at a multiplicity of <0.01. Clones containing retroviral integrations were identified by selection with G418 or hygromycin, respectively. After 3 weeks of selection, individual clones were isolated by limiting dilution and assayed for β-galactosidase (β-gal) expression in situ. Cells were fixed and stained with 5-bromo-4-chloro-3 indolyl-β-d-galactopyranoside (X-Gal; Sigma). All infection and staining procedures were performed in parallel on the various lines and repeated at least three times.
Drug Treatment of Cells.
Cells were grown in medium containing different concentrations (5–20 μM) of 1-β-d-arabinofuranosylcytosine or (1–10 μM) of 5-aza-C (Sigma) for 72 hr. Drug-treated cells for DNA and RNA isolation were grown in selective media containing 5 μM 5-aza-C.
PCR and Transcription–Translation Reactions.
Primer pairs used for genomic amplifications of β-gal sequences were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAC CAC CAT GGT TAC GGA TTC GGA TCC-3′ with 5′-CTG TTT ACC TTG TGG AGC G-3′ and 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAC CAC CAT GGC TGT GCC GAA ATG GTC C-3′ with 5′-CCG GAG AAC CTG CGT GC-3′, yielding products 2030 bp and 1995 bp in length, respectively. PCR products were used as templates in coupled transcription–translation reactions (Promega). The resultant 35S-labeled proteins were separated by SDS/polyacrylamide gel electrophoresis (Tris–glycine gel) as described by Powell et al. (24).
Southern Blot Analysis.
DNA samples (5 μg) were separated by electrophoresis on a 0.7% agarose gel and transferred to a Zeta-Probe membrane (Bio-Rad). The blot was probed with a gel-purified 3594-bp BamHI fragment of the β-gal gene derived from the plasmid pCMVβ (CLONTECH), labeled by random priming with [32P]dCTP (25).
RNA Isolation and Northern Blot Analysis.
Total RNA was prepared by CsCl gradient ultracentrifugation of guanidine isothiocyanate-lysed cells. Northern blot analysis was performed as described (26), except that QuickHyb (Stratagene) was used for hybridization. As a quantitation control, a gel-purified 300-bp PstI/HindIII fragment of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene derived from the plasmid pHcGAP (ATCC) (27) was used for rehybridization after stripping filters with 0.1× SSC/0.1% SDS at 100°C for 20 min.
Bisulfite Genomic Sequencing.
Bisulfite treatment of genomic DNA was performed as described by Frommer et al. (28) and modified by Clark et al. (29). For PCR amplification of the bisulfite-modified integrated viral 5′ long terminal repeat (LTR), the primers 5′-GAA TAG AAA AGT TTA GAT TAA GG-3′ and 5′-CAA AAC TAA TTA ACT AAC TAA TAC-3′ (DNAgency) were used, resulting in a 668-bp product. The original sequences corresponding to these primers did not contain 5′-CpG-3′ dinucleotides. Amplified DNA was gel purified and blunt-end-ligated into the EcoRV-digested plasmid pZErO-1 (Invitrogen). After transformation, individual colonies were picked, and genomic DNA was prepared and sequenced using the internal primer 5′-CCC ART AAT AAA TCA ATA ATC C-3′. The analysis was performed on HCT116–1 cells before and after 5-aza-C treatment, and on HT29–6 cells. At least 12 DNA clones, each of whose deoxycytosines (when not in a CpG context) were completely converted to uridines, were sequenced for each line.
RESULTS
Expression of Retroviral Sequences.
Previous studies of DNA methylation in cancer have largely evaluated sequences already present in the genome of cancer cells. Because methylation is heritable, such studies cannot be used to determine if the process of de novo methylation is abnormal at the time of testing. To develop an assay for this purpose, we chose to examine sequences introduced into the cancer cell by retroviral infection. Once integrated, such viral sequences can be methylated (30–32), and preliminary experiments in our laboratory suggested that the expression of such sequences in human cancer cell lines was variable, perhaps reflecting differences in de novo methylation (unpublished data). The virus used for these experiments contained the β-gal reporter driven by a retroviral LTR and the G418 resistance gene (NeoR) under the control of the simian virus 40 early promoter.
Ten colorectal cancer cell lines were infected at low multiplicity, and G418-resistant clones were selected and stained in situ with X-Gal to evaluate the expression of β-gal. HCT116 and HT29 cells exemplify the two distinct patterns of expression observed. Virtually all HT29 clones stained intensely with X-Gal, indicating that the reporter was expressed in the selected cells. In contrast, many HCT116 clones were not stained at all, and even in those colonies which did exhibit staining, only a small fraction of the cells in each clone was stained (Fig. 1). This observation was repeated six times in the two lines, with virtually identical results. All clones contained the retroviral genome, including the Neo and β-gal sequences, as indicated by the G418 resistance and confirmed by Southern blots (see later).
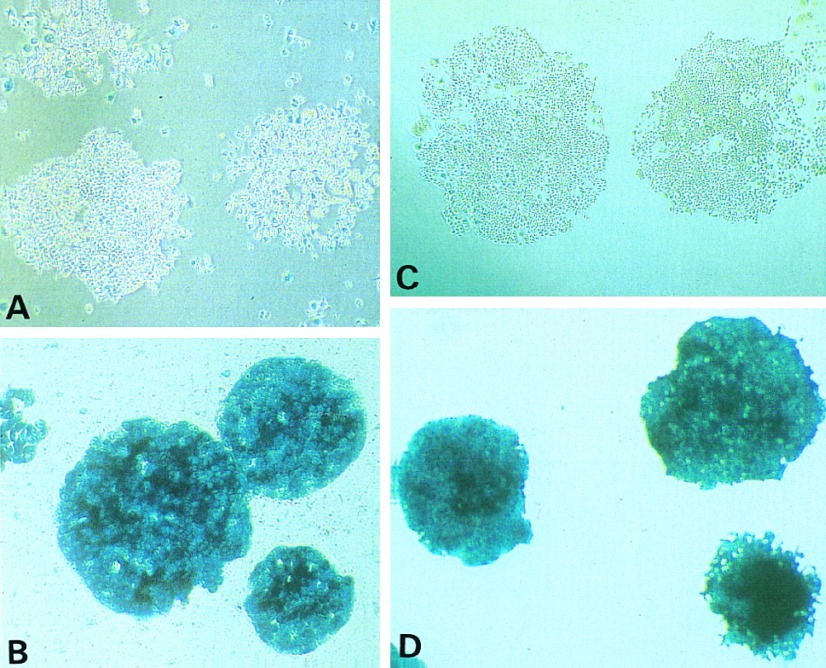
Figure 1.
Histochemical detection of β-gal expression. (A and B) After infection with the retrovirus Gal/Neo and selection with G418, cells were stained in situ with X-Gal to confirm the expression of β-gal. Whereas the majority of cells from individual clones of cell line HCT116 was not stained (A), virtually all cells of HT29 stained intensely blue (B). (C and D) Cells were stained in situ with X-Gal after retroviral infection with Gal/Hyg and selection with hygromycin. Whereas virtually all HT29 cells stained blue (D), the majority of cells from individual clones of HCT116+ch3 were not stained (C). (All ×10.)
Each of the ten lines studied showed one of these two easily distinguishable patterns of β-gal expression (Table 1, top 10 lines). In five lines (HT29, SW480, SW837, Colo 205, and Caco 2), the great majority of cells resistant to G418 were stained. In the other five lines (HCT116, LoVo, SW48, RKO, and DLD1) most cells were unstained. A striking correlation was noted between the propensity of the cells to express exogenous sequences and their proficiency in mismatch repair. All five non-expressing lines were mismatch repair (MMR) deficient, while the five β-gal-expressing lines were normal in this regard. Previous studies have demonstrated that LoVo has an inactivating mutation of the mutS homolog hMSH2 (19, 21), DLD1 has an inactivating mutation of the hMSH2 partner GTBP (17), SW48 and HCT116 have mutations of the mutL homolog hMLH1 (16, 21), and RKO is defective in either hMLH1 or its heterodimeric partner hPMS2 (J. Drummond and P. Modrich, personal communication).
Table 1.
Expression of β-gal gene
| Gene transfer agent | Cell line | Expression, % of clones
|
No. of clones evaluated | MMR status | |||
|---|---|---|---|---|---|---|---|
| With 80–100% blue cells | With 50–80% blue cells | With 20–50% blue cells | With <20% blue cells | ||||
| Gal/Neo virus | HT29 | 93 | 4 | 1 | 2 | 190 | Proficient |
| SW480 | 91 | 7 | 0 | 2 | 122 | Proficient | |
| SW837 | 95 | 2 | 0 | 3 | 500 | Proficient | |
| Colo205 | 45 | 20 | 20 | 15 | 285 | Proficient | |
| Caco2 | 60 | 19 | 5 | 16 | 74 | Proficient | |
| HCT116 | 4 | 7 | 8 | 81 | 84 | Deficient | |
| LoVo | 2 | 2 | 6 | 90 | 214 | Deficient | |
| SW48 | 10 | 6 | 12 | 72 | 405 | Deficient | |
| RKO | 10 | 8 | 12 | 70 | 318 | Deficient | |
| DLD1 | 26 | 20 | 20 | 34 | 328 | Deficient | |
| Gal/Neo plasmid | HT29 | 12 | 7 | 8 | 73 | 464 | Proficient |
| HCT116 | 0 | 0 | 1 | 99 | 254 | Deficient | |
| Gal/Hyg virus | HT29 | 95 | 1 | 1 | 3 | 2602 | Proficient |
| HCT116 | 13 | 13 | 27 | 47 | 557 | Deficient | |
| HCT116+ch3 | 11 | 11 | 20 | 58 | 670 | Proficient | |
MMR-proficient and MMR-deficient cell lines were infected with the retrovirus Gal/Neo (lines 1–10) or were transfected with the plasmid used for Gal/Neo virus production (lines 11 and 12) and were selected with G418. Alternatively, cell lines were infected with the retrovirus Gal/Hyg (lines 13–15) and selected with hygromycin. Individual clones were stained with X-Gal to detect β-gal expression. For each clone the percentage of cells expressing β-gal (blue cells) was determined.
To examine the time course of β-gal expression, HT29 and HCT116 cells were examined at various intervals after retroviral infection. A similar number of cells in both lines (1.3–1.6%) were stained 22 hr after infection, demonstrating similar efficiencies of viral infection. Differences became apparent as early as 2 days after infection, and they were magnified over the time course of the experiment. After selection, virtually all surviving HT29 cells expressed β-gal, while only 4–7% of HCT116 cells expressed the gene (data not shown).
The decreased expression of the exogenously introduced β-gal gene in MMR-deficient lines was not restricted to genes introduced by retroviral transduction. It was also observed after DNA transfection with the plasmid used for virus production. Staining of transfected clones with X-Gal 3 weeks after G418 selection demonstrated β-gal expression in only 1 of 254 HCT116 clones analyzed, while 27% of HT29 clones contained a substantial number of blue cells (Table 1, lines 11 and 12).
Analysis of Retroviral Integrates.
To test the possibility that the differences in expression of the exogenously introduced sequences between the MMR-proficient and -deficient cell lines were related to mutational processes affected by mismatch repair, we examined the organization and sequence of the retroviral integrates. Upon Southern blotting, each of 8 clones analyzed from four MMR-deficient lines (nonexpressors) and each of 14 clones from three MMR-proficient lines (expressors) was found to contain only one or two β-gal gene fragments, each of the intensity expected for single-copy genes. The absence of expression was therefore not due to a gross alteration of β-gal sequences in MMR-deficient cell lines.
The low fraction of β-galactosidase-expressing cells within G418-resistant clones seemed inconsistent with inactivation by point mutation. Even in MMR-deficient cells, inactivating mutations occur at a rate of only 10−4 to 10−5 per gene per generation (18, 33, 34), while over 80% of the selected MMR-deficient cells did not express β-gal (Table 1, top 10 lines). To formally exclude very high rates of mutation within β-gal sequences, we searched for truncating mutations. The β-gal genes in 13 clones from MMR-deficient cells were amplified with PCR, then transcribed and translated in vitro. Twelve of 13 clones generated polypeptides of the expected full-length size. Only one clone, LoVo-1, exhibited a truncated β-gal protein (data not shown). Interestingly, this clone exhibited absolutely no X-Gal-staining cells, while in each of the other 12 clones, a small fraction of cells exhibited such staining. Thus, nonsense mutations of β-gal were not likely the reason that some lines failed to express β-gal, suggesting an epigenetic basis.
Methylation and Transcription of β-gal Genes.
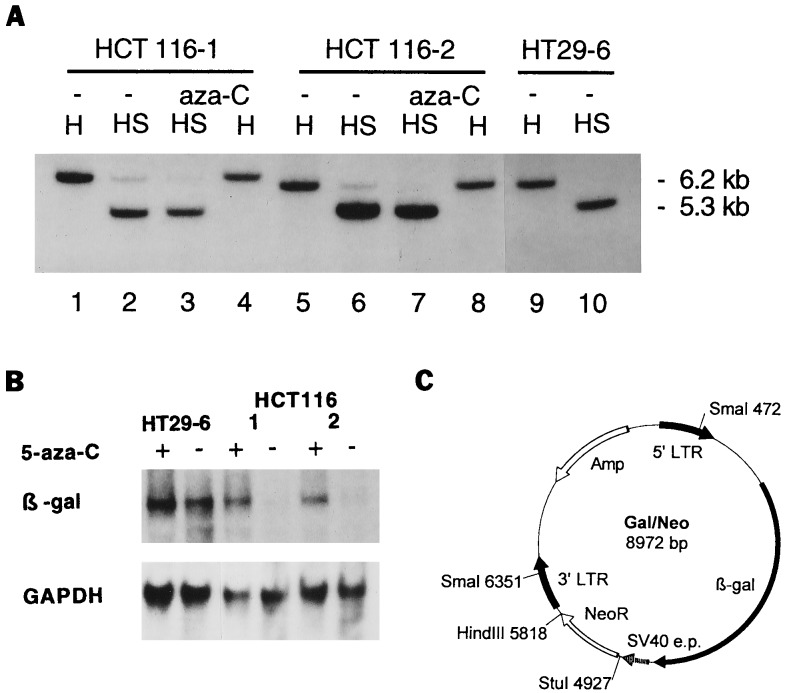
To determine whether the integrated retroviral sequences were methylated differentially, Southern blots of SmaI-digested genomic DNA from the various clones were tested. SmaI is a methylation-sensitive restriction endonuclease that does not cleave DNA when the central cytosine of its recognition sequence, 5′-CCCGGG-3′, is methylated (35). DNA was digested with HindIII alone to reveal the restriction fragment containing the β-gal gene, or with HindIII plus SmaI to test methylation of this fragment. Comparison of the intensity of the HindIII fragment with that of the HindIII/SmaI fragment revealed that few, if any, of the HT29–6 cells were methylated at the SmaI site. In contrast, the SmaI digest of HCT116–1 DNA revealed methylation of the SmaI site, as demonstrated by its partial resistance to the restriction endonuclease (Fig. 2A).
Figure 2.
Methylation of β-gal gene after retroviral infection. (A) Analysis of 5′LTR promoter by methylation-sensitive restriction enzyme. DNA from untreated (−) or 5-aza-C-treated cells was digested with HindIII alone (H) or with HindIII plus SmaI (HS). The blotted gel was probed with a [32P]dCTP-labeled fragment of the β-gal gene. The HindIII fragment containing β-gal sequences was 6.2 kb in clone HT29–6 (lane 9), and SmaI reduced this fragment to 5.35 kb (lane 10), reflecting the invariant size separating the HindIII and SmaI sites in the provirus Gal/Neo. The SmaI digest of the HCT116 clones (lane 2 and 6) revealed two bands, indicating methylation of the SmaI site in these cells. Treatment of the HCT116 clones with 5-aza-C resulted in the removal of methyl groups, so that only a 5.35-kb fragment was detected after SmaI plus HindIII digestion (lanes 3 and 7). (B) Transcription of β-gal gene. Total RNA of the clones HCT116–1, HCT116–2, and HT29–6 was probed with a [32P]dCTP-labeled fragment of the β-gal gene. The level of expression of β-gal mRNA in clones HCT116–1 and 2 was low (lanes 4 and 6) but increased substantially after 5-aza-C treatment (lanes 3 and 5). The 5-aza-C treatment had no effect on β-gal mRNA of clone HT29–6 (lanes 1 and 2). Rehybridization of the filter with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe confirmed that equal amounts of RNA were loaded. (C) Scheme of retroviral vector Gal/Neo. The Gal/Neo vector contains the β-gal reporter driven by a retroviral LTR and the G418-resistance gene (NeoR) under the control of the simian virus 40 early promoter (SV40 e.p.). Positions of the two SmaI and the single HindIII restriction sites are indicated. The restriction sites HindIII and StuI were used for the construction of the provirus Gal/Hyg.
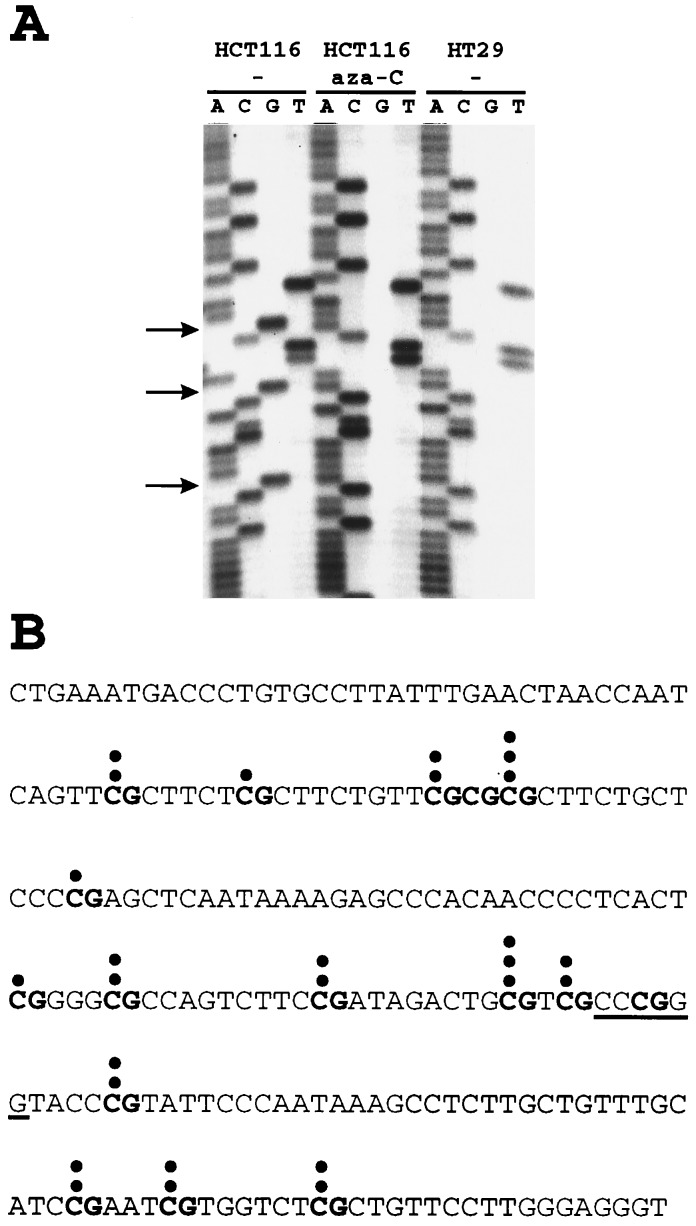
Though methylation of the SmaI site was different in HCT116 cells than in the HT29 cells, the relatively low level of SmaI resistance did not seem to explain the dramatic difference in β-gal expression between these lines. However, as observed in previous studies of rodent cells (36, 37), the SmaI site is only one of several potential methylation sites within the viral 5′LTR. To get a more comprehensive view of the methylation of sequences surrounding the SmaI site, we sequenced genomic templates treated with bisulfite. The 214 bp surrounding the SmaI site contain 76% of the CpG dinucleotides in the 5′LTR, and we therefore concentrated on this region. The bisulfite genomic sequencing technique allows methylated cytosines to be distinguished through their resistance to bisulfite-mediated deamination; unmethylated cytosines are converted to uridine by bisulfite (28). There were 16 CpG sites within the 214 bp surrounding the SmaI site. In HT29 cells, no methylation at any of these CpG sites was found. In contrast, the 5′LTRs in HCT116 cells were associated with an average of 2.33 methylated CpG sites (range 0–9 sites per LTR; P < 0.025 by Student’s t test). The distribution of methylated CpG residues in the 12 genomic 5′LTR’s analyzed is shown in Fig. 3B, and examples of the sequencing data are in Fig. 3A. As a technical control for the efficiency of bisulfite treatment, it was noted that each of 52 deoxycytosines not in CpG contexts was completely converted to uridines in all of the HCT116 and HT29 clones evaluated.
Figure 3.
Bisulfite genomic sequencing of 5′LTR. Genomic DNAs of infected cell lines were treated with bisulfite, converting deoxycytosine but not 5-methylcytosine residues into uracil through deamination. After treatment, the viral 5′LTR region was PCR-amplified, the PCR products were cloned, and a 214-bp region surrounding the SmaI site (see Fig. 2C) was sequenced. (A) Examples of clones HCT116–1 (before and after 5-aza-C treatment) and HT29–6 are shown. Arrows mark the locations of methylated CpG dinucleotides in HCT116–1. (B) Distribution of methylated CpG dinucleotides within the 5′LTR promotor of HCT116–1. The DNA sequence of the 214-bp region surrounding the SmaI site (underlined) is shown. Its 16 CpG dinucleotides are typed in boldface. The shaded area represents the region that is shown on the sequencing gel in A. Each dot represents a 5-methylcytosine residue that was not converted into uracil in one of the 12 cloned PCR products.
Reversal of Methylation.
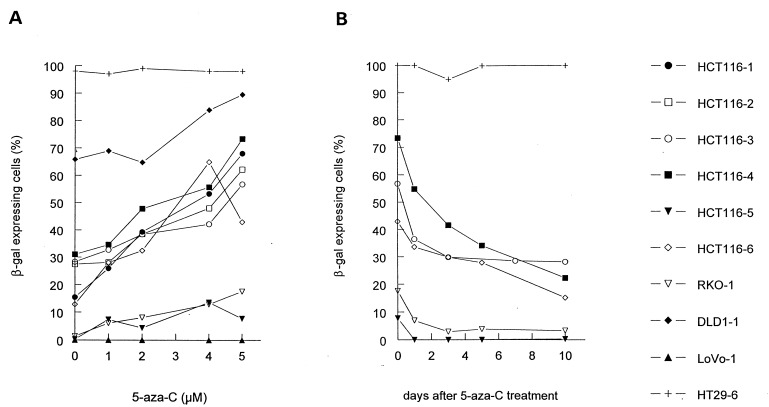
The documentation of methylation of 5′LTR sequences reveals a correlation with β-gal expression that could not necessarily be regarded as causative. To obtain functional evidence that differences in methylation were responsible for the expression differences, we treated cells with 5-aza-C. This compound inactivates methyltransferase activity, reverses methylation, and restores retroviral expression in retrovirally infected rodent cells (8–10, 38). In 8 of 9 nonexpressor clones tested, 5-aza-C significantly increased the fraction of cells expressing β-gal in a dose-dependent manner (Fig. 4A). In contrast, in three expressor clones, the high level of β-gal expression was not affected by 5-aza-C (example in Fig. 4A). The single nonexpressor clone in which 5-aza-C had no effect was LoVo-1 (Fig. 4A). As mentioned above, LoVo-1 had a truncating mutation in the β-gal gene that should not be reversible by drug treatment.
Figure 4.
Reversal of β-gal inactivation by 5-aza-C. (A) Nine MMR-deficient clones (HCT116–1, HCT116–2, HCT116–3, HCT116–4, HCT116–5, HCT116–6, RKO-1, DLD1–1, and LoVo-1) and the MMR-proficient clone HT29–6 were treated with different concentrations (0, 1, 2, 4, or 5 μM) of 5-aza-C for 72 hr and stained with X-Gal. In 8 of 9 MMR-deficient clones, 5-aza-C significantly increased the fraction of cells expressing β-gal in a dose-dependent manner. The single MMR-deficient clone in which 5-aza-C had no effect was LoVo-1. LoVo-1 had a truncating mutation in the β-gal gene (see text). (B) To determine whether the reversal of inactivation of β-gal expression by 5-aza-C was permanent, the drug (5 μM) was removed from the medium, and β-gal expression was measured at subsequent times. The clones reverted to the same basal level of β-gal expression found before 5-aza-C treatment.
Treatment with 5-aza-C resulted in the removal of methyl groups, so that only a 5.3-kb fragment was found after SmaI plus HindIII digestion in clone HCT116–1 (Fig. 2A). Similar degrees of methylation of the SmaI site were observed in three different HCT116 clones tested, all reversible with 5-aza-C. No resistance to SmaI, either before or after 5-aza-C treatment, was observed in three HT29 clones similarly tested (example in Fig. 2A). This reduction in methylation was confirmed and quantitated by bisulfite genomic sequencing. The average number of methylated CpG dinucleotides was reduced by 84% after 5-aza-C treatment of HCT116 cells (0.37 methylated CpG sites per 5′LTR, range 0–3, P < 0.020 by Student’s t test; example in Fig. 3A).
To determine whether the reversal of inactivation of β-gal expression by 5-aza-C was permanent, we removed the drug from the medium and measured β-gal expression at subsequent times. As shown in Fig. 4B, the clones reverted to the same basal level of β-gal expression found before 5-aza-C treatment. To ensure that the reversion was not simply the result of cellular toxicity due to 5-aza-C treatment, we treated cells with another cytidine analog, the antimetabolite 1-β-d-arabinofuranosylcytosine. This drug exhibited toxicity at 10 μM as high as that observed with 6 μM 5-aza-C, but treatment with 1-β-d-arabinofuranosylcytosine at concentrations of 5, 10, or 20 μM for 4 days did not increase the fraction of cells expressing β-gal (data not shown).
Finally, to ensure that the drug affected expression at the RNA level, we performed Northern blot analysis. The level of expression of β-gal mRNA in each of three HCT116 clones tested was low but increased substantially after 5-aza-C treatment. The 5-aza-C treatment had no effect on β-gal mRNA in three HT29 clones tested (examples in Fig. 2B).
Methylation and Failure of MMR.
The observations recorded above suggested that the absence of a MMR system might in some way lead to the methylation of exogenously introduced sequences. To test this hypothesis, we tested the HCT116 derivative (HCT116+ch3) in which the MMR deficiency had been corrected by transfer of a normal chromosome 3 containing the hMLH1 gene missing in HCT116 cells (39). Western blot analysis showed, as expected, that the hMLH1 protein was expressed in the HCT116+ch3 cell line but missing in HCT116 (data not shown). Because the transferred chromosome 3 was tagged with a G418-resistance marker, the retrovirus Gal/Neo (Fig. 2C) could not be used and a new retroviral vector, Gal/Hyg, was constructed by replacing the NeoR gene of Gal/Neo with a hygromycin-resistance gene (HygR). The expression of β-gal in HT29 and HCT116 cells after infection with this virus mimicked that resulting from Gal/Neo infection (Fig. 1D and the last three lines of Table 1). The pattern of β-gal expression in the HCT116+ch3 line was very low, similar to the pattern obtained by infection of HCT116 and the other MMR-deficient lines (Fig. 1C and Table 1). These data demonstrate that de novo methylation of exogenously introduced sequences in the MMR-deficient lines is not due to the absence of MMR activity.
DISCUSSION
The methylation of exogenously introduced retroviral sequences is not a new observation (30–32, 36). The novelty of the observations reported here lies in the differences in methylation and expression of foreign genes in MMR-proficient versus MMR-deficient cell lines. This observation was completely unexpected and highly statistically significant (P < 0.00001 by χ2). Some previous studies have suggested that methylation can be affected in various ways by DNA repair abnormalities, though the results were somewhat contradictory and no specific effect of MMR has been delineated (40, 41). Our experiments showed that MMR could not be directly responsible for the methylation differences, as correction of the MMR deficiency in HCT116 cells did not alter its ability to methylate exogenously introduced sequences.
Previous studies showing that the methylation of specific DNA sequences was different in tumor cells than in bulk preparations of “normal cells” could be interpreted in many ways. The tumor cells could have originated from stem cells that had different patterns of methylation than the bulk of the normal, differentiated cells used for comparison. Additionally, the tumor cells could have passed through some bottleneck in which methylation was altered, and this pattern could then have been transmitted to daughter cells through normal maintenance methylation. In contrast, the fact that methylation of exogenous sequences in colorectal cancer lines differed dramatically among the cell lines we tested shows that methylation abnormalities are not just historical events, but represent continuing physiologic differences that persist throughout the lifetime of the tumor cells.
Which of the two methylation patterns observed here represents the “normal” type likely to be found in nonneoplastic stem cell precursors? Though this question cannot be answered definitively at present, we feel it is likely that the methylation competence of the MMR-deficient tumor cells represents the physiologically normal state. This judgment is based on prior observations demonstrating that retroviral gene expression is often extinguished over time in normal tissues and that this extinction is associated with a methylation of the retroviral LTR similar to that described here (30, 31, 36, 38). According to this hypothesis, the MMR-deficient lines are methylation proficient (MMR−, MET+), while the MMR-proficient lines are methylation deficient (MMR+, MET−).
What is the basis upon which cells with a MMR+, MET− phenotype are selected during tumorigenesis? We propose the following speculative explanation. It has long been suspected that some sort of genetic instability is necessary for a tumor to accumulate the numerous genetic alterations that accompany its development (42–44). There appear to exist two pathways of genetic instability in colorectal cancer. The first is found in about 15% of colorectal tumors and involves point mutations, microdeletions, and microinsertions associated with MMR deficiency (45, 46). The second is found in MMR-proficient cells and involves gains and losses of whole chromosomes (47). We suggest that methylation abnormalities are intrinsically and directly involved in the generation of the second type of instability, thus allowing for the selection of MET− cells during the clonal evolution of tumors. This hypothesis is supported by the observation that demethylation is associated with chromosomal aberrations, including mitotic dysfunction and translocation (48, 49), and is consistent with the innovative hypothesis relating methylation and aneuploidy recently put forward by Thomas (50).
Acknowledgments
The authors thank S. Baylin for critical reading of the manuscript and Genetic Therapy Inc. for providing the retrovirus G1BgSVNa. This work was supported by grants from the Fonds zur Förderung der wissenschaftlichen Forschung (C.L.), the Clayton Foundation, and National Institutes of Health Grants CA 35494 and CA 62924. B.V. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- 5-aza-C
5-azacytidine
- β-gal
β-galactosidase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactopyranoside
- LTR
long terminal repeat, MMR, mismatch repair
References
- 1.Bird A. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 2.Noyer-Weldner M, Trautner T A. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhauser; 1993. pp. 39–108. [Google Scholar]
- 3.Yeivin A, Razin A. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhauser; 1993. pp. 523–568. [Google Scholar]
- 4.Bird A. Nature (London) 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 5.Gama-Sosa M A, Slagel V A, Trewyn R W, Ehrlich M. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goelz S E, Vogelstein B, Hamilton S P, Feinberg A P. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A P, Gehrke C W, Kuo K C, Ehrlich M. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 8.Santi D V, Garrett C E, Barr P J. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 9.Santi D V, Normen A, Garrett C E. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jüttermann R, Li E, Jaenisch R. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landolph J R, Jones P A. Cancer Res. 1982;42:817–823. [PubMed] [Google Scholar]
- 12.Baylin S B, Makos M, Wu J J, Yen R W, de Bustros A, Vertino P, Nelkin B D. Cancer Cells. 1991;3:383–390. [PubMed] [Google Scholar]
- 13.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 14.Jones P A. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 15.Laird P W, Jackson-Grusby L, Fazeli A, Dickinson S L, Jung W E, Li E, Weinberg R A, Jaenisch R. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Peterson G M, Watson P, Lynch H T, Peltomaki P, Mecklin J P, de la Chapelle A, Kinzler K W, Vogelstein B. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 17.Papadopoulos N, Nicolaides N C, Liu B, Parsons R E, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K V, Kinzler K W, Jiricny J, Vogelstein B. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 18.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 19.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 20.da Costa L T, Liu B, El-Deiry W S, Hamilton S R, Kinzler K W, Vogelstein B. Nat Genet. 1995;9:10–11. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Nicolaides N C, Markowitz S, Willson J K V, Parsons R E, Jen J, Papadopoulos N, Peltomaki P, de la Chapelle A, Hamilton S R, Kinzler K W, Vogelstein B. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 22.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell S M, Petersen G M, Krush A J, Booker S, Jen J, Giardiello F M, Hamilton S R, Vogelstein B, Kinzler K W. N Engl J Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 26.El-Deiry W S, Nelkin B D, Celano P, Yen R-W C, Falco J P, Hamilton S R, Baylin S B. Proc Natl Acad Sci USA. 1991;88:3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tso J Y, Sun X H, Kao T H, Reece K S, Wu R. Nucleic Acids Res. 1985;13:2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark S J, Harrison J, Paul C L, Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbers K, Schnieke N, Stuhlmann H, Jahner D, Jaenisch R. Proc Natl, Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. Nature (London) 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 32.Bednarik D P. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhauser; 1993. pp. 300–329. [Google Scholar]
- 33.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eshleman J R, Lang E Z, Bowerfind G K, Parsons R, Vogelstein B, Willson J K, Veigl M L, Sedwick W D, Markowitz S D. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 35.Roberts R. Nucleic Acids Res. 1990;18:S2331–S2365. [PMC free article] [PubMed] [Google Scholar]
- 36.Challita P-M, Kohn D B. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challita P-M, Skelton D, El-Khoueiry A, Yu X-J, Weinberg K, Kohn D B. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoeben R C, Migchielsen A A J, van der Jagt R C M, van Ormondt H, van der Eb A J. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Cancer Res. 1994;54:4302–4312. [PubMed] [Google Scholar]
- 40.Branch P, Hampson R, Karran P. Cancer Res. 1995;55:2304–2309. [PubMed] [Google Scholar]
- 41.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 42.Loeb L. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 43.Hartwell L. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 44.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 45.Modrich P. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 46.Marra G, Boland C R. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 47.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature (London), in press. [DOI] [PubMed]
- 48.Schmid M, Grunert D, Haaf T, Engel W. Cytogenet Cell Genet. 1983;36:554–561. doi: 10.1159/000131972. [DOI] [PubMed] [Google Scholar]
- 49.Schmid M, Haaf T, Grunert D. Hum Genet. 1984;67:257–263. doi: 10.1007/BF00291352. [DOI] [PubMed] [Google Scholar]
- 50.Thomas J H. Proc Natl Acad Sci USA. 1995;92:480–482. doi: 10.1073/pnas.92.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]