Abstract
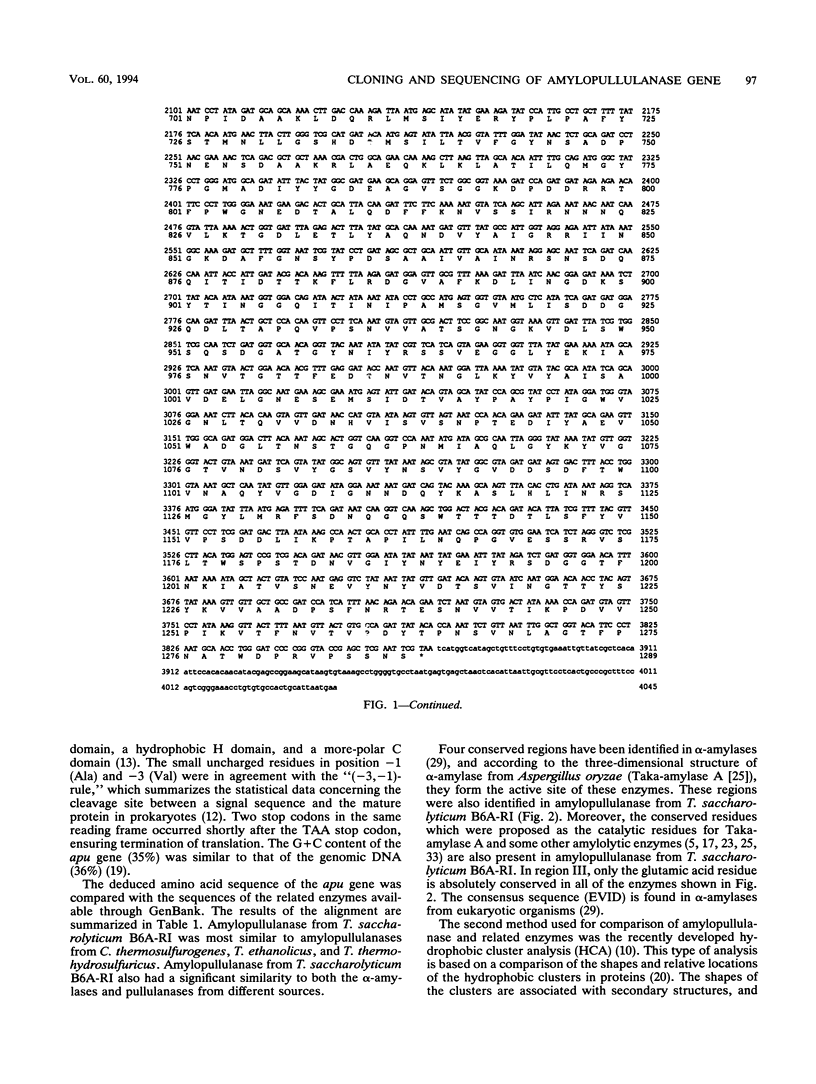
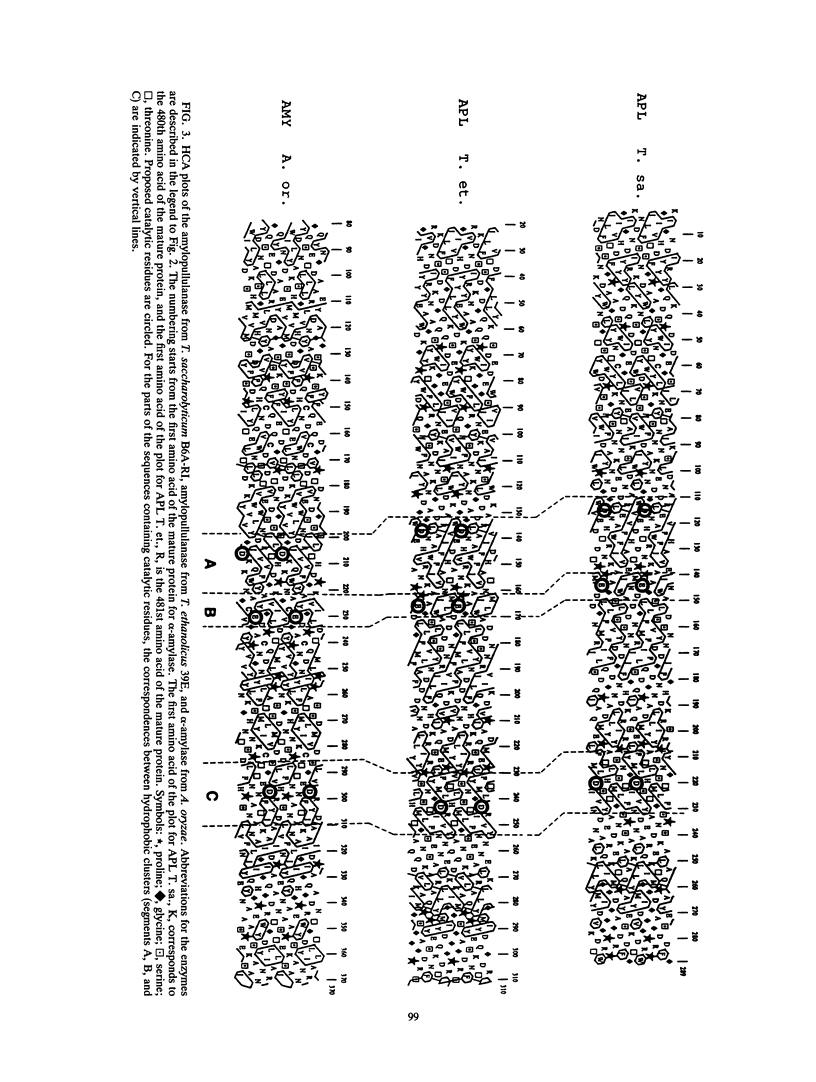
The amylopullulanase gene (apu) of the thermophilic anaerobic bacterium Thermoanaerobacterium saccharolyticum B6A-RI was cloned into Escherichia coli. The complete nucleotide sequence of the gene was determined. It encoded a protein consisting of 1,288 amino acids with a signal peptide of 35 amino acids. The enzyme purified from E. coli was a monomer with an M(r) of 142,000 +/- 2,000 and had same the catalytic and thermal characteristics as the native glycoprotein from T. saccharolyticum B6A. Linear alignment and the hydrophobic cluster analysis were used to compare this amylopullulanase with other amylolytic enzymes. Both methods revealed strictly conserved amino acid residues among these enzymes, and it is proposed that Asp-594, Asp-700, and Glu-623 are a putative catalytic triad of the T. saccharolyticum B6A-RI amylopullulanase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buisson G., Duée E., Haser R., Payan F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J. 1987 Dec 20;6(13):3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R. D., Yang S. S., McAlister M. P. Cloning of the debranching-enzyme gene from Thermoanaerobium brockii into Escherichia coli and Bacillus subtilis. J Bacteriol. 1987 Sep;169(9):4302–4307. doi: 10.1128/jb.169.9.4302-4307.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Katsuragi N., Takizawa N., Murooka Y. Entire nucleotide sequence of the pullulanase gene of Klebsiella aerogenes W70. J Bacteriol. 1987 May;169(5):2301–2306. doi: 10.1128/jb.169.5.2301-2306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Schulz G. E. Structure of cyclodextrin glycosyltransferase refined at 2.0 A resolution. J Mol Biol. 1991 Feb 20;217(4):737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989 Jun;135(6):1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Takata H., Okada S., Imanaka T. Analysis of the active center of Bacillus stearothermophilus neopullulanase. J Bacteriol. 1991 Oct;173(19):6147–6152. doi: 10.1128/jb.173.19.6147-6152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemesle-Varloot L., Henrissat B., Gaboriaud C., Bissery V., Morgat A., Mornon J. P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990 Aug;72(8):555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- Mathupala S. P., Lowe S. E., Podkovyrov S. M., Zeikus J. G. Sequencing of the amylopullulanase (apu) gene of Thermoanaerobacter ethanolicus 39E, and identification of the active site by site-directed mutagenesis. J Biol Chem. 1993 Aug 5;268(22):16332–16344. [PubMed] [Google Scholar]
- Mathupala S., Saha B. C., Zeikus J. G. Substrate competition and specificity at the active site of amylopullulanase from Clostridium thermohydrosulfuricum. Biochem Biophys Res Commun. 1990 Jan 15;166(1):126–132. doi: 10.1016/0006-291x(90)91920-n. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Melasniemi H., Paloheimo M. Cloning and expression of the Clostridium thermohydrosulfuricum alpha-amylase-pullulanase gene in Escherichia coli. J Gen Microbiol. 1989 Jun;135(6):1755–1762. doi: 10.1099/00221287-135-6-1755. [DOI] [PubMed] [Google Scholar]
- Melasniemi H., Paloheimo M., Hemiö L. Nucleotide sequence of the alpha-amylase-pullulanase gene from Clostridium thermohydrosulfuricum. J Gen Microbiol. 1990 Mar;136(3):447–454. doi: 10.1099/00221287-136-3-447. [DOI] [PubMed] [Google Scholar]
- Melasniemi H. Purification and some properties of the extracellular alpha-amylase-pullulanase produced by Clostridium thermohydrosulfuricum. Biochem J. 1988 Mar 15;250(3):813–818. doi: 10.1042/bj2500813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero L., Mazzéi-Pierron Y., Abadie B., Chicheportiche Y., Marchis-Mouren G. Complete amino acid sequence and location of the five disulfide bridges in porcine pancreatic alpha-amylase. Biochim Biophys Acta. 1986 Jan 30;869(2):147–157. doi: 10.1016/0167-4838(86)90289-x. [DOI] [PubMed] [Google Scholar]
- Podkovyrov S. M., Burdette D., Zeikus J. G. Analysis of the catalytic center of cyclomaltodextrinase from Thermoanaerobacter ethanolicus 39E. FEBS Lett. 1993 Feb 15;317(3):259–262. doi: 10.1016/0014-5793(93)81288-b. [DOI] [PubMed] [Google Scholar]
- Podkovyrov S. M., Zeikus J. G. Structure of the gene encoding cyclomaltodextrinase from Clostridium thermohydrosulfuricum 39E and characterization of the enzyme purified from Escherichia coli. J Bacteriol. 1992 Aug;174(16):5400–5405. doi: 10.1128/jb.174.16.5400-5405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B. C., Lamed R., Lee C. Y., Mathupala S. P., Zeikus J. G. Characterization of an endo-Acting Amylopullulanase from Thermoanaerobacter Strain B6A. Appl Environ Microbiol. 1990 Apr;56(4):881–886. doi: 10.1128/aem.56.4.881-886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B. C., Mathupala S. P., Zeikus J. G. Purification and characterization of a highly thermostable novel pullulanase from Clostridium thermohydrosulfuricum. Biochem J. 1988 Jun 1;252(2):343–348. doi: 10.1042/bj2520343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi N., Furukawa M., Nagaba H., Kirita N., Tsuboi A., Udaka S. Isolation of a cDNA encoding Aspergillus oryzae Taka-amylase A: evidence for multiple related genes. Gene. 1989 Dec 14;84(2):319–327. doi: 10.1016/0378-1119(89)90506-4. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock S., Mornon J. P., Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992 Oct;5(7):629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]
- Yang S. S., Coleman R. D. Detection of pullulanase in polyacrylamide gels using pullulan-reactive red agar plates. Anal Biochem. 1987 Feb 1;160(2):480–482. doi: 10.1016/0003-2697(87)90079-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zacharius R. M., Zell T. E., Morrison J. H., Woodlock J. J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal Biochem. 1969 Jul;30(1):148–152. doi: 10.1016/0003-2697(69)90383-2. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Ben-Bassat A., Hegge P. W. Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol. 1980 Jul;143(1):432–440. doi: 10.1128/jb.143.1.432-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Abrahmsén L. Species-specific variation in signal peptide design. Implications for protein secretion in foreign hosts. FEBS Lett. 1989 Feb 27;244(2):439–446. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]



