Abstract
The toluene metabolic pathway of Pseudomonas mendocina KR1 is chromosomally encoded, but the pathway could be transferred by conjugation from strain KR1 to the chromosome of P. aeruginosa or P. putida. Such transconjugants utilized toluene, p-cresol, and p-hydroxybenzaldehyde. However, transconjugants were unable to further transfer toluene genes to other recipients unless Pseudomonas sex factor R68.45 was present in trans. Although the genes encoding the upper pathway for toluene metabolism in P. mendocina KR1 are sufficiently linked to permit their coordinate mobilization, they were found to be encoded in three independently regulated units: one encoding toluene-4-monooxygenase, a second encoding p-cresol methylhydroxylase and p-hydroxybenzaldehyde dehydrogenase, and a third encoding p-hydroxybenzoate hydroxylase. The last two regulatory units were cloned from the chromosome of a P. aeruginosa transconjugant onto a plasmid designated pRO1999. Analysis of pRO1999 showed that genes encoding p-cresol methylhydroxylase and p-hydroxybenzaldehyde dehydrogenase are organized as an operon; the gene encoding p-hydroxybenzaldehyde dehydrogenase is transcribed first, and this is followed by transcription of the gene encoding p-cresol methylhydroxylase. This operon is regulated by a positively acting regulator. The P. mendocina KR1 gene encoding p-hydroxybenzoate hydroxylase was linked to, but independently regulated from, the genes encoding toluene-4-monooxygenase, p-cresol methylhydroxylase, and p-hydroxybenzaldehyde dehydrogenase.
Full text
PDF


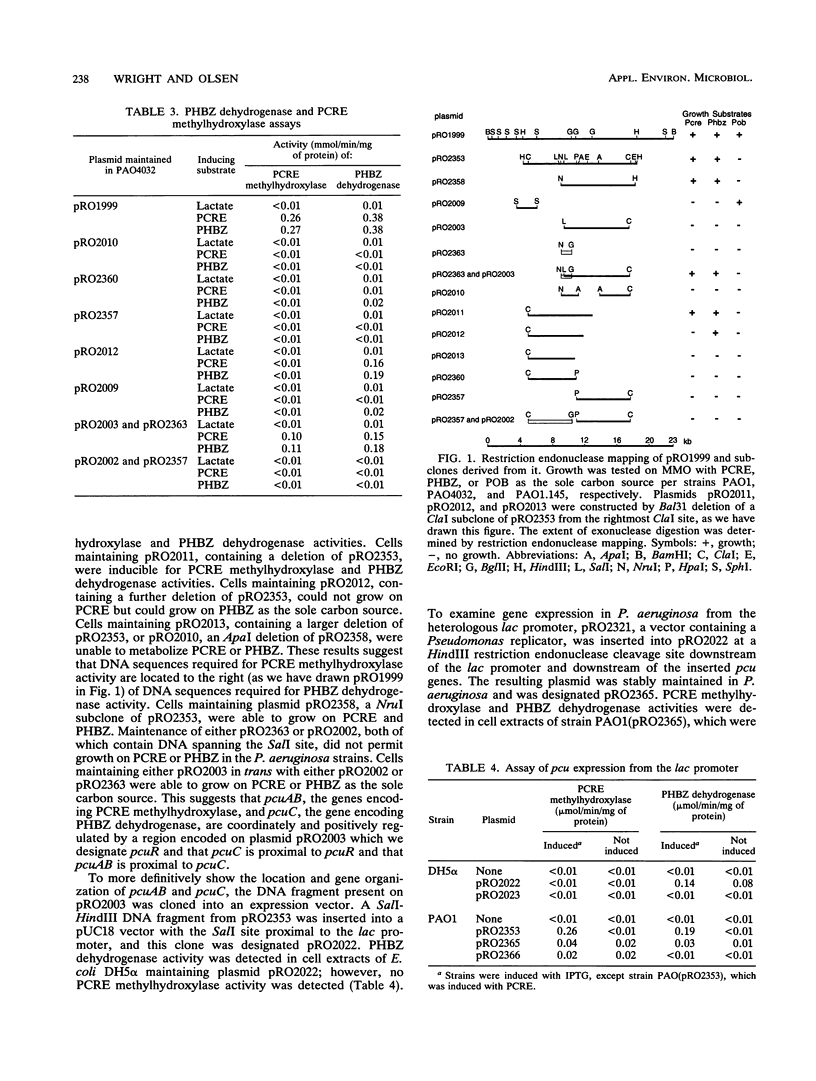
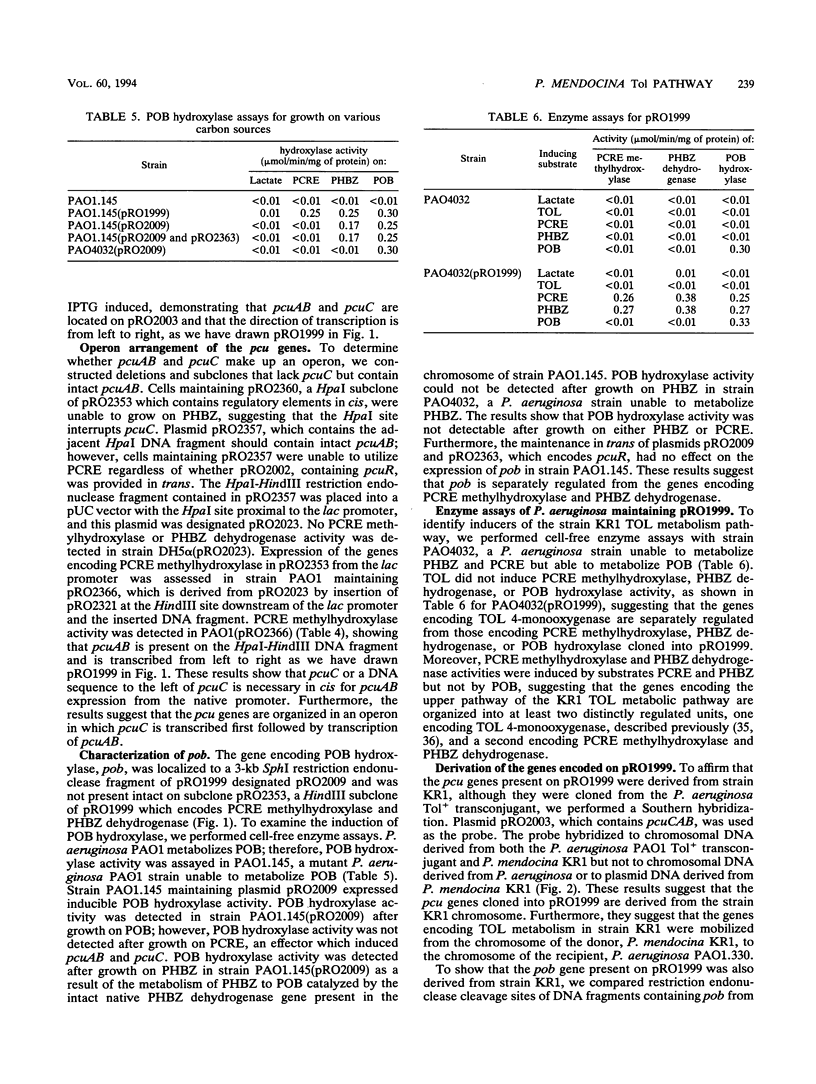



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arvidsson R. H., Nordling M., Lundberg L. G. The azurin gene from Pseudomonas aeruginosa. Cloning and characterization. Eur J Biochem. 1989 Jan 15;179(1):195–200. doi: 10.1111/j.1432-1033.1989.tb14540.x. [DOI] [PubMed] [Google Scholar]
- Brandsch R., Bichler V. Studies in vitro on the flavinylation of 6-hydroxy-D-nicotine oxidase. Eur J Biochem. 1986 Oct 15;160(2):285–289. doi: 10.1111/j.1432-1033.1986.tb09969.x. [DOI] [PubMed] [Google Scholar]
- Burlage R. S., Hooper S. W., Sayler G. S. The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol. 1989 Jun;55(6):1323–1328. doi: 10.1128/aem.55.6.1323-1328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskey S. M., Peccoraro V., Olsen R. H. Initial catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO: pathway description, mapping of mutations, and cloning of essential genes. J Bacteriol. 1987 Jun;169(6):2398–2404. doi: 10.1128/jb.169.6.2398-2404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Entsch B., Nan Y., Weaich K., Scott K. F. Sequence and organization of pobA, the gene coding for p-hydroxybenzoate hydroxylase, an inducible enzyme from Pseudomonas aeruginosa. Gene. 1988 Nov 30;71(2):279–291. doi: 10.1016/0378-1119(88)90044-3. [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Pasloske B. L., Paranchych W. Expression of the Pseudomonas aeruginosa PAK pilin gene in Escherichia coli. J Bacteriol. 1986 Feb;165(2):625–630. doi: 10.1128/jb.165.2.625-630.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D., Holloway B. W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Mar 30;144(3):243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewetson L., Dunn H. M., Dunn N. W. Evidence for a transmissible catabolic plasmid in Pseudomonas putida encoding the degradation of p-cresol via the protocatechuate ortho cleavage pathway. Genet Res. 1978 Nov;32(3):249–255. doi: 10.1017/s0016672300018747. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Hopper D. J. Incorporation of [18O]water in the formation of p-hydroxybenzyl alcohol by the p-cresol methylhydroxylase from Pseudomonas putida. Biochem J. 1978 Oct 1;175(1):345–347. doi: 10.1042/bj1750345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Jones M. R., Causer M. J. Periplasmic location of p-cresol methylhydroxylase in Pseudomonas putida. FEBS Lett. 1985 Mar 25;182(2):485–488. doi: 10.1016/0014-5793(85)80359-8. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Kemp P. D. Regulation of enzymes of the 3,5-xylenol-degradative pathway in Pseudomonas putida: evidence for a plasmid. J Bacteriol. 1980 Apr;142(1):21–26. doi: 10.1128/jb.142.1.21-26.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson B. G., Pascher T., Nordling M., Arvidsson R. H., Lundberg L. G. Expression of the blue copper protein azurin from Pseudomonas aeruginosa in Escherichia coli. FEBS Lett. 1989 Mar 27;246(1-2):211–217. doi: 10.1016/0014-5793(89)80285-6. [DOI] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H. Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol. 1990 Aug;172(8):4624–4630. doi: 10.1128/jb.172.8.4624-4630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A. Molecular chaperones and protein translocation across the Escherichia coli inner membrane. Mol Microbiol. 1991 Jan;5(1):19–22. doi: 10.1111/j.1365-2958.1991.tb01821.x. [DOI] [PubMed] [Google Scholar]
- McIntire W., Edmondson D. E., Hopper D. J., Singer T. P. 8 alpha-(O-Tyrosyl)flavin adenine dinucleotide, the prosthetic group of bacterial p-cresol methylhydroxylase. Biochemistry. 1981 May 26;20(11):3068–3075. doi: 10.1021/bi00514a013. [DOI] [PubMed] [Google Scholar]
- McIntire W., Hopper D. J., Singer T. P. p-Cresol methylhydroxylase. Assay and general properties. Biochem J. 1985 Jun 1;228(2):325–335. doi: 10.1042/bj2280325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire W., Singer T. P. Resolution of p-cresol methylhydroxylase into catalytically active subunits and reconstitution of the flavocytochrome. FEBS Lett. 1982 Jul 5;143(2):316–318. doi: 10.1016/0014-5793(82)80124-5. [DOI] [PubMed] [Google Scholar]
- Mercer A. A., Loutit J. S. Transformation and transfection of Pseudomonas aeruginosa: effects of metal ions. J Bacteriol. 1979 Oct;140(1):37–42. doi: 10.1128/jb.140.1.37-42.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T. P., McIntire W. S. Covalent attachment of flavin to flavoproteins: occurrence, assay, and synthesis. Methods Enzymol. 1984;106:369–378. doi: 10.1016/0076-6879(84)06039-0. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Whited G. M., Gibson D. T. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol. 1991 May;173(9):3017–3020. doi: 10.1128/jb.173.9.3017-3020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited G. M., Gibson D. T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991 May;173(9):3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Karl M. R., Blatt L. M., Simon M. J., Winter R. B., Fausset P. R., Lu H. S., Harcourt A. A., Chen K. K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991 Sep;173(17):5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Karl M. R. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1992 Nov;174(22):7253–7261. doi: 10.1128/jb.174.22.7253-7261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., McCombie W. R., Gibson D. T., Finette B. A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988 Jun;54(6):1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Olsen R. H., Ballou D. P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989 Nov;171(11):5907–5914. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., de Vos W. M., Harayama S., Zehnder A. J. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992 Dec;56(4):677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



